Abstract
As the name implies, Stimulated by Retinoic Acid 8 is an early retinoic acid (RA) responsive gene pivotal for the beginning of meiosis in female and male germ cells. Its expression is strictly time-dependent and cell-specific (pre-meiotic germ cells) and likely requires a complex mechanism of regulation. In this study, we demonstrate a direct negative control of SOHLH1 and SOHLH2, 2 germ cell specific bHLH transcription factors, on Stra8 expression. We observed a negative correlation between STRA8 and SOHLH1 expression in prepuberal differentiating mouse KIT+ spermatogonia and found that SOHLH1 and SOHLH2 were able to directly and cooperatively repress STRA8 expression in cell lines in vitro through binding to its promoter. We also identified 2 canonical E-Box motives in the Stra8 promoter that mediated the negative regulation of SOHLH1 and SOHLH2 on these gene both in the cell lines and KIT+ spermatogonia. We hypothesize that this novel negative activity of SOHLH1and SOHLH2 in male cooperates with that of other transcription factors to coordinate spermatogonia differentiation and the RA-induced meiosis and in female ensures STRA8 down-regulation at mid-end stages of meiotic prophase I.
Keywords:
Abbreviations
| STRA8 | = | Stimulated by Retinoic Acid 8 |
| EC | = | Embyonal Carcinoma |
| RA | = | retinoic acid |
| PGC | = | primordial germ cell |
| RARE | = | Retinoic Acid Response Element |
| TF | = | transcription factor |
| ChIP | = | chromatin immunoprecipitation |
| dpc | = | days post coitum |
| dpp | = | days postpartum |
Introduction
Stimulated by Retinoic Acid Gene 8 (STRA8) protein has been proposed as the molecular effector of Retinoic Acid (RA) involved in meiosis-inductive activity. The mouse Stra8 gene, originally identified in Embryonal Stem (ES) cells and germ cell-derived Embryonal Carcinoma (EC) cells after RA treatment,Citation1,2 is expressed at relatively high levels in male and female premeiotic germ cells. In female embryos lacking Stra8, the specification and the initial development of the primordial germ cells (PGCs), the precursor of the adult gametes, appear normal, but they fail to undergo premeiotic DNA replication and meiotic chromosome condensation.Citation3,4 In prenatal spermatogonia of Stra8 deficient mice, the premeiotic DNA replication is conserved and spermatocytes are able to partly condense chromosomes and initiate meiotic recombination;Citation5,6 spermatocytes fail, however, to regularly continue over the leptotene stage of meiotic prophase I.Citation5
Although these studies support the pivotal role of Stra8 in gametogenesis, the molecular function/s of the protein remain unknown. First described as a cytoplasmic protein,Citation2 it has been recently demonstrated in our laboratory, that it actually shuttles between the nucleus and cytoplasm.Citation7 It was also shown that STRA8 can bind to DNA and possesses a transcriptional activation domain in its C-terminal region.Citation7,8
Although in several of developmental processes the basic molecular mechanisms underlying the RA action have been identified, its action in promoting entry into meiosis through Stra8 induction has yet to be clarified. Actually it is known that in responding cells, RA usually serves as a ligand for 2 families of nuclear receptors, the RA receptors (RARα, β, or γ) and the retinoid X receptors (RXRα, β, or γ). RAR–RXR heterodimers bind to specific genomic DNA sequences, known as Retinoic Acid Response Elements (RARE), and regulate gene expression (for a review see refs. 9–10 and references here in). A number of studies showing that specific antagonists or agonists of the RARs were able to influence the onset of meiosis and the expression of Stra8 in cultured germ cells, suggested a canonical mode of RA action on the germ cell meiotic entry (for a review see ref. 11). This notion was initially also supported by the report that the Stra8 promoter contains 2 RARE, corresponding to DR2 and IR5 type, located in a 400 bp region upstream of the Stra8 transcriptional start site (TSS).Citation12 However, the importance of such elements for Stra8 regulation remains controversial. In particular, it was reported that in the fetal ovary of Raldh2−/− mice, that lack RA