Abstract
Eukaryotic DNA replication is initiated through stepwise assembly of evolutionarily conserved replication proteins onto replication origins, but how the origin DNA is unwound during the assembly process remains elusive. Here, we established a site-specific origin on a plasmid DNA, using in vitro replication systems derived from Xenopus egg extracts. We found that the pre-replicative complex (pre-RC) was preferentially assembled in the vicinity of GAL4 DNA-binding sites of the plasmid, depending on the binding of Cdc6 fused with a GAL4 DNA-binding domain in Cdc6-depleted extracts. Subsequent addition of nucleoplasmic S-phase extracts to the GAL4-dependent pre-RC promoted initiation of DNA replication from the origin, and components of the pre-initiation complex (pre-IC) and the replisome were recruited to the origin concomitant with origin unwinding. In this replication system, RecQ4 is dispensable for both recruitment of Cdc45 onto the origin and stable binding of Cdc45 and GINS to the pre-RC assembled plasmid. However, both origin binding of DNA polymerase α and unwinding of DNA were diminished upon depletion of RecQ4 from the extracts. These results suggest that RecQ4 plays an important role in the conversion of pre-ICs into active replisomes requiring the unwinding of origin DNA in vertebrates.
Introduction
Chromosomal DNA replication in all living organisms initiates through the stepwise assembly of initiator proteins onto replication origins in the chromosome. The initiation reaction involves the interplay between initiator proteins and origin DNA, which leads to the conversion of double-stranded DNA (dsDNA) at the origin regions into single-stranded DNA (ssDNA) for the assembly of the replication machinery, the replisome.Citation1,2 In most bacteria, DNA replication initiates from a single origin on a chromosome; the detailed molecular process for the initial changes in DNA structure and protein assembly has been disclosed by using an in vitro replication system. In eukaryotes, replication initiates from numerous origins distributed on chromosomes; intensive studies with various eukaryotes have revealed the conserved mechanisms for the initiation of eukaryotic chromosomal replication. The first step of initiation is the assembly of the pre-replicative complex (pre-RC) containing inactive replicative helicase, the hexameric Mcm2-7 complex, at each replication origin from the end of M to G1 phase.Citation3 The assembly of pre-RC licenses the genome for subsequent DNA replication in S phase.Citation4 Upon entry into S phase, pre-RC is converted into the pre-initiation complex (pre-IC), which contains various initiator proteins formed on the unwound origin. Active replisomes are then formed from pre-IC, following activation of the helicase, through yet unknown mechanisms. The use of a budding yeast replication origin with a defined short sequence is advantageous for investigating such a conversion process. The sequential assembly of initiator proteins onto yeast origins has been proposed,Citation5 although the sequence specificity of origin DNA appears to be relaxed and diversified in metazoans.Citation6 Replication in Xenopus eggs in particular shows little sequence specificity of the origin and the initiation reaction has been difficult to analyze at a specific site on the template DNA.
The reconstitution of pre-RC on plasmid DNA has been successfully conducted with budding yeast and Xenopus systems, and the molecular process of pre-RC assembly, which requires the cooperative action of the origin recognition complex (ORC), Cdc6, and Cdt1, has been investigated.Citation7-10 The key feature of pre-RC assembly is the formation of a head-to-head double-hexameric Mcm2-7 encircling dsDNA at the origins.Citation11-13 Recent studies further show the essential role of ATPase activity of Mcm2-7 in the assembly process.Citation14-15 At the onset of S phase, pre-RC is activated by the action of 2 protein kinases, cyclin-dependent kinase (CDK) and Dbf4/Drf1-dependent kinase, and converted into pre-IC, which contains replisome components such as Cdc45, Mcm2-7, and GINS but not DNA polymerase α (Polα). It is not yet known how origin unwinding occurs before Polα loading and how inactive helicase in pre-IC is converted into active helicase, which provides templates for primer synthesis by Polα.
Initiator proteins involved in the activation of pre-RC have been extensively investigated with various organisms, showing that most proteins are well conserved beyond phyla.Citation16 In particular, identification of the CMG complex consisting of Cdc45, Mcm2-7, and GINS as an active DNA helicaseCitation17-18 leads to a common belief that the basic mechanism for the activation of pre-RC is conserved in eukaryotes. However, limited conservations of yeast initiator proteins such as Sld2, Sld3, and Dpb11 with metazoan RecQ4, Treslin, and TopBP1, respectively, suggest that there might be some additional and/or distinct roles for these metazoan replication initiators.Citation19-21 For example, vertebrate TopBP1 has 8 BRCT domains, whereas yeast Dpb11/Cut5 retains only 4. The first 3 BRCTs are sufficient for vertebrate replication,Citation22 but all 4 BRCTs are essential and each has distinct role in yeast replication; the first 2 BRCTs interact with Sld3 and the remaining 2 are required for interaction with Sld2/Drc1,Citation23-25 which forms the pre-loading complex with GINS and DNA polymerase ϵ.Citation26 In metazoans, however, direct interaction of RecQ4 with GINS has not been reported. Instead, a recent study shows that Dpb11 and TopBP1 retain conserved binding site for GINS, which is apparently required for an efficient initiation reaction.Citation27 Therefore, RecQ4 may have a different role in the initiation reaction from that of yeast Sld2. Indeed, RecQ4 is dispensable for the chromatin association of GINS and Cdc45, but is involved in the loading step of RPA and Polα onto chromatin,Citation28-29 in stark contrast to the essential role of Sld2 in the recruitment of GINS to pre-RC. Suppression of RPA loading upon RecQ4 depletion further suggests that RecQ4 is involved in the unwinding of origin DNA.Citation28 However, the exact role of RecQ4 in the initiation of metazoan DNA replication remains unknown.
To gain molecular insight into the initial conversion steps, it is important to examine both the structural changes in origin DNA and the assembly of proteins onto the origin. For this purpose, an in vitro replication system of plasmid DNA with a defined origin should be beneficial. Indeed, with the budding yeast in vitro replication system, recent studies have shown that pre-RC assembled on the plasmids could be activated in S-phase extracts of yeast cells, but that the replication efficiency was a small percentage of input DNA.Citation30-32 Xenopus egg extracts, a unique in vitro replication system, provide us with efficient DNA replication (nearly 100% efficiency),Citation33 and nucleoplasmic extract (NPE) prepared from the nuclei assembled with the extracts allowed us to examine this efficient plasmid DNA replication in vitro.Citation34 However, it has been difficult to investigate structural changes in origin DNA and the assembly of proteins at the origins because of the lack of a specific origin sequence in the Xenopus replication system.Citation35
Here, we have constructed a site-specific origin on a plasmid DNA with Xenopus egg extracts to investigate the structural changes in origin DNA and accompanying assemblies of proteins at the origins. Previous studies with Xenopus eggs reported the successful specification of the origin on a plasmid DNA by restricting the binding of ORC through DNA methylationCitation36 or by recruiting a transcription complex.Citation37 However, these studies mainly aimed to specify replication origins and used indirect methods to specify the origin. Our strategy was inspired by a report demonstrating that the recruitment of ORC or Cdc6 onto the specific site of a plasmid is sufficient to initiate site-specific DNA replication in mammalian cells.Citation38 By tethering Cdc6 at the GAL4-binding sites on a plasmid DNA, we succeeded in assembling pre-RC preferentially at a defined site on a plasmid DNA through a 2-step assembly protocol. Further addition of NPE led to the initiation of DNA replication from the site-specific origin, where preferential DNA unwinding was indicated by P1 nuclease digestion. Taking advantage of this in vitro replication system, we found that RecQ4 is dispensable for recruitment of Cdc45 onto the origin and stable binding of Cdc45 and GINS to the pre-RC assembled plasmid. However, both origin binding of Polα and unwinding of DNA were diminished upon the depletion of RecQ4 from the extracts. These results suggest that RecQ4 plays an important role in the conversion of pre-ICs into active replisomes in vertebrates.
Results
In vitro pre-RC formation by tethering GAL4-Cdc6 at a specific site on a plasmid
The interphase extracts of Xenopus eggs allowed us to recapitulate the initiation events of DNA replication by using sperm chromatin or plasmid DNA as a template. However, replication origins, the sites of pre-RC formation, are apparently distributed at random site(s) along the template DNA. Therefore, it has been difficult to analyze the assembly of initiator proteins and local DNA structures at origins. To overcome this problem, we tried to construct a site-specific origin with the egg extracts. We used the strategy of recruiting one of the pre-RC assembly factors to a defined site on a plasmid, which allows initiation from an artificial replication origin in human cells.Citation38 We first purified recombinant Xenopus Cdc6 fused with the GAL4 DNA-binding domain (GAL4-Cdc6, Fig. S1A). Recombinant GAL4-Cdc6 promoted the loading of Mcm2-7 onto sperm chromatin in Cdc6-depleted extracts at similar concentrations to that of endogenous Cdc6 (Fig. S1B). In addition, Mcm2-7 bound to chromatin was resistant to high-salt wash, as previously observed for untreated egg extractsCitation39 (Fig. S1C). These results confirm that GAL4-Cdc6 fully complements the function of endogenous Cdc6 for the loading of Mcm2-7 onto chromatin.
We next analyzed pre-RC formation by GAL4-Cdc6 tethered to a plasmid. We prepared a plasmid containing 0, 1, or 6 copies of the GAL4 DNA-binding site and, to isolate the plasmid, we randomly biotinylated it (on average 1 biotinylated site per plasmid) in order to attach streptavidin-conjugated magnetic beads. After incubating the bead-coupled plasmids with GAL4-Cdc6, we recovered them by using the magnetic beads and washing with a buffer containing high salt to reduce non-specific binding of GAL4-Cdc6. The washed plasmids were then incubated in Cdc6-depleted extracts, which retained all factors required for the loading of Mcm2-7 except for endogenous Cdc6. This stepwise protocol enabled us to avoid non-specific binding of endogenous Cdc6 to other sites on the plasmids (). After isolating the bead-coupled plasmids from the extracts, we examined the pre-RC components bound to the plasmids. Immunoblot analysis of GAL4-Cdc6 showed that the amount of GAL4-Cdc6 bound to the plasmids increased with increasing copy numbers of the DNA-binding site (), indicating that GAL4-Cdc6 is preferentially retained at the defined site on the plasmid. Mcm2, one of the components of Mcm2-7, was recruited to GAL4-Cdc6 bound plasmids, and the amount of Mcm2 increased with increasing copy numbers of the binding site (). Binding of Mcm2 to the plasmid depends on GAL4-Cdc6 but not on GAL4-GST bound to the plasmid (, compare lanes 2 and 4). These results suggest that GAL4-Cdc6 bound to the DNA-binding site facilitates Mcm2-7 loading. In the following experiments, we used plasmids containing 6 tandem copies of the GAL4 DNA-binding site.
Figure 1. Assembly of pre-RC onto a plasmid DNA, depending on GAL4 DNA-binding sites. (A) Schematic of pre-RC assembly onto a plasmid DNA, depending on the binding sites. Plasmids attached to magnetic beads were used for the assembly of pre-RC. (B) Binding of GAL4 Cdc6, Mcm2, and Orc2 to the plasmids. Plasmids with various copy numbers of the GAL4 DNA-binding site (6×, 1×, 0×) were attached to the magnetic beads and incubated in a binding buffer with GAL4-Cdc6 on ice for 10 min. After incubation, the bead-coupled plasmids were isolated, washed with a high-salt buffer containing 350 mM NaCl and 100 mM KCl, and incubated in Cdc6-depleted egg extracts (ΔCdc6 LSS) at 23°C for 30 min. The plasmids were then isolated and washed with a low-salt buffer containing 100 mM KCl. Proteins bound to the plasmids were separated by SDS-PAGE and analyzed by immunoblotting. (C) Mcm2 binding to the plasmids, depending on the bound GAL4-Cdc6. The bead-coupled plasmids with or without the DNA-binding sites were pre-incubated in a binding buffer containing GAL4-Cdc6 or GAL4-GST on ice for 10 min. The washed plasmids were then incubated in ΔCdc6 LSS at 23°C for 30 min to assemble pre-RC on the plasmid. Proteins bound to the plasmids were analyzed. (D) Inhibition of Mcm2 binding to the plasmids by geminin. The bead-coupled plasmids bound with GAL4-Cdc6 were incubated in ΔCdc6 LSS in the presence or absence of 1 μM GST-geminin. The plasmids without GAL4-binding sites were used as a negative control (lane 1). (E) ORC-dependent Mcm2 binding to the plasmids bound with GAL4-Cdc6. The bead-coupled plasmids bound with GAL4-Cdc6 were incubated in Cdc6-depleted (ΔCdc6) or Cdc6 and Orc2 double-depleted (ΔOrc2ΔCdc6) LSS at 23°C for 30 min. The proteins in the extracts (extract) and bound to the plasmids (plasmid) were analyzed. (F) High-salt-wash-resistant binding of Mcm2 to the plasmids bound with GAL4-Cdc6. The bead-coupled plasmids incubated in ΔCdc6 LSS were isolated and washed with either the low-salt (100 mM KCl; L) or the high-salt (250 mM KCl; H) buffer after the GAL4 pre-RC was assembled on the plasmids. Proteins bound to the plasmids were analyzed.
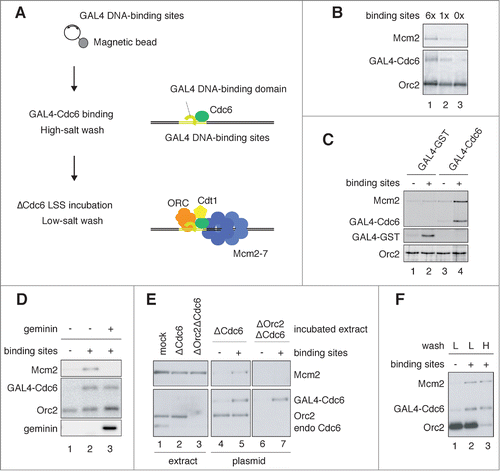
To understand the physiological features of Mcm2-7 loading via GAL4-Cdc6, we examined Mcm2 bound to the plasmids under different conditions. First, we found that the binding of Mcm2 was completely inhibited by a licensing inhibitor, geminin, which binds to Cdt1 and inhibits Mcm2-7 loading (, compare lanes 2 and 3).Citation40,41 Second, Mcm2 failed to bind the plasmids in extracts depleted of both Orc2 and Cdc6 (, lane 7), showing that ORC is required for GAL4-Cdc6-mediated binding of Mcm2. Third, Mcm2 bound to the plasmids was resistant to high-salt wash, whereas most of the bound Orc2 was removed (, compare lanes 2 and 3). Taken together, these results suggest that GAL4-Cdc6-mediated Mcm2 binding retains physiologically relevant characteristics to conventional pre-RC assembled on sperm chromatin in egg extracts. Here, we call GAL4-Cdc6-mediated loading of Mcm2-7 “GAL4 pre-RC.”
Assembly of GAL4 pre-RC proximal to the GAL4 DNA-binding sites
We used a chromatin immunoprecipitation (ChIP) assay combined with quantitative PCR to analyze the localization of components of GAL4 pre-RC assembled on the plasmid DNA. Four primer sets to detect proximal and distal sites from the GAL4 DNA-binding sites were used for the PCR (sites X, Y, Y′, and Z) (). ChIP with anti-Cdc6 antibody resulted in significant enrichment of DNA proximal to the binding sites (, Cdc6, site X). This enrichment was dependent on both the GAL4 DNA-binding sites and GAL4-Cdc6 (, Cdc6). These results confirm the stable association of GAL4-Cdc6 at the DNA-binding sites in the extracts.
Figure 2. Assembly of GAL4 pre-RC on the plasmids proximal to GAL4 DNA-binding sites. (A) Schematic diagram of the 7.6 kbp plasmid. The location of 4 primer sets (X, Y, Y′, and Z) used for the ChIP analyses, and 6× GAL4 DNA-binding sites (light green) are indicated. (B) ChIP analysis of pre-RC components bound to the plasmids. The bead-coupled plasmids with (+) and without (-) the DNA-binding sites were incubated with GAL4-Cdc6, and the pre-RC was assembled as described in . The localization of pre-RC components was examined by ChIP. Antibodies used for immunoprecipitation are indicated at the top of each panel. The amount of target DNA in each IP sample was determined by qPCR, and relative enrichment was estimated by dividing the amount obtained with each IP sample by that of the mock IP sample using control antibody. Average data are shown as a bar graph with standard deviation (SD) indicated by bars from 3 independent PCR reactions of each ChIP sample. (C) Effect of geminin on the assembly of GAL4 pre-RC on the plasmids. The bead-coupled plasmids with the DNA-binding sites were pre-incubated with GAL4-GST (GST) or GAL4-Cdc6 (Cdc6), and the assembly of pre-RC components in the presence (+) and absence (−) of GST-geminin (1 μM) was analyzed as in (B).
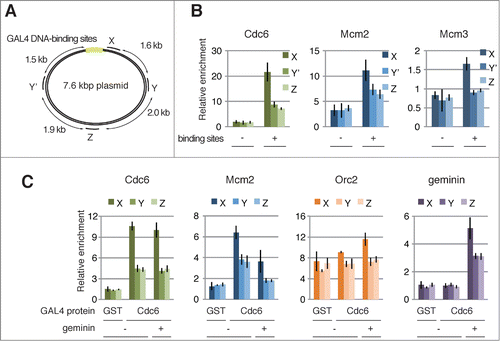
We next examined the localization of Mcm2 and Mcm3 by ChIP (, Mcm2 and Mcm3). As expected, Mcm2 and Mcm3 were enriched at site X, proximal to the binding sites, whereas their enrichment was decreased at site Y or Y′, which are located about 1.5 kb from site X in the clockwise and counterclockwise direction, respectively. Again, we observed no significant enrichment at site X on the control plasmid without the binding site (, Mcm2 and Mcm3 binding sites) or on the GAL4-GST bound plasmid (, Mcm2, GST). We further confirmed the preferential association of Mcm2 and GAL4-Cdc6 at a proximal region of the DNA-binding sites in a different plasmid backbone, indicating that the enrichment is specific to the binding sites (Fig. S2A). Therefore, we concluded that GAL4-Cdc6 bound at the GAL4 DNA-binding sites facilitates the recruitment of Mcm2-7 to form GAL4 pre-RC at a proximal region of the DNA-binding sites.
This notion is further supported by the fact that the association of Mcm2 at site X was decreased in the presence of geminin (, Mcm2, geminin +), whereas the association of GAL4-Cdc6 was not affected (, Cdc6, geminin +). In accord with the reduced binding of Mcm2, geminin appears to be preferentially located in the vicinity of the binding sites (, geminin, +). We also noted that geminin reduced the association of Mcm2 at distal sites, Y and Z, to background levels (, Mcm2, compare GST and Cdc6).