synthesis, Stra8 expression occurs normally.Citation13 In line with this, Le Bouffant et al.Citation14 showed that in embryonic mouse ovary the ablation of the homeobox Msh-like (Msx) genes 1 and 2 resulted in downregulation of Stra8. Moreover, Doublesex and mab-3 related transcription factor 1 (Dmrt1) and 6 (Dmrt6) appear to play a crucial role in regulating meiosis and Stra8 expression.Citation15-18 In particular, DMRT1 acts in a opposite sex-dependent way. Both in fetal female germ cells and postnatal male germ cells, DMRT1 is able to bind to the Stra8 promoter. Citation15-18 While in female germ cells the ablation of Dmrt1 results in lack of Stra8 expression,Citation16 in male germ cells it caused uncontrolled meiotic entry.Citation15 In these latter, DMRT1 regulates the responsiveness to RA repressing directly the transcription of Stra8 and delaying the enter into meiosis until spermatogonia are not completely competent.Citation15
Stra8 expression appears also under the control of epigenetic events. Feng et al.Citation19 have recently reported that in the female mouse germ cells, NOTCH signaling is probably necessary for maintaining the epigenetic state of Stra8 in a way suitable for RA stimulation. In F9 EC cells, Trichostatin-A (TSA), an inhibitor of class I/II histone deacetylase (HDAC), was found to mimic and synergize the inductive effect of RA on Stra8 expression.Citation20 In ES cells, the expression of Stra8 was shown to be regulated by CBP and p300, 2 histone acetyltransferase (HAT)-activity-containing proteins, while RA itself was reported to increase histone acetylation at the Stra8 promoter.Citation21 In these cells, the most prominent binding sites for CTCFL (BORIS), a key coordinator factor of the 3-dimensional chromatin structure, are localized on the first intronic region of Stra8 and when CTCFL was overexpressed, Stra8 was induced.Citation22
Finally, recent results suggest that, like other genes transcriptionally activated by RA at early times, in female germ cells Stra8 resides in bivalent chromatin region and the Polycomb repressive complex 1 (PRC1) functions as a gatekeeper that control the responsiveness of premeiotic germ cells to the RA signaling.Citation23
SOHLH1 and SOHLH2, 2 testis and ovary-specific bHLH transcription factors (TFs), essential for both spermatogenesisCitation24-27 and oogenesis,Citation28,29 might also play a role in the regulation of Stra8 expression. In the mouse, SOHLH1 is expressed in Aal, A1–A4, Intermediate spermatogonia and at lower level in B spermatogoniaCitation24,27,30 and is up-regulated by RA treatment.Citation30 SOHLH2 is expressed in all A spermatogonia (As–Aal and A1–A4) and absent in B spermatogonia.Citation25-27,Citation30 Moreover, Sohlh1- and Sohlh2-knockout mice share common phenotypes, which in homozygosity leads to sterility both in testes and ovary.Citation24-29 STRA8 is upregulated either in female or in male germ cells of both mutants,Citation26-29 suggesting that SOHLH1 and SOHLH2 might exert an inhibitory action on Stra8 expression. Actually, here we report a novel repressive role of SOHLH1 and SOHLH2 on Stra8 expression through direct binding to 2 E-box motives in the -1,400 bp region of its promoter.
Results
Immunolocalization of STRA8 and SOHLH1 in postnatal mouse testis
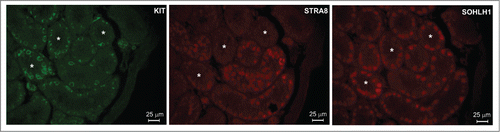
It is well known that spermatogonia differentiation is a RA-dependent event accompanied by the acquisition of Kit expressionCitation31 and that both undifferentiated (KIT−) and differentiating (KIT+) spermatogonia constitute an heterogeneous cell population. Previous results showed that populations of KIT+ spermatogonia express SOHLH1Citation27,30 and STRA8,Citation32 while KIT− spermatogonia express primarily SOHLH2Citation27,30 but not STRA8. In order to determine in single spermatogonia whether there is a correlation between the expression of SOHLH1 and STRA8, we immunoocalized these proteins in testes from 7 dpp mice. We observed that STRA8 and SOHLH1 were expressed in a subset of both KIT+ and KIT− spermatogonia and that the majority (about 70%) of the SOHLH1+ cells were negative for STRA8 and vice versa (), raising the possibility of a functional negative relationship between the 2 proteins.