Initiation of DNA replication depending on GAL4 pre-RC
We next examined whether GAL4 pre-RC is capable of initiating DNA replication. To this end, we used the NPE of Xenopus eggs, which recapitulates the S-phase nuclear environment and thus promotes DNA replication without nuclear formation.Citation34,42 In order to replicate plasmids, pre-RC should be pre-assembled before addition of NPE. For this purpose, we prepared plasmids assembled with either GAL4 pre-RC as explained above, or with conventional pre-RC that was assembled by incubating the bead-coupled plasmids in the egg extracts containing endogenous Cdc6. After we assembled the pre-RCs, NPE was added to the extracts, and the time course of incorporation of 32P-dATP into the plasmid DNA was examined (). The plasmids recovered from the magnetic beads were separated by neutral agarose gel electrophoresis. We detected 1 major DNA band without the addition of NPE at about 3 kb (, at 0 min), which may represent a nicked or relaxed form of the plasmid DNA. Upon addition of NPE, smaller DNA at about 2 kb became more visible, which may correspond to supercoiled DNA, possibly generated by nucleosome formation in NPE.Citation42 Initial incorporation of 32P-dATP was observed at much higher molecular masses than these 2 major forms of DNA, which can likely be attributed to the formation of the theta form of replication intermediates.Citation43,42 At later times, the lower 2 bands were detected in the autoradiograph, suggesting the completion of plasmid replication. Notably, the plasmid with GAL4 pre-RC was replicated as efficiently as that with conventional pre-RC (). Only a background level of DNA synthesis was detected with the plasmid in the absence of pre-RCs (, -pre-RC). Moreover, nucleotide incorporation was inhibited in the presence of a CDK inhibitor, roscovitine (, roscovitine +). These results show that GAL4 pre-RC assembled on the plasmid DNA is functional for DNA replication.
Figure 3. Replication of the plasmid DNA assembled with pre-RCs in NPE. (A) DNA replication products analyzed by agarose gel electrophoresis. Conventional pre-RC and GAL4 pre-RC were assembled by incubating the bead-coupled plasmids (3.0 kbp) at 23°C for 30 min in mock-depleted and Cdc6-depleted HSS, respectively. We then added 2 vol of NPE containing [α32P]-dATP to 1 vol of the assembly mixture, and the reaction was terminated at the indicated time by isolating DNA. The purified DNA was separated by agarose gel electrophoresis, and the replication products were visualized by autoradiography. The plasmid without the GAL4-binding sites was used as a negative control (-preRC) and 500 μM roscovitine was added to inhibit the initiation of DNA replication (+). (B) Graphic presentation of quantified data from (A). Total amounts of [α32P]-dATP incorporated into DNA were measured and plotted against incubation time in NPE.
![Figure 3. Replication of the plasmid DNA assembled with pre-RCs in NPE. (A) DNA replication products analyzed by agarose gel electrophoresis. Conventional pre-RC and GAL4 pre-RC were assembled by incubating the bead-coupled plasmids (3.0 kbp) at 23°C for 30 min in mock-depleted and Cdc6-depleted HSS, respectively. We then added 2 vol of NPE containing [α32P]-dATP to 1 vol of the assembly mixture, and the reaction was terminated at the indicated time by isolating DNA. The purified DNA was separated by agarose gel electrophoresis, and the replication products were visualized by autoradiography. The plasmid without the GAL4-binding sites was used as a negative control (-preRC) and 500 μM roscovitine was added to inhibit the initiation of DNA replication (+). (B) Graphic presentation of quantified data from (A). Total amounts of [α32P]-dATP incorporated into DNA were measured and plotted against incubation time in NPE.](/cms/asset/f874d33e-ee93-4a3d-9560-df14befe99b6/kccy_a_1007003_f0003_b.gif)
Assembly of pre-IC and replisome proximal to the GAL4 DNA-binding sites
In accord with the replication of plasmid DNA assembled with either conventional or GAL4 pre-RC, as shown in , components of the replicative CMG helicase (Cdc45, Psf2) and DNA polymerases (Polα, Polϵ) were similarly recruited to the plasmids after addition of NPE (). We observed that the recruitment of these proteins was suppressed by inhibiting CDK (, compare +/− roscovitine), which shows that their loading is CDK-dependent. In addition, RecQ4 and Treslin, putative components of pre-IC, were recruited in the presence of both conventional and GAL4 pre-RC (, lanes 4 and 7). Interestingly, Treslin was recruited in the presence of roscovitine (lanes 5 and 8), suggesting CDK-independent loading of the protein.Citation29,22 These results further support our notion that GAL4 pre-RC is functional for initiating DNA replication.
Figure 4. Site-specific assembly of the pre-IC and replisome components and initiation of DNA replication. (A) Recruitment of initiator proteins onto the bead-coupled plasmids assembled with pre-RCs. The plasmids assembled with the conventional or the GAL4 pre-RC were incubated in NPE supplemented with 1 ng/μl aphidicolin at 23°C for 30 min, and 500 μM roscovitine was used for inhibiting the initiation of DNA replication (+ roscovitine). The plasmids bound with GAL4-GST were used as a negative control of GAL4 pre-RC assembly (-pre-RC). Proteins bound to the plasmids (plasmid) or in the extracts (extract) were analyzed by immunoblotting. (B) Localization of the proteins bound to the plasmids proximal and distal to the DNA-binding sites. The bead-coupled plasmids bound with GAL4-GST (-pre-RC) or GAL4-Cdc6 (GAL4 pre-RC) were incubated in Cdc6-depleted HSS at 23°C for 30 min and further incubated for 30 min after addition of NPE containing 1 ng/μl aphidicolin. We added 1 μM p21 to inhibit the initiation of DNA replication (p21 +). ChIP analysis was performed using antibodies as indicated at the top of the panels. The amount of target DNA in each IP sample was determined by qPCR using primer sets at the proximal (X) and distal site (Z), and relative enrichment was estimated as described in . (C) Identification of the initiation sites by BrdUTP incorporation into the plasmids assembled with GAL4 pre-RC. The plasmids with or without pre-RC (GAL4 pre-RC or -pre-RC, respectively) were incubated in NPE supplemented with 1 ng/μl aphidicolin and 200 μM BrdUTP at 23°C for 15 min. Roscovitine (500 μM) was added to inhibit the initiation of DNA replication (roscovitine +). The incorporation reaction was terminated by isolating DNA and the isolated DNA was then fragmented, followed by immunoprecipitation with anti-BrdU antibody. Enrichment of DNA fragments incorporated with BrdUTP was determined by qPCR with 3 primer sets (X, Y, and Z). The amounts of target DNA in each IP sample relative to the mock IP sample, both of which were determined by qPCR, were taken as relative enrichment. Average data are shown as bar graphs with standard deviation (SD) indicated by bars from 3 independent PCR reactions of each IP sample.
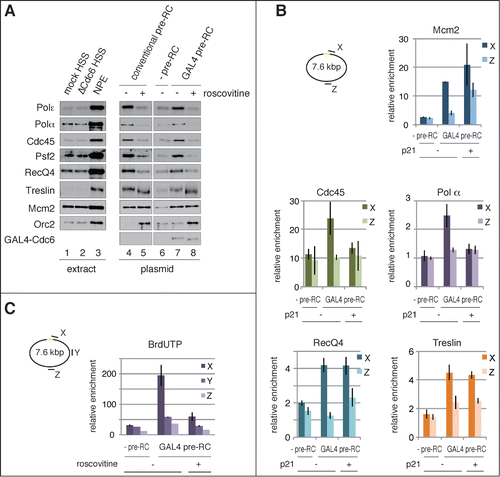
To define localization of the pre-IC and replisome components on the plasmid, we performed ChIP analysis by using 2 primer sets to detect sites X and Z (, also shown in ). We found that Cdc45 and Polα components of the replisome were preferentially recruited to the proximal site (site X), but not to the opposite of the DNA-binding sites (site Z) (, Cdc45 and Polα). In accord with the results shown in , association of Cdc45 and Polα with the site X was inhibited in the presence of a CDK inhibitor, p21 (, Cdc45 and Polα p21). We also found that RecQ4 and Treslin were preferentially recruited at site X. Consistent with the results of immunoblotting (, GAL4 pre-RC), Treslin binding at site X was apparently independent of CDK activity (, Treslin + p21). Since the binding of Treslin appears to be dependent on GAL4 pre-RC (, compare pre-RC and GAL4 pre-RC), Treslin may be recruited to Gal4 pre-RC by interacting with its components. Specific binding of replication initiation proteins on the plasmid further supports the view that GAL4 pre-RC assembled in the vicinity of the GAL4 DNA-binding sites is converted into active replisome through pre-IC formation.