SOHLH1 and SOHLH2 down-regulate Stra8 expression
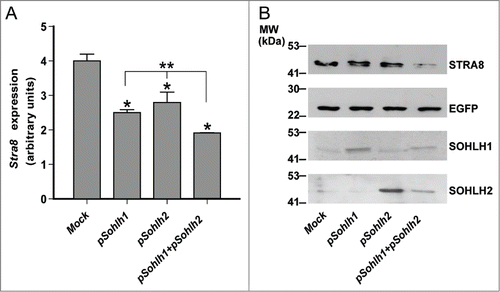
Sohlh1 and Sohlh2 are able to form homo-and heterodimers and bind E-Box motives in DNA of their target genes.Citation26-28,Citation30 They have been mainly described as positive transcriptional regulators and their role as transcriptional repressors is not well characterized.Citation26-28 To verify whether SOHLH1 and SOHLH2 might negatively regulate Stra8 expression, we transfected pcDNA3-Sohlh1, pcDNA3-Sohlh2 or both expression vectors in the germ cells-derived P19 Embryonal Carcinoma (EC) cells, that express Stra8 upon RA stimulation. We observed that the overexpression of either Sohlh1 or Sohlh2 significantly down-regulated Stra8 levels stimulated by RA both at RNA and protein level while they did not affect the expression of the co-transfected control Egfp reporter (). Moreover Stra8 downregulation resulted higher following the co-expression of either transcription factors (TFs) (). As expected,Citation30 overexpression of SOHLH1 and/or SOHLH2 in P19EC cells stimulated endogenous KIT expression (Fig. S1).
Figure 2. Overexpression of SOHLH1 and/or SOHLH2 inhibits Retinoic Acid (RA)-stimulated Stra8 expression in P19EC cells. (A) qRT-PCR analysis of Stra8 expression in P19EC cells electropored with the indicated plasmids and treated, after 4 hr of culture, with 1 μM RA for additional 24 hr. Each qRT-PCR was carried out in triplicate. Bars indicate the mean ± SEM, n = 3 different transfection. *P ≤ 0.01 respect to the Mock; **P ≤ 0.01 for Stra8 expression levels in Sohlh1 plus Sohlh2 co-transfected P19EC cells versus single transfectants. (B) Western blot analysis of STRA8 levels in P19EC cells electropored with limiting concentration of pEgfp expressing vector and pcDNA3-Sohlh1, pcDNa3-Sohlh2, alone or together and treated as above.
SOHLH1 and SOHLH2 down-regulate the Stra8 promoter activity through E-Box DNA binding motif
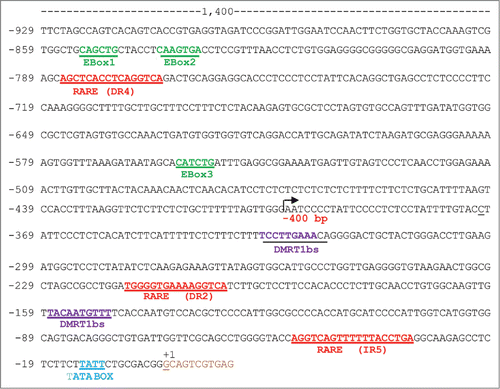
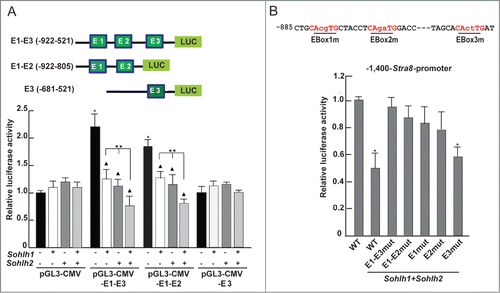
Sequence analysis of the −1,400 bp regulatory region of the mouse Stra8 gene for the presence of canonical E-Box motif (namely CANNTG), revealed the presence of 3 putative E-Box sites at positions −558 bp (CATCTG;) (Ebox-3, proximal), −841 bp (CAAGTG) (Ebox-2, middle) and −853 bp (CAGCTG) (Ebox-1, distal), relative to the transcriptional start site (TSS) (). The −1,400 bp region, located upstream from TSS (+1), includes the TATA promoter sequence and it is known to drive the expression of Stra8 in the testisCitation32-33 and fetal ovary.Citation35
Figure 3. Partial schematic representation of the −1,400 Stra8 promoter region. +1 indicates the start of transcription. The TATA-box like sequence (blue), the putative binding sites (RARE, red) for retinoic acid receptor, for DMRT1 (purple) and the 3 E-Boxes (green) are indicated. It is also indicated the begin of the −400-Stra8-promoter construct ( ).