Initiation of DNA replication from the site-specific origin
We next attempted to determine the initiation site of DNA replication. For this purpose, the incorporation of 5-bromo-2′-deoxyuridine 5′-triphosphate (BrdUTP) into the plasmids was analyzed by immunoprecipitation of fragmented plasmid DNA with anti-BrdU antibody ( and Fig. S3). We used, as a negative control for GAL4 pre-RC, GAL4-GST-bound plasmid, which did not support pre-RC formation (, lane 2). In order to restrict the incorporation of BrdUTP during the initial stage of DNA replication, we performed the reaction in the presence of 1 ng/μl aphidicolin, which slows the elongation of DNA replication. Three primer sets (site X, Y, and Z) were used to detect the immunoprecipitated DNA fragments (). We found that the incorporation of BrdUTP was enriched at site X, proximal to the DNA-binding site, but not at the distal sites Y and Z. The incorporation of BrdUTP at sites Y and Z was at a background level because it was comparable to that observed in the presence of CDK inhibitor (GAL4 pre-RC, +roscovitine), or in the absence of pre-RC (-pre-RC). In addition, we found symmetric enrichment of BrdUTP incorporation in close proximity to the DNA-binding sites: a similar level of enrichment was observed with the primer sets to detect the left and right side of the DNA-binding sites (Fig. S3, X and X′). These results demonstrate that DNA replication was initiated in the vicinity of the DNA-binding sites where GAL4 pre-RC had been assembled () and replisome components had been recruited after addition of NPE (). In other words, a site-specific origin was established by tethering Cdc6 to the GAL4 DNA-binding sites.
Unwinding of site-specific origin DNA upon activation of GAL4 pre-RC
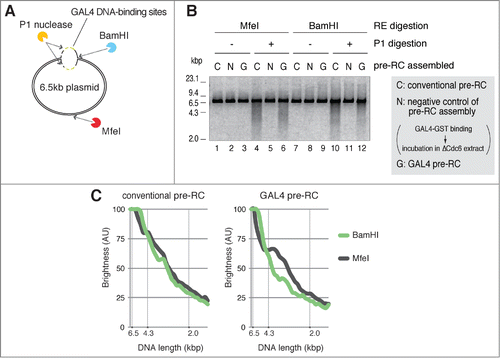
The results showed that replication initiation and initial association of replisome components occur in the proximity of the GAL4 DNA-binding sites. These findings prompted us to directly examine the unwinding of dsDNA in the vicinity of the binding sites. In order to detect unwound DNA, we used P1 nuclease, which specifically digests ssDNA without affecting dsDNA. P1 nuclease has been successfully used to detect the unwinding of origin DNA with an in vitro bacterial DNA replication system.Citation44,45 We first examined whether the plasmid DNA assembled with conventional pre-RC could be digested by P1 nuclease, depending on the assembly of the replisome that unwinds DNA (Fig. S4). The unwinding of DNA is initiated by adding NPE in the presence of high-concentration aphidicolin, which blocks DNA synthesis and thus promotes accumulation of ssDNA regions of the plasmids. The plasmids were then isolated from the extracts and incubated in a buffer containing P1 nuclease to digest ssDNA. Subsequently, digested plasmids were separated by electrophoresis. Smear DNA fragments were observed only when the replisome was established from conventional pre-RC (Fig. S4, lane 6), suggesting that unwound DNA was digested by P1 nuclease. Without the incubation in NPE, P1 nuclease treatment scarcely affected circular and supercoiled forms of the plasmids (Fig. S4, lane 1). Incubation in NPE slightly increased the supercoiled forms, which may be due to the assembly of nucleosomes in NPE (lanes 2, 3, and 4, see also ). However, smear DNA fragments were not generated by the P1 nuclease treatment in the absence of replisome formation, such as without pre-RC formation (lane 5, +geminin) or CDK activity (lane 7, +p21). Further digestion by a restriction enzyme, BamHI, cleaved the plasmid at a unique site and generated linear DNA (Fig. S4, lanes 8 to 14, +BamHI). Again, smear DNA fragments were detected only after treatment with P1 nuclease (Fig. S4, lane 13). These results show that DNA unwinding can be detected by P1 nuclease treatment, depending on the activation of pre-RC in NPE.
We next examined whether the site-specific origin is preferentially unwound when GAL4 pre-RC is activated in NPE. To distinguish the unwound region of the plasmids, we used 2 restriction enzymes, BamHI and MfeI, which recognize either the proximal or the opposite side of the GAL4 DNA-binding sites, respectively (). When the plasmids with conventional pre-RC were incubated in NPE and treated with P1 nuclease, similar smear DNA fragments were generated upon further digestion with MfeI or BamHI (, lanes 4 and 10). Densitometric scanning of fragmented DNA after electrophoresis shows a nearly identical distribution of DNA fragments with both restriction enzymes (, conventional pre-RC). In contrast, when the plasmids with GAL4 pre-RC were incubated in NPE, the digestion of P1-treated DNA with MfeI and BamHI resulted in different distributions of the fragmented DNA (, lanes 6 and 12, see also Fig. S5). MfeI-digested fragments showed a peak at about 3 to 4 kbp, suggesting that these fragments were generated through digestion at the GAL4 DNA-binding sites and the MfeI site by P1 nuclease and MfeI, respectively ( and Fig. S5, GAL4 pre-RC, MfeI). BamHI-digested fragments, in contrast, showed enrichment of nearly full-length DNA and reduction of shorter length smear DNA ( and C, GAL4 pre-RC, BamHI). The different DNA fragmentation pattern obtained with BamHI digestion can be explained by assuming that the BamHI site, located in the vicinity of the binding sites, may be abolished because of the unwinding of these regions followed by P1 nuclease digestion, or that the unwound region is located near the BamHI site. Of note is that the ratio of smear DNA fragments to total DNA by treatment with restriction enzymes varied from one experiment to the other (cf. Fig. S5). This may be due to the difficulty in controlling the initiation efficiency and hence the extent of the unwound region of DNA. Nonetheless, the fragmentation patterns obtained with either MfeI or BamHI are reproducible, i.e., enrichment of about 3 to 4 kb fragments is observed only by MfeI digestion. Thus, we conclude that the proximal regions to the GAL4 DNA-binding sites are preferentially unwound and susceptible to P1 nuclease digestion upon origin firing.
Figure 5. Unwinding of a site-specific origin upon activation of GAL4 pre-RC. (A) Schematic of P1 nuclease assay for unwinding of plasmid DNA after activation of pre-RCs in NPE. P1-treated DNA was further digested with BamHI or MfeI that recognizes different unique digestion sites of the plasmid. BamHI and MfeI sites are located at the proximal side (at the edge; 0 kb distance) and opposite side (3.0 or 3.3 kb distance) of the DNA-binding sites, respectively. (B) The bead-coupled plasmids assembled with conventional (C) or GAL4 pre-RC (G) were incubated in NPE containing 67 ng/μl aphidicolin at 23°C for 30 min to allow unwinding of dsDNA. Plasmids bound with GAL4-GST were used as a negative control of pre-RC assembly (N). After incubation, the plasmids were isolated and treated with P1 nuclease to digest ssDNA regions of the plasmids. P1-treated DNA was then purified and further digested with BamHI or MfeI. The purified DNA from the restriction enzyme digestions was subjected to agarose gel electrophoresis and visualized. (C) The distribution of DNA fragments obtained after P1 nuclease followed by the restriction enzyme digestions from a single experiment. The amount of DNA was estimated from the staining of DNA in agarose gel with the aid of the Image J program and shown as brightness. The peak values of each sample were taken as 100%. Reproducibility of the data is shown in Figure S5.
RecQ4-dependent unwinding of DNA after pre-IC formation
Previous studies suggest that the formation of the CMG complex is a crucial step for the activation of Mcm2-7 helicase, but it remains unclear whether the formation of CMG is sufficient for the unwinding of origin DNA.Citation46,47 Here we focused on the role of RecQ4 because it is required for the efficient recruitment of RPA and Polα to chromatin, prerequisite events for DNA synthesis after unwinding of origin DNA, but is not required for chromatin binding of Cdc45 or GINS, essential components of the CMG complex.Citation28,29 By taking advantage of the in vitro system, we investigated the role of RecQ4 in the origin unwinding step. We first examined whether the activation of pre-RC in NPE recapitulates the event observed with chromatin binding after nuclear formation. Upon depletion of RecQ4 from both high-speed supernatant (HSS) (pre-RC formation) and NPE (initiation of DNA replication), we found that RecQ4 is required for efficient DNA binding of Polα and DNA replication activity, but is dispensable for the binding of Cdc45 and Sld5, components of the CMG (, lanes 5 and 7, and 6B, compare mock and ΔRecQ4, −). We also confirmed that the N-terminal fragment of RecQ4 (termed N2),Citation29 which shares homology with Sld2, rescued the replication activity and DNA binding of Polα in RecQ4-depleted extracts (, compare lanes 7 and 8, and 6B, ΔRecQ4, compare − and +N2). These results are in accord with the results of previous studies using sperm chromatin.Citation28,29
Figure 6 (See previous page). RecQ4-dependent assembly of replisome components, initiation of replication, and unwinding of dsDNA. (A) RecQ4-dependent and -independent binding of initiator proteins to the plasmids. The bead-coupled plasmids were incubated in mock- or RecQ4-depleted HSS at 23°C for 30 min to allow pre-RC assembly. Subsequently, 2 vol of mock-depleted NPE with or without 1 μM p21, or RecQ4-depleted NPE with or without 530 nM N-terminal fragment of RecQ4 (N2), was added to 1 vol of each corresponding HSS and further incubated for 30 min. Proteins bound to the plasmids (plasmid) or in the extracts (extract) were analyzed by immunoblotting. (B) RecQ4 promotes replication of the plasmids in NPE. The bead-coupled plasmids (3.0 kbp) assembled with pre-RC were incubated in NPE as shown in , except for the presence of [α32P]-dATP. The reactions were terminated at the indicated time and the incorporation of [α32P]-dATP into DNA was detected after separating DNA by agarose gel electrophoresis. (C) RecQ4-dependent and -independent recruitment of replication proteins to the site-specific origin. The bead-coupled plasmids (7.6 kbp) bound with GAL4-Cdc6 were incubated at 23°C for 30 min in Cdc6-depleted or Cdc6 and RecQ4 double-depleted HSS. After the incubation, mock-depleted or RecQ4-depleted NPE containing 1 ng/μl aphidicolin was added to the corresponding HSS and further incubated for 30 min. The localization of proteins bound to the plasmids was examined by ChIP analysis as described in , except for using primer sets at X and Z, proximal and distal to the origin, respectively. Antibodies used for ChIP are indicated at the top of each panel. (D) Stable binding of Cdc45, Mcm2-7, and GINS to the plasmid both in the presence and absence of RecQ4. The bead-coupled plasmids were incubated in mock- or RecQ4-depleted extracts as described in . After incubation in the mock- or RecQ4-depleted NPE, plasmid-beads were isolated and washed with the buffer containing various concentrations of NaCl. Proteins bound to the plasmids (plasmid) and in the extracts (extract) were analyzed by immunoblotting. (E) RecQ4 promotes unwinding of DNA in NPE. The bead-coupled plasmids (7.6 kbp) were incubated at 23°C for 30 min in RecQ4-depleted HSS, and then 2 vol of RecQ4-depleted NPE was added with and without a 720 nM N-terminal fragment of RecQ4 (N2) in the presence of 67 ng/μl aphidicolin. After 30 min incubation, the plasmids were isolated and treated with P1 nuclease at 38°C for 10 min. Total DNA isolated from the mixture was further digested with BamHI, and the purified DNA was separated by agarose gel electrophoresis. As a positive control, mock-depleted extracts were used for the unwinding assay (mock).