To verify whether SOHLH1 and SOHLH2 were able to modulate the promoter activity of Stra8, we generated 2 expression vectors in which the −1,400 bp and −400 bp regulatory regions drive the expression of the Luciferase genes (pGL3−1,400-Stra8-Luc and pGL3-400−Stra8-Luc vectors) and Egfp (−1,400-Stra8-Egfp; -400-Stra8-Egfp vectors). These plasmids were transiently transfected in P19 EC cells (not shown) and HEK293T (used primarily in such experiments for their higher transfection efficiency) along with Sohlh1 or Sohlh2 or both expression vectors or with an empty vector as a control. Renilla luciferase was used as an internal control for normalization of transfection efficiency. Lysates were obtained 48 hr after transfection and after 24 hr from the addition of 1 μM RA. The results in (insert) showed that the plasmid construct containing the −1,400 bp-Stra8 regulatory region upstream the Luciferase gene [(containing the DR4 (according to Raverdeau et al.,Citation36 DR2 and IR5 RARE sequences) was responsive to RA treatment. When the HEK293T or P19 EC cells (data not shown) were transfected with Sohlh1 or Sohlh2 or both plasmids, Stra8 directed luciferase activity was significantly inhibited. Although either TFs showed a significant inhibitory activity, we found that co-transfection of both Sohlh1 and Sohlh2 maximally inhibited Stra8-mediated luciferase activity. We found that the −400 bp region of the Stra8 promoter that contains the TATA box and the 2 putative RARE elements (DR2 and IR5) but not the E-Boxes, was not responsive to RA stimulation in HEK293T or P19 EC (, insert). As expected, luciferase activity was not influenced by Sohlh1 or Sohlh2 or both plasmids co-transfection following RA stimulation. We also used Egfp as a reporter present in 2 Stra8 promoter plasmids (p-1,400-Stra8-Egfp, p−400-Stra8-Egfp). Co-transfection of each plasmid with Myc-Sohlh1 or Myc-Sohlh2 or both in HEK293T cells was followed by stimulation with RA for 24 hr and the levels of EGFP evaluated by Western blot analysis. To normalize for the transfection efficiency, we introduced in all samples a vector expressing a Myc-Tyr fragment, a construct that was previously validated to be constantly expressed and inert.Citation30,37 As shown in , we found that SOHLH1 or SOHLH2 were able to significantly inhibit EGFP expression when compared to mock-transfected cells. Also in this case, EGFP down-regulation was stronger when both TFs were simultaneously introduced into the cells (). As for the luciferase reporter assays, EGFP levels were not affected by the SOHLHs when the −400 bp region of Stra8 promoter was transfected (). To understand if the E-boxes present in the Stra8 promoter mediate SOHLHs action, we constructed pGL3-CMV-Luc vectors containing all the 3 E-boxes (E1-E3; -922-521 bp), the first and second (E1-E2; -922-805 bp) and only the third (E3; -681-521 bp) (, upper panel) and assayed their transcriptional activities in HEK293T cells in the presence of Sohlh1 or Sohlh2 or both expressing plasmids. The transcriptional assays showed that the region containing the 3 E-Boxes functioned as enhancer element and promoted higher basal transcriptional activity (). When Sohlh1 and Sohlh2 were transfected alone or in combination, the luciferase activity was significantly inhibited when the E1 and E2 sequences were present, while E3 was dispensable (). To determine whether the regulatory action of SOHLHs on the Stra8 promoter was mediated by the E-Boxes, we mutated the 2 internal bases in E1, E2 and E3 motives of the pGL3−1,400-Stra8-Luc construct (CAcgTG, CAgaTG and CActTG respectively) and expressed them in reporter assays with Sohlh1 and Sohlh2 (). Mutation of either E1 or E2 strongly reduced the inhibitory effect of both SOHLH1 and SOHLH2, while the E3-mutated consensus did not influence the inhibitory activity of the TFs (). Comparable results with an E1/E2/E3 triple-mutant promoter were obtained (), thus suggesting that the adjacent E1 and E2 E-boxes were the most important site of SOHLH1 and SOHLH2 binding.