![Figure 6 (See previous page). RecQ4-dependent assembly of replisome components, initiation of replication, and unwinding of dsDNA. (A) RecQ4-dependent and -independent binding of initiator proteins to the plasmids. The bead-coupled plasmids were incubated in mock- or RecQ4-depleted HSS at 23°C for 30 min to allow pre-RC assembly. Subsequently, 2 vol of mock-depleted NPE with or without 1 μM p21, or RecQ4-depleted NPE with or without 530 nM N-terminal fragment of RecQ4 (N2), was added to 1 vol of each corresponding HSS and further incubated for 30 min. Proteins bound to the plasmids (plasmid) or in the extracts (extract) were analyzed by immunoblotting. (B) RecQ4 promotes replication of the plasmids in NPE. The bead-coupled plasmids (3.0 kbp) assembled with pre-RC were incubated in NPE as shown in Figure 6A, except for the presence of [α32P]-dATP. The reactions were terminated at the indicated time and the incorporation of [α32P]-dATP into DNA was detected after separating DNA by agarose gel electrophoresis. (C) RecQ4-dependent and -independent recruitment of replication proteins to the site-specific origin. The bead-coupled plasmids (7.6 kbp) bound with GAL4-Cdc6 were incubated at 23°C for 30 min in Cdc6-depleted or Cdc6 and RecQ4 double-depleted HSS. After the incubation, mock-depleted or RecQ4-depleted NPE containing 1 ng/μl aphidicolin was added to the corresponding HSS and further incubated for 30 min. The localization of proteins bound to the plasmids was examined by ChIP analysis as described in Figure 2, except for using primer sets at X and Z, proximal and distal to the origin, respectively. Antibodies used for ChIP are indicated at the top of each panel. (D) Stable binding of Cdc45, Mcm2-7, and GINS to the plasmid both in the presence and absence of RecQ4. The bead-coupled plasmids were incubated in mock- or RecQ4-depleted extracts as described in Figure 6A. After incubation in the mock- or RecQ4-depleted NPE, plasmid-beads were isolated and washed with the buffer containing various concentrations of NaCl. Proteins bound to the plasmids (plasmid) and in the extracts (extract) were analyzed by immunoblotting. (E) RecQ4 promotes unwinding of DNA in NPE. The bead-coupled plasmids (7.6 kbp) were incubated at 23°C for 30 min in RecQ4-depleted HSS, and then 2 vol of RecQ4-depleted NPE was added with and without a 720 nM N-terminal fragment of RecQ4 (N2) in the presence of 67 ng/μl aphidicolin. After 30 min incubation, the plasmids were isolated and treated with P1 nuclease at 38°C for 10 min. Total DNA isolated from the mixture was further digested with BamHI, and the purified DNA was separated by agarose gel electrophoresis. As a positive control, mock-depleted extracts were used for the unwinding assay (mock).](/cms/asset/6a708186-563d-4a3b-a989-f34d2aead138/kccy_a_1007003_f0006_oc.gif)
We next examined the localization of RecQ4, Mcm2, Cdc45, and Polα on the plasmids with GAL4 pre-RC in the presence or absence of RecQ4. ChIP analysis showed that RecQ4 depletion from the extracts abolished the specific association of RecQ4 and Polα at site X (, RecQ4 and Polα), consistent with the results obtained by immunoblot analysis shown in (lane 7). Intriguingly, association of Mcm2 and Cdc45 with site X appears to be independent of the presence of RecQ4 in the extracts (, Mcm2 and Cdc45). This is consistent with DNA binding of these proteins (, lanes 5 and 7), suggesting that RecQ4 is not required for the association of Cdc45 with Mcm2-7 located in the vicinity of the binding sites.
Binding of Cdc45 and Sld5 to the plasmid, along with ChIP analysis of Mcm2 and Cdc45 in the absence of RecQ4, may suggest that RecQ4 is not required for the assembly of the CMG complex. To test this possibility further, we examined the effect of a high-salt wash on Cdc45 and Sld5 bound to the plasmid in the presence and absence of RecQ4 in the extracts. shows that the binding of Polα and PCNA, a loading clamp for replicative polymerases, was diminished by depleting RecQ4 from the extracts (compare lanes 3 and 6), but that Mcm2, Cdc45, and Sld5 were similarly bound to the plasmids irrespective of RecQ4 depletion (compare lanes 3 and 6). The binding of both Polα and PCNA to the plasmids was diminished after a high-salt wash, and most proteins were dissociated from the plasmids after an 800 mM NaCl wash (mock, compare lanes 3, 4, and 5). However, binding of Mcm2, Cdc45, and Sld5 to the plasmids was only slightly decreased after the high-salt wash, whether the plasmids were incubated in mock or RecQ4-depleted extracts (compare lanes 4, 5 and 7, 8). These results show that Cdc45, Mcm2-7, and GINS are tightly bound to the plasmid even in the absence of RecQ4.
We further examined whether RecQ4 promotes DNA unwinding. As shown in , smear DNA fragments were generated by P1 nuclease treatment when the plasmids with conventional pre-RC were incubated in the mock-depleted extracts (, lane 4). This was greatly reduced when the plasmids were incubated in the RecQ4-depleted extracts (, lane 5). Importantly, the reduction of smear DNA fragments observed in the absence of RecQ4 was partially recovered by adding the recombinant N2 protein back to the depleted extracts (, compare lanes 5 and 6). Consistent with this result, a similar recovery of DNA replication activity was observed when the RecQ4-depleted extracts were supplemented with recombinant N2 (, ΔRecQ4, compare – and +N2). These results suggest that RecQ4 promotes DNA unwinding for subsequent recruitment of RPA and Polα.
Discussion
The replication origin is a landmark of the genome for the assembly of replication machinery and the initiation of DNA replication in proliferating cells, from bacteria to humans.Citation48 Here we have established an in vitro replication system of plasmid DNA with a site-specific origin. By tethering Cdc6 to the GAL4 DNA-binding sites of a plasmid, we succeeded in assembling pre-RC on the plasmid DNA in Xenopus egg extracts. ChIP analysis of pre-RC components suggests the assembly of pre-RC in the vicinity of the binding sites. Rather weak enrichment of Mcm2 or Mcm3 in the vicinity of the binding sites may be due to the non-specific loading of Mcm2-7 catalyzed by residual Cdc6 in the depleted extracts or by GAL4 Cdc6 non-specifically bound to the plasmid. Consistent with this finding, geminin reduced the association of Mcm2 at the distal sites, Y and Z, to background levels (, Mcm2, compare GST and Cdc6). However, the assumption that a large amount of Mcm2-7 was non-specifically loaded onto the plasmids contradicts the present results, which show minimal GAL4-Cdc6 independent binding of Mcm2 to the plasmid () and specific incorporation of BrdUTP into the vicinity of—but not distal to—the binding sites ( and S3). Therefore, we prefer to assume that Mcm2-7 is loaded in the vicinity of the binding sites and that the bound Mcm2-7 may interact non-specifically with the other region of the plasmid DNA to be stabilized after cross-linking of DNA and proteins in ChIP analysis. With a smaller plasmid (3 kbp in Fig. S1), we noticed less enrichment of Mcm2 and Cdc6 in the vicinity of the binding sites. This may either be in accord with our assumption, or the technical difficulty in fragmenting smaller plasmids by sonication may explain the rather weak enrichment of pre-RC near the binding sites. In any case, the present results fit well with a previous study showing that tethering ORC or Cdc6 to the target site of plasmids is sufficient to create an artificial origin in mammalian cells.Citation38
We did not observe, in contrast to the results of the previous study, specific enrichment of Orc2 at the GAL4 DNA-binding sites on the plasmid DNA (, Orc2). The tethering of GAL4-Cdc6 did not significantly affect Orc2 localization on the plasmid. This result is in accord with the data showing that the amount of Orc2 bound to the plasmid is not affected by the presence or absence of Cdc6 tethered to the plasmid ( and C). Since metazoan ORC has no significant sequence preference in its DNA binding,Citation49 our data likely reflect sequence-independent, random DNA binding of Xenopus ORC. Intriguingly, however, Orc2 was enriched at the DNA-binding sites when geminin was added to the extracts (, Orc2, +geminin). This result suggests that geminin stabilizes a transient Mcm2-7 loading complex that includes ORC, and in the absence of geminin, ORC is quickly dissociated from the binding sites after the loading of Mcm2-7.
In contrast to ORC, Mcm2-7 appears to stay on the loaded site. Using ChIP analysis, we tested the possible translocation of loaded Mcm2-7 along DNA without origin firing. If Mcm2-7 could freely slide along dsDNA, loaded Mcm2-7 might move away from the DNA-binding sites where it is recruited. As a consequence, enrichment of Mcm2-7 at site X may be reduced when GAL4 pre-RC is incubated in the extracts for a longer period. Since the loading of Mcm2-7 on the plasmid reached a maximal level within 30 min (Fig. S2B), we compared the distributions of Mcm2 on the plasmid incubated for 30 and 90 min in the extracts (Fig. S2C). No significant reduced association of Mcm2 at site X was observed even after 90 min of incubation, suggesting that Mcm2-7 stably associates at the initially loaded site. Stable association of Mcm2-7 with DNA may explain the different localization of Mcm2-7 and ORC bound to chromatin in HeLa cell nuclei.Citation50 Different localization of Mcm2-7 and ORC might be due to the rapid dissociation of ORC from the loading site of Mcm2-7 and rebinding to DNA in a sequence non-specific manner.
By using NPE, we demonstrated that GAL4 pre-RC is converted into the replisome and that DNA replication is initiated from the vicinity of the GAL4 DNA-binding sites. In addition, the P1 nuclease assay suggests unwinding of plasmid DNA from the vicinity of the binding sites. Taken together, the results of the present study show that the site of pre-RC assembly coincides with the site of replisome assembly, origin unwinding, and initiation of DNA replication. In addition, establishment of the in vitro system with the egg extracts enables us to study molecular mechanisms for the initiation of DNA replication in higher eukaryotes. One possible drawback of the present system is that we used the plasmids coupled to magnetic beads, which may contribute to a lower efficiency of replication. The previous study with NPE suggests nearly 100% replication efficiency of naked template DNA,Citation34 and we found that the replication activity of the bead-coupled plasmid was as much as 40% of that of free plasmid by incubating the conventional pre-RC in NPE (Fig. S6).
Topological constraints of the bead-coupled plasmids may also contribute to the apparent absence of uncoupling of DNA polymerase and helicase in the present study. A previous study shows that the plasmid DNA in NPE undergoes extensive topological changes during DNA replication.Citation42 In the presence of aphidicolin, DNA structures are rapidly converted into U-form DNA, and the authors suggest the extensive unwinding of DNA, exposing several kilobases of the ssDNA region in the presence of aphidicolin. In the present study, ChIP analysis suggests that replisome components such as Polα and Cdc45 are assembled in the vicinity of the GAL4 DNA-binding sites in the presence of aphidicolin. In addition, the different fragmentation patterns of plasmids treated with P1 nuclease followed by the restriction enzyme suggest the limited unwinding of DNA. If extensive unwinding of DNA occurs in these experiments, the observed fragmentation patterns after digestion by 2 different restriction enzymes would be no different with the GAL4 pre-RC assembled plasmid. The apparent failure to detect the extensive unwinding of DNA in NPE could be because we used plasmid DNA attached to the beads.
The successful establishment of the in vitro replication system with the defined origin enabled us to study the assembly of replication initiation factors such as Treslin and RecQ4, which have diverged from their putative homologs in yeast (Sld3 and Sld2/Drc1, respectively). We found that RecQ4 localizes to the site-specific origin, which is in accord with the previous observation that RecQ4 associates with replication origins during the G1/S phase in human cells.Citation51 Furthermore, our study revealed that Treslin is localized in the vicinity of the origin after incubating the pre-RC in NPE, consistent with its role in the assembly of pre-IC.Citation22 In contrast, we found that Xenopus RecQ4 is not directly involved in the loading of Cdc45 onto the replication origin, but is important for recruitment of Polα to the origin. Considering the interdependent chromatin binding of Cdc45 and GINS,Citation52 as well as the tight binding of Cdc45, Mcm2-7, and GINS on the plasmid in the absence of RecQ4, it is reasonable to speculate that RecQ4 functions after the formation of pre-IC containing “inactive” CMG complex to convert it into replisomes containing “active” CMG complex. RecQ4-dependent unwinding of DNA () and the recruitment of Polα to the origin () support the view that RecQ4 is involved in the unwinding of the origin to initiate DNA replication, but how RecQ4 promotes DNA unwinding remains unknown. Recent studies with yeasts suggest that Mcm10 is required for the functioning of the CMG complex,Citation53-55 presumably after pre-IC formation. In human cells, RecQ4 is reported to interact directly with Mcm10, and human RecQ4 seems to have an important role in the assembly of the CMG complex onto chromatin.Citation56,57 It would therefore be interesting to test whether RecQ4 activates helicase through Mcm10 in Xenopus egg extracts. In conclusion, we have established a metazoan in vitro replication system with a site-specific origin, and with this system we suggest that RecQ4 functions at a step after the assembly of the CMG complex. Further examination with this in vitro replication system will give us fundamental insights into the molecular mechanism for the initiation of DNA replication in higher eukaryotes.
Materials and Methods
Assembly of pre-RCs on the plasmids and incubation of pre-RCs in NPE
Incubation of bead-coupled plasmids in extracts was performed as describedCitation58 with some modification. To assemble the GAL4 pre-RC, we washed 200 ng of bead-coupled plasmids 2 times with 100 μl of ice-cold EB (100 mM KCl, 2.5 mM MgCl2, and 50 mM HEPES-KOH, pH 7.5) supplemented with 0.25% Nonidet P-40 (NP40). The plasmids were then mixed with 210 nM GAL4-Cdc6 in 10 μl binding buffer (EB supplemented with 400 mM NaCl and 0.02% NP40) and incubated for 10 min on ice. After incubation, the beads were washed 3 times with 100 μl of ice-cold EB supplemented with 350 mM NaCl and 0.25% NP40 and once with EB supplemented with 0.25% NP40. The beads were then mixed with 10 μl of Cdc6-depleted low-speed supernatant (LSS) (40 ng plasmid/μl) and incubated at 23°C for 30 min to assemble the GAL4 pre-RC. For assembling the conventional pre-RC, the bead-coupled plasmids were washed 2 times with 100 μl of ice-cold EB supplemented with 0.25% NP40 and incubated in LSS (40 ng plasmid/μl). The reaction was stopped by washing the beads 3 times with 150 μl of ice-cold EB supplemented with 0.25% NP40 and 1.5 mM ATP. For analyzing proteins bound to the plasmids, the washed beads were resuspended in SDS-PAGE sample buffer containing 4% 2-mercaptoethanol and heated at 95°C for 5 min before being subjected to SDS-PAGE.
For incubation of pre-RCs in NPE, we used membrane-depleted egg extracts (HSS) instead of LSS to assemble pre-RCs, and the beads assembled with the GAL4 or the conventional pre-RC were incubated at 23°C for various times after adding 2 vol of NPE supplemented with and without aphidicolin to 1 vol of HSS containing the beads. Immunoblotting of proteins bound to the plasmids was performed as described previously.Citation59
Chromatin immunoprecipitation (ChIP) assay
The ChIP assay was performed as follows. Bead-coupled plasmids were incubated in the egg extracts to allow protein binding to the plasmid DNA. After incubation in the extracts, the plasmids were briefly washed with EB supplemented with 0.25% NP40 and 1.5 mM ATP; resuspended with 200 μl of EB supplemented with 0.25% NP40, 1.5 mM ATP, and 1% formaldehyde; and incubated for 10 min at room temperature. The cross-linked products were washed 3 times with 150 μl of EB containing 0.25% NP40 and 2 times with 150 μl of lysis buffer (140 mM NaCl, 1% TritonX-100, 20 mM β-glycerophosphate, 50 mM NaF, 0.1 mM Na3VO4, 0.5 mM sodium pyrophosphate, 1 mM EDTA, and 50 mM HEPES-KOH, pH 7.5), and then resuspended with 130 μl of sonication buffer (lysis buffer supplemented with 1× complete Mini (Roche) and 1 mM PMSF) and transferred into a microTUBE (Covaris). The plasmid DNA was sheared to ∼500 bp average size using Covaris S220 (Covaris, USA) with 30 cycles of 15 s sonication at 30 W. The sonicated samples were diluted to give a final concentration of 1 ng DNA/μl, with the sonication buffer containing 5 mg/ml BSA and 0.4 mg/ml salmon sperm, and the magnetic beads were removed with a magnet before immunoprecipitation. The samples were then divided into several aliquots for immunoprecipitation with the respective antibody-coupled beads (IP sample) and also for use as an input sample. For the preparation of antibody-coupled beads, Dynabeads protein A (Veritus) was incubated overnight with the appropriate volume of antiserum at 4°C. Antibody-coupled beads were washed 4 times with PBS containing 5 mg/ml BSA. Each IP sample containing 35 ng DNA was subjected to 300 μg of protein A conjugated beads bound with the antibody and incubated for 3 h at 4°C. After incubation, the beads were washed 3 times with the lysis buffer, 3 times with the lysis buffer containing 0.36 M NaCl, and 3 times with wash buffer (250 mM LiCl, 0.5% NP40, 0.5% (w/v) sodium deoxycholate, 1 mM EDTA, and 10 mM Tris-HCl, pH 8.0). The washed beads, or the input sample, were diluted to 100 μl by adding elution buffer (10 mM EDTA, 1% SDS, and 10 mM Tris-HCl, pH 8.0) and incubated for 10 min at 65°C. The supernatant was supplemented with 200 mM NaCl, 100 ng RNase, and 1% SDS and reverse cross-linked at 65°C by overnight incubation. The DNA was extracted using the QIAquick PCR Purification Kit (QIAGEN) or the FastGene Gel/PCR Extraction Kit (NIPPON Genetics) after a 2 h incubation with 0.1 μg of Proteinase K at 37°C. Purified DNA fragments were analyzed by quantitative real-time PCR (Chromo4, Bio-Rad), and PCR reactions were performed using the Thunderbird SYBR qPCR mix (Toyobo). The cycle threshold value was calculated by using Opticon Monitor software (Bio-Rad). The amounts of target DNA in the IP samples relative to the mock IP sample were determined by qPCR, and 3 PCR reactions for each ChIP sample were performed to obtain the standard deviation. The reproducibility of all ChIP data was confirmed by separate experiments. For the IP analysis against BrdUTP incorporation, the plasmids assembled with the GAL4 pre-RC were incubated in NPE supplemented with 200 μM BrdUTP, and immunoprecipitation was performed without cross-linking plasmids. Before the immunoprecipitation, the plasmid DNA was sheared by sonication, and the bead-coupled plasmids were heated at 95°C for 10 min to dissociate the magnetic beads from the DNA. Dynabeads protein G (Veritas) conjugated with anti-BrdU (B44, BD) or anti-myc (9E10, Santa Cruz) monoclonal antibody was used for immunoprecipitation from the sample containing 50 ng sheared DNA.