Figure 4. SOHLH1 and SOHLH2 repress the Stra8 promoter activity. (A) HEK293T cells were co-transfected with 200 ng of reporter pGL3-1,400-Stra8-Luc (upper panel) and with different amounts of pcDNA3-Sohlh1- and/or pcDNA3-Sohlh2-expressing vectors as indicated in the Materials and Methods. The cells were treated with 1 μM RA 24 hr after transfection. After 48 hr, cells were harvested and lysed, and luciferase activity were determined and normalized by Renilla luciferase. The results are shown as relative luciferase activity (± SEM) (*P <0.01). The transfections were repeated 3 times. Insert shows the effect of RA treatment in mock- transfected cell . (B) Sohlh1 and/or Sohlh2-expressing vectors were co-transfected in HEK293T cells with the deletion mutant pGL3-400-Stra8-Luc (upper panel) and the luciferase assay was performed as above (mean ± SEM from three different experiments). Relative luciferase activity in mock-transfected cells treated with RA is shown in the insert. (C) Western blot analysis of EGFP expression in HEK293T cells co-transfected with 500 ng of p1,400-Stra8-Egfp reporter, shown in the upper panel, and with Myc-Sohlh1- and/or Myc-Sohlh2-expressing vectors (500 ng or 250 ng). The Myc-Tyr-expressing plasmid was co-transfected in all cases to normalize the transfection efficiency. 1 μM RA was added to the medium 24 hr after transfection and the cells were lysed 48 hr after transfection. The relative ratio of the EGFP expression levels were estimated compared with the reporter expression levels without Sohlhs. (D) Western blot analysis was performed as above using the p400-Stra8-Egfp reporter shown in the upper panel.
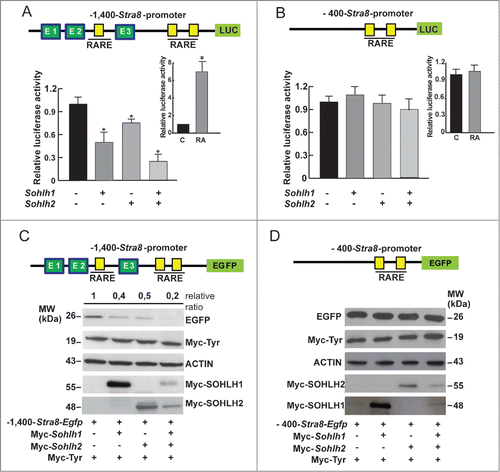
Figure 5. SOHLH1 and SOHLH2 down-regulate the Stra8 promoter activity through E-Box binding motif. (A) Upper panel: schematic representation of the different portions of the Stra8 promoter used in this reporter experiments that contain canonical E-box consensus sequences indicated in green. Lower panel: co-transfection of the different reporter constructs with Sohlh1- and/or Sohlh2-expressing vectors into HEK293T cells. Luciferase activities were measured 48 hr after transfection and normalized to the Renilla activity. Bars depict the means ± SEM of 3 experiments. *P <0.01 respect to the pGLG3-CMV Mock; ▴ P < 0.01 respect to the Mock. **P < 0.01 in Sohlh1 plus Sohlh2 co-transfected cells vs. single transfectants. (B). Upper panel: illustration of mutated E-Box sequences, in each of which the 2 internal bases were replaced as indicated. Lower panel: mutational analysis of the 3 E-Boxes in the Stra8 promoter in the presence of overexpressed Sohlh1 and Sohlh2. HEK293T cells were co-transfected with 200 ng of WT and mutant pGL3-1,400-Stra8-Luc reporter constructs and 400 ng of pcDNA3-Sohlh1 and pcDNA3-Sohlh2 and treated with 1 μM RA after 24 hr. Reporter assays were performed as above.
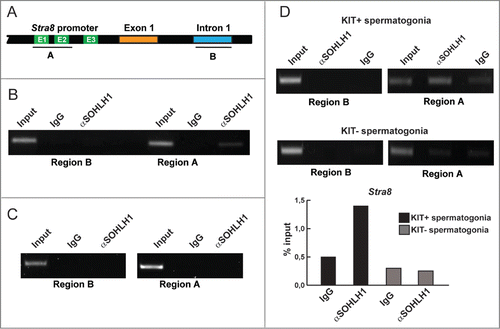
SOHLH1 binds in vivo the upstream regulatory region of Stra8 gene in prepuberal spermatogonia
In order to verify whether that SOHLH1 was able to bind the E-Boxes-containing-Stra8 regulatory region, we performed ChIP assays by immunoprecipitating SOHLH1 from formaldehyde cross-linked chromatin obtained from 7 dpp spermatogonia. We designed primer pair that specifically amplify the genomic fragments containing the first 2 E-box sequences (region A) and primer pair that amplify an intronic region in which E-Boxes were not included (region B) (). Using PCR, we found that only the region containing the E1 and E2 E-boxes sequences was significantly enriched in SOHLH1-immunoprecipitated DNA, while region B, which did not contain E-boxes, was not amplified from SOHLH1-immunoprecipitated chromatin (). We used the mouse NIH3T3 cell line that did not express SOHLHs as a negative control and as expected, no amplicon was present in the immunoprecipitation when using both primer pairs (). In addition, we repeated ChIP experiments on chromatin from both KIT− and KIT+ spermatogonia. The results reported in showed that a significant binding of SOHLH1 was achieved mostly to the chromatin region A obtained from KIT+ spermatogonia.
Figure 6. SOHLH1 binds to the Stra8 promoter of mainly KIT+ differentiating spermatogonia. (A) Stra8 regulatory region flanking exon 1 is displayed showing region A that contains the E-boxes (E1-E3) and region B in the first intron used as negative control. (B) Chromatin immunoprecipitation (ChIP) assay on spermatogonia prepared from 7 dpp mouse testes. The antibody against SOHLH1 precipitates genomic DNA of the Stra8 promoter containing E-Boxes. Input was the PCR product from 2% of the DNA used before IP. (C) ChIP experiment on NIH3T3 cells used as a negative control. (D) ChIP assays on KIT+ and KIT− spermatogonia isolated from 7 dpp mouse testes. Lower panel: densitometric quantification of SOHLH1 binding (% input) from the above ChIP results was performed by determining the amount of SOHLH1-specific signal compared with input for KIT+ or KIT− spermatogonia.
Discussion
A number of studies indicate that Stimulated by Retinoic Acid 8 (STRA8) is crucial for the beginning of meiosis both in embryonic female and postnatal male germ cells.Citation3-6 However, as reported in the Introduction, the regulation of the Stra8 gene by RA, at least in the embryonal ovary, is controversialCitation38 while the role of the encoded protein remains a mystery.
A key event of spermatogenesis, is the transition from spermatogonial mitotic proliferation and differentiation to meiotic entry by spermatocyte. This complex differentiation program is under the control, among others, of RA (for a review see ref. 39). Although several genes that control the early stage of spermatogenesis have been identified, how spermatogonia differentiation is regulated and coordinated with the beginning of meiosis remains poorly understood. (for a review see ref. 40). Several transcription factors have been shown to control spermatogonia maturation. Among these, DMRTs and SOHLHs play a pivotal role. DMRT1 and DMRT6 cooperate in repressing or activating genes involved in spermatogonia differentiation and mitotic/meiotic shift.Citation15,17 Downstream DMRTs are SOHLH1 and SOHLH2 that suppress genes involved in the maintenance of spermatogonial stem cells and induce genes important for spermatogonia differentiation.Citation24-27,Citation30 In the present study, we described in details a novel negative regulatory role exerted by SOHLH1 and SOHLH2 on the Stra8 gene expression.