P1 nuclease assay
Bead-coupled plasmids were incubated in HSS at 40 ng DNA/μl at 23°C for 30 min to assemble the pre-RCs, and 2 vol of NPE supplemented with 100 ng/μl aphidicolin was added to 1 vol of the incubation mixture and further incubated for 30 min to promote unwinding of dsDNA. The beads were then washed 3 times with EB supplemented with 0.25% NP40 and 1.5 mM ATP and finally suspended in 24 μl of P1 reaction buffer (400 mM NaCl, 8 mM Mg(OAc)2, 0.1 mM Zn(OAc)2, 5 mM ATP, 0.32 mg/ml BSA, 30% glycerol, and 60 mM HEPES-KOH, pH 7.5). P1 nuclease treatment was initiated by adding 1 μl of P1 nuclease (5 units, Wako). After 10 min incubation at 38°C, the reaction was stopped by diluting samples with 75 μl stop buffer (20 mM EDTA, 1% SDS) containing 0.4 mg/ml proteinase K and further incubated for 2 h. DNA was then released from the beads by incubating the mixtures with 100 μl phenol-chloroform at 65°C for 1.5 h. DNA was further purified by phenol-chloroform extraction and ethanol precipitation, and the DNA obtained was further digested by a restriction enzyme. P1 and restriction enzyme-digested DNA were separated by agarose gel electrophoresis, and DNA was visualized by staining with gel red (Wako). The stained DNA band was quantified using LAS-1000 (FUJIFILM) with the aid of ImageJ software (NIH).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Author Contributions
YS performed all the experiments presented in the manuscript, MK contributed to the ChIP analysis, and TT contributed to experiments using NPE. YS, YK, SM, and HT contributed to the design and interpretation of the experiments. All authors contributed to the writing of the manuscript.
Acknowledgments
We are grateful to Shou Waga for technical advice on manipulating magnetic beads coupled with plasmid DNA and to Hiroyuki Araki and Masaru Ishii for utilizing Covaris S220 for the ChIP assay.
Funding
This work was supported by Research Fellowships of the Japan Society for the Promotion of Science for Young Scientists, and by the NIG Collaborative Research Program (2012-B-15, 2013-A-24).
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- Li Y, Araki H. Loading and activation of DNA replicative helicases: the key step of initiation of DNA replication. Genes Cells 2013; 18:266-77; PMID:23461534; http://dx.doi.org/10.1111/gtc.12040
- O’Donnell M, Langston L, Stillman B. Principles and concepts of DNA replication in bacteria, archaea, and eukarya. Cold Spring Harb Perspect Biol. 2013; 5: a010108; PMID:23818497; http://dx.doi.org/10.1101/cshperspect.a010108
- Diffley JFX. Regulation of early events in chromosome replication. Curr Biol 2004; 14: R778-86; PMID:15380092; http://dx.doi.org/10.1016/j.cub.2004.09.019
- Blow JJ, Dutta A. Preventing re-replication of chromosomal DNA. Nat Rev Mol Cell Biol 2005; 6:476-86; PMID:15928711; http://dx.doi.org/10.1038/nrm1663
- Araki H. Initiation of chromosomal DNA replication in eukaryotic cells; contribution of yeast genetics to the elucidation. Genes Genet Syst 2011; 86:141-9; PMID:21952204; http://dx.doi.org/10.1266/ggs.86.141
- Méchali M. Eukaryotic DNA replication origins: many choices for appropriate answers. Nat Rev Mol Cell Biol 2010; 11:728-38; PMID:20861881; http://dx.doi.org/10.1038/nrm2976
- Costa A, Hood I V, Berger JM. Mechanisms for initiating cellular DNA replication. Annu Rev Biochem 2013; 82:25-54; PMID:23746253; http://dx.doi.org/10.1146/annurev-biochem-052610-094414
- Siddiqui K, On KF, Diffley JF. Regulating DNA replication in eukarya. Cold Spring Harb Perspect Biol 2013; 5: a012930; PMID:23838438; http://dx.doi.org/10.1101/cshperspect.a012930
- Riera A, Tognetti S, Speck C. Helicase loading: How to build a MCM2-7 double-hexamer. Semin Cell Dev Biol 2014; 30:104-109; PMID:24637008; http://dx.doi.org/10.1016/j.semcdb.2014.03.008
- Yardimci H, Walter JC. Prereplication-complex formation: a molecular double take? Nat Struct Mol Biol 2014; 21:20-5; PMID:24389553; http://dx.doi.org/10.1038/nsmb.2738
- Evrin C, Clarke P, Zech J, Lurz R, Sun J, Uhle S, Li H, Stillman B, Speck C. A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc Natl Acad Sci U S A 2009; 106:20240-5; PMID:19910535; http://dx.doi.org/10.1073/pnas.0911500106
- Remus D, Beuron F, Tolun G, Griffith JD, Morris EP, Diffley JFX. Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell 2009; 139:719-30; PMID:19896182
- Gambus A, Khoudoli GA, Jones RC, Blow JJ. MCM2-7 form double hexamers at licensed origins in Xenopus egg extract. J Biol Chem 2011; 286:11855-64; PMID:21282109; http://dx.doi.org/10.1074/jbc.M110.199521
- Kang S, Warner M, Bell S. Multiple Functions for Mcm2–7 ATPase Motifs during Replication Initiation. Mol Cell 2014; 55:1-11.
- Coster G, Frigola J, Beuron F, Morris E, Diffley J. Origin Licensing Requires ATP Binding and Hydrolysis by the MCM Replicative Helicase. Mol Cell 2014; 55:1-12.