The observations that DMRT1 regulated spermatogonial commitment to meiosis inducing Sohlh1 and suppressing Stra8 expressionCitation15 and that Stra8 was up-regulated either in female and male germ cells of Sohlh1 and Sohlh2 knockout mice,Citation26-29 prompted us to investigate whether SOHLH1 and SOHLH2 actually may directly suppress Stra8 expression. A first positive confirmation of this hypothesis came from the immunolocalization of the SOHLH1 and STRA8 proteins in single prepuberal spermatogonia. We observed that in some seminiferous tubules con-Kit+ spermatogonia were both STRA8 positive or STRA8 negative. Moreover, in other tubules, STRA8 was clearly present in KIT− spermatogonia, probably corresponding to the B spermatogonia or preleptotene spermatocyte in which Kit starts to be downregulated.Citation41 The heterogeneity of the spermatogonia population was even more evident observing the contemporary expression of KIT, STRA8 and SOHLH1 that are all markers of differentiating spermatogonia regulated by RA.Citation30,32,41 In particular, we noticed that, as a rule, the expression of SOHLH1 and STRA8 was mutually exclusive. This observation parallels the distribution of SOHLH1 in the adult testis reported in previous studies in which SOHLH1 in KIT+ differentiating spermatogonia was shown to gradually disappears in positive intermediate and B spermatogoniaCitation27 that are STRA8 positive.Citation32,42
Subsequent analyses in P19 Embryonal Carcinoma (EC) cell line responsive to RA and transfected with suitable probes, confirmed the repressive action of both SOHLH1 and SOHLH2 on the Stra8 gene expression. Moreover, they indicated that both TFs exerted a cooperative direct inhibition of the gene by binding to 2 canonical E-Box motives within the −1,400 bp regulatory region of the Stra8 promoter known to drive the expression of the gene both in testis Citation33,34 and fetal ovary.Citation35 The cooperation between SOHLH1 and SOHLH2 in repressing Stra8 is not surprising since, as other bHLH transcription factors, they function in spermatogonia by forming homo-or heterodimersCitation27,30,43 and in this last configuration, they are more efficient, for example in stimulating Kit expression.Citation30 We speculate that the cooperation between SOHLH1 and SOHLH2 in repressing Stra8 might ensure a more stringent negative regulation in those KIT+ not fully differentiated spermatogonia in which SOHLH2 is still present and SOHLH1 starts to be expressed.
The finding that in a ChIP assay, SOHLH1 bounds the E1 and E2 Box motives of the Stra8 promoter in the chromatin obtained from KIT+ spermatogonia, strongly supports a regulative role of this transcription factor on the Stra8 gene expression in such cells.
Since STRA8 is pivotal for the beginning of meiotic prophase, it is not surprising that its expression must be finely regulated. In this contest, our data suggest that SOHLH1 and SOHLH2, directly repressing Stra8 expression in the presence of RA, contribute together with DMRT1 and DMRT6 to ensure that meiosis starts only when spermatogonia reached the appropriate differentiation.
In the fetal ovary, in contrast to the testis, DMRT1 is a positive direct regulator of Stra8 expression,Citation16 while 2 homeobox proteins, MSX1 and MSX2, appear necessary for the maintenance of Stra8 level in the meiotic oocytes.Citation14 Moreover, the Polycomb repressive complex (PRC1) is important to maintain Stra8 repressed until the female PGCs do not acquire the meiotic competence.Citation23 Interestingly, in these cells the Stra8 promoter is characterized by a bivalent chromatin state, in which an activating (H3K4me3) and a repressive (H3K27me3) modification of the histones are contemporary present.Citation23 In the female germ line, SOHLH2, but not SOHLH1, was expressed as early as 12.5 dpc and remains expressed at least up to 15.5 dpc.Citation30 Since Stra8 begins to be expressed around 12.5 dpc and is completely down-regulated 3–4 d later, a repressive action of SOHLH2 on Stra8 during this window time can be excluded. This is, however, possible during later meiotic prophase I stages, since in Sohlh1 and Sohlh2-knockout oocytes, Stra8 is the only meiotic gene to be abnormally upregulated at perinatal stages.Citation26,28 At this stages, SOHLH1 has been demonstrated to transcriptionally stimulate through binding to canonical E-box sequences, the promoter activity of Lhx8 and Zp1, 2 genes crucial for the progression of primordial follicles into primary follicles.Citation28
Actually until now, SOHLH1 and SOHLH2 were known as positive regulators of gene expression both in male and female germ cells.Citation26-28,Citation30 Here we show, for the first time, that by binding to E-Boxes, SOHLH1 and SOHLH2 are able to negatively modulate gene transcription. Such double activatory and inhibitory action is common to other members of the bHLH TF family. For example, myogenic regulatory factors, such as MyoD, Myf5, myogenin, and MRF4, all members of the same bHLH TF family, can either activate or repress the transcription of target genes.Citation44 The posttranslational modifications,Citation45 the epigenetic modification of chromatin, the ability to associate with both histone acetylase or deacetylase and recruitment of additional proteins to the transcriptional complex are all involved in the specific mode of action of these TFs (for a review see refs. 46, 47).