- Makarova KS, Koonin E V. Archaeology of eukaryotic DNA replication. Cold Spring Harb Perspect Biol 2013; 5: a012963; PMID:23881942
- Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci U S A 2006; 103: 10236-41; PMID:16798881; http://dx.doi.org/10.1073/pnas.0602400103
- Ilves I, Petojevic T, Pesavento JJ, Botchan MR. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol Cell 2010; 37:247-58; PMID:20122406; http://dx.doi.org/10.1016/j.molcel.2009.12.030
- Sanchez-Pulido L, Diffley JFX, Ponting CP. Homology explains the functional similarities of Treslin/Ticrr and Sld3. Curr Biol 2010; 20: R509-10; PMID:20620901; http://dx.doi.org/10.1016/j.cub.2010.05.021
- Masai H. RecQL4: a helicase linking formation and maintenance of a replication fork. J Biochem 2011; 149:629-31; PMID:21436139; http://dx.doi.org/10.1093/jb/mvr031
- Wardlaw C, Carr A, Oliver A. TopBP1: A BRCT-scaffold protein functioning in multiple cellular pathways. DNA Repair 2014; 22:165-174; PMID:25087188; http://dx.doi.org/10.1016/j.dnarep.2014.06.004
- Kumagai A, Shevchenko A, Shevchenko A, Dunphy WG. Treslin collaborates with TopBP1 in triggering the initiation of DNA replication. Cell 2010; 140:349-59; PMID:20116089; http://dx.doi.org/10.1016/j.cell.2009.12.049
- Kamimura Y, Masumoto H, Sugino A, Araki H. Sld2, which interacts with Dpb11 in Saccharomyces cerevisiae, is required for chromosomal DNA replication. Mol CellBiol 1998; 18:6102-9; PMID:9742127
- Zegerman P, Diffley JFX. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature 2007; 445:281-5; PMID:17167417; http://dx.doi.org/10.1038/nature05432
- Fukuura M, Nagao K, Obuse C, Takahashi TS, Nakagawa T, Masukata H. CDK promotes interactions of Sld3 and Drc1 with Cut5 for initiation of DNA replication in fission yeast. Mol Biol Cell 2011; 22:2620-33; PMID:21593208
- Muramatsu S, Hirai K, Tak Y-S, Kamimura Y, Araki H. CDK-dependent complex formation between replication proteins Dpb11, Sld2, Pol ϵ, and GINS in budding yeast. Genes Dev 2010; 24:602-12; PMID:20231317; http://dx.doi.org/10.1101/gad.1883410
- Tanaka S, Komeda Y, Umemori T, Kubota Y, Takisawa H, Araki H. Efficient initiation of DNA replication in eukaryotes requires Dpb11/TopBP1-GINS interaction. Mol Cell Biol 2013; 33:2614-22; PMID:23629628; http://dx.doi.org/10.1128/MCB.00431-13
- Sangrithi MN, Bernal JA, Madine M, Philpott A, Lee J, Dunphy WG, Venkitaraman AR. Initiation of DNA replication requires the RECQL4 protein mutated in Rothmund-Thomson syndrome. Cell 2005; 121:887-98; PMID:15960976; http://dx.doi.org/10.1016/j.cell.2005.05.015
- Matsuno K, Kumano M, Kubota Y, Hashimoto Y, Takisawa H. The N-terminal noncatalytic region of Xenopus RecQ4 is required for chromatin binding of DNA polymerase alpha in the initiation of DNA replication. Mol Cell Biol 2006; 26:4843-52; PMID:16782873; http://dx.doi.org/10.1128/MCB.02267-05
- Heller RC, Kang S, Lam WM, Chen S, Chan CS, Bell SP. Eukaryotic origin-dependent DNA replication in vitro reveals sequential action of DDK and S-CDK kinases. Cell 2011; 146:80-91; PMID:21729781; http://dx.doi.org/10.1016/j.cell.2011.06.012
- Gros J, Devbhandari S, Remus D. Origin plasticity during budding yeast DNA replication in vitro. EMBO J 2014; 33:621-36; PMID:24566988; http://dx.doi.org/10.1002/embj.201387278
- On KF, Beuron F, Frith D, Snijders AP, Morris EP, Diffley JFX. Prereplicative complexes assembled in vitro support origin-dependent and independent DNA replication. EMBO J. 2014; 33:605-20; PMID:24566989; http://dx.doi.org/10.1002/embj.201387369
- Blow JJ, Laskey R. Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell 1986; 47:577-87; PMID:3779837; http://dx.doi.org/10.1016/0092-8674(86)90622-7
- Walter J, Sun L, Newport J. Regulated Chromosomal DNA Replication in the Absence of a Nucleus. Cell 1998; 1:519-529; PMID:9660936
- Méchali M, Kearsey S. Lack of specific sequence requirement for DNA replication in Xenopus eggs compared with high sequence specificity in yeast. Cell 1984; 38:55-64; PMID:6380762; http://dx.doi.org/10.1016/0092-8674(84)90526-9
- Harvey KJ, Newport J. CpG Methylation of DNA Restricts Prereplication Complex Assembly in Xenopus Egg Extracts. Mol Cell Biol 2003; 23:6769-6779; PMID:12972597; http://dx.doi.org/10.1128/MCB.23.19.6769-6779.2003
- Danis E, Brodolin K, Menut S, Maiorano D, Girard-Reydet C, Méchali M. Specification of a DNA replication origin by a transcription complex. Nat. Cell Biol. 2004; 6:721-30; PMID:15247921; http://dx.doi.org/10.1038/ncb1149
- Takeda DY, Shibata Y, Parvin JD, Dutta A. Recruitment of ORC or CDC6 to DNA is sufficient to create an artificial origin of replication in mammalian cells. Genes Dev 2005; 19:2827-36; PMID:16322558; http://dx.doi.org/10.1101/gad.1369805
- Rowles a, Tada S, Blow JJ. Changes in association of the Xenopus origin recognition complex with chromatin on licensing of replication origins. J Cell Sci 1999; 112:2011-8; PMID:10341218
- Wohlschlegel JA, Dwyer BT, Dhar SK, Cvetic C, Walter JC, Dutta A. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science 2000; 290:2309-12; PMID:11125146; http://dx.doi.org/10.1126/science.290.5500.2309
- Tada S, Li A, Maiorano D, Méchali M, Blow J. Repression of origin assembly in metaphase depends on inhibition of RLF-B / Cdt1 by geminin. Nat Cell Biol 2001; 3:107-113; PMID:11175741; http://dx.doi.org/10.1038/35055000
- Walter J, Newport J. Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase alpha. Mol Cell 2000; 5:617-27; PMID:10882098
- Li J, Kelly T. Simian virus 40 DNA replication in vitro. Proc Natl Acad Sci U S A 1984; 81:6973-6977; PMID:6095264; http://dx.doi.org/10.1073/pnas.81.22.6973
- Bramhill D, Kornberg A. Duplex opening by dnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell 1988; 52:743-55; PMID:2830993; http://dx.doi.org/10.1016/0092-8674(88)90412-6
- Keyamura K, Abe Y, Higashi M, Ueda T, Katayama T. DiaA dynamics are coupled with changes in initial origin complexes leading to helicase loading. J Biol Chem 2009; 284:25038-50; PMID:19632993; http://dx.doi.org/10.1074/jbc.M109.002717
- Boos D, Frigola J, Diffley JFX. Activation of the replicative DNA helicase: breaking up is hard to do. Curr Opin Cell Biol 2012; 24:423-30; PMID:22424671; http://dx.doi.org/10.1016/j.ceb.2012.01.011
- Tanaka S, Araki H. Helicase activation and establishment of replication forks at chromosomal origins of replication. Cold Spring Harb Perspect Biol. 2013; 5: a010371; PMID:23881938
- Leonard AC, Méchali M. DNA replication origins. Cold Spring Harb Perspect Biol 2013; 5: a010116; PMID:23838439
- Vashee S, Cvetic C, Lu W, Simancek P, Kelly TJ, Walter JC. Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes Dev. 2003; 17:1894-908; PMID:12897055; http://dx.doi.org/10.1101/gad.1084203
- Ritzi M, Baack M, Musahl C, Romanowski P, Laskey RA, Knippers R. Human minichromosome maintenance proteins and human origin recognition complex 2 protein on chromatin. J Biol Chem 1998; 273:24543-9; PMID:9733749; http://dx.doi.org/10.1074/jbc.273.38.24543
- Thangavel S, Mendoza-Maldonado R, Tissino E, Sidorova JM, Yin J, Wang W, Monnat RJ, Falaschi A, Vindigni A. Human RECQ1 and RECQ4 helicases play distinct roles in DNA replication initiation. Mol Cell Biol 2010; 30:1382-96; PMID:20065033; http://dx.doi.org/10.1128/MCB.01290-09
- Kubota Y, Takase Y, Komori Y, Hashimoto Y, Arata T, Kamimura Y, Araki H, Takisawa H. A novel ring-like complex of Xenopus proteins essential for the initiation of DNA replication. Genes Dev. 2003; 17:1141-52; PMID:12730133; http://dx.doi.org/10.1101/gad.1070003
- Watase G, Takisawa H, Kanemaki MT. Mcm10 plays a role in functioning of the eukaryotic replicative DNA helicase, Cdc45-Mcm-GINS. Curr Biol 2012; 22:343-9; PMID:22285032; http://dx.doi.org/10.1016/j.cub.2012.01.023
- Van Deursen F, Sengupta S, De Piccoli G, Sanchez-Diaz A, Labib K. Mcm10 associates with the loaded DNA helicase at replication origins and defines a novel step in its activation. EMBO J. 2012; 31:2195-206; PMID:22433841; http://dx.doi.org/10.1038/emboj.2012.69
- Kanke M, Kodama Y, Takahashi TS, Nakagawa T, Masukata H. Mcm10 plays an essential role in origin DNA unwinding after loading of the CMG components. EMBO J 2012; 31:2182-94; PMID:22433840; http://dx.doi.org/10.1038/emboj.2012.68
- Im J-S, Ki S-H, Farina A, Jung D-S, Hurwitz J, Lee J-K. Assembly of the Cdc45-Mcm2-7-GINS complex in human cells requires the Ctf4/And-1, RecQL4, and Mcm10 proteins. Proc Natl Acad Sci U S A 2009; 106:15628-32; PMID:19805216; http://dx.doi.org/10.1073/pnas.0908039106
- Xu X, Rochette PJ, Feyissa EA, Su T V, Liu Y. MCM10 mediates RECQ4 association with MCM2-7 helicase complex during DNA replication. EMBO J 2009; 28:3005-14; PMID:19696745
- Waga S, Zembutsu A. Dynamics of DNA binding of replication initiation proteins during de novo formation of pre-replicative complexes in Xenopus egg extracts. J Biol Chem 2006; 281:10926-34; PMID:16497662; http://dx.doi.org/10.1074/jbc.M600299200
- Ode KL, Fujimoto K, Kubota Y, Takisawa H. Inter-origin cooperativity of geminin action establishes an all-or-none switch for replication origin licensing. Genes Cells 2011; 16:380-96; PMID:21426446; http://dx.doi.org/10.1111/j.1365-2443.2011.01501.x
- Coleman TR, Carpenter PB, Dunphy WG. The Xenopus Cdc6 protein is essential for the initiation of a single round of DNA replication in cell-free extracts. Cell 1996; 87:53-63; PMID:8858148; http://dx.doi.org/10.1016/S0092-8674(00)81322-7