We do not known how SOHLH1 and SOHLH2 by binding to E-Boxes, are able to exert opposite effects in spermatogonia, negative on the Stra8 promoter and positive on Kit promoterCitation30 and Sohlh1 promoter itself.Citation43 In the last case, it is the ability of SOHLH1 and SOHLH2 to form a ternary complex with SP1 that binds a GC rich sequence adjacent to the E-Boxes, that results in strong promoter activation. Since the repressive action of SOHLHs occurs in the presence of elevated level of RA, interaction with the RAR/RXR and/or repressive epigenetic modification of the chromatin are possible and will be investigated in future studies.
Materials and Methods
Immunohistochemistry
For immunohistochemistry, serial 5 μm thick sections were obtained from testes of 7 dpp CD1 mice, fixed in buffered formalin and paraffin embedded. Slides were dewaxed, rehydrated, and microwaved in 10 mM sodium citrate buffer, pH 6 for 10 min. After blocking with 10% goat serum, sections were incubated with preimmune serum and with rabbit polyclonal anti-KIT (1:100 AF1356, R&D System), anti-SOHLH1 (1:200 Ab41520, Abcam) and anti-STRA8 (1:400 Ab49405, Abcam) antibodies diluted in PBS/0.5% BSA at 4°C overnight. 1:500 goat anti-rabbit (Alexafluor 568, Thermo Fisher Scientific Inc.) were used as secondary antibodies for 1 hr incubation at RT. Samples were visualized under a Leica CTR600 microscope with a 40X objective.
DNA constructs
The coding region of mouse Sohlh1 (GenBank NM_001001714) and Sohlh2 (GenBank NM_028937) was amplified by RT-PCR from 1 microgram of total RNA obtained from 10 dpp mouse testis using primers 1–4 listed in and cloned in pcDNA3 vector. Plasmids expressing the fusion protein Myc-SOHLH1 and Myc–SOHLH2 were constructed subcloning the coding sequence of mouse Sohlh1 and Sohlh2 within the N-terminus of pCDNA3-N2Myc by using restriction enzymes EcoRI and XhoI for Sohlh1 and BamHI and XhoI for Sohlh2 (primers 1–2 and 3–4 respectively, ). The −1,400-Stra8-Egfp plasmid was generously provided by Prof. JL. Tilly (University of Boston, USA). The −400-Stra8-Egfp vector was obtained subcloning the corresponding region (-400 bp +11 bp) amplified by PCR using the -1,400-Stra8-Egfp as template and primers 5–6 listed in . Plasmids used for luciferase reporter assays were obtained by subcloning the Stra8-promoter regions in pGL3-basic-vector (Promega) by using restriction enzymes XhoI/NcoI (pGL3-1,400-Stra8-Luc; pGL3-400-Stra8-Luc). The deletion mutants containing the E-Boxes E1-E3 (−922-521 bp), E1-E2 (−922-805 bp) and E3 (−652-521 bp) were obtained amplifying the corresponding fragments by PCR by using 1,400-Stra8-Egfp as template and primers 7–10 as indicated in and cloning them in pGL3-CMV-vector (Promega). Mutation of E-Box binding sites in pGL3-1,400-Stra8-Luc were introduced by QuickChange Lightning Multi Site-Directed Mutagenesis Kit (#210513, Agilent Technologies) using primers designed with QuickChange Primer Design Program and listed in . 100 ng of pGL3-1,400-Stra8-Luc were used as template with 100 ng of mutagenic primers and the synthesis reaction was cycled as indicated in instruction manual. All cloned constructs and mutants were confirmed by DNA sequencing (BMR, University of Padova, Italy).
Table 1. Sequences of the primers used in this study
Cell transfection
Mouse P19 embryonic carcinoma cells (ECP19, ATCC CRL-1825) were grown on feeder-free, gelatine-coated plates in D-MEM (Life Technologies) supplemented with 2 mM glutamine, 100 U/ml penicillin/streptomycin, 1 mM sodium pyruvate, 1× nonessential amino acids (all from Sigma-Aldrich) and with 10% fetal bovine serum (FBS; Life Technologies). For STRA8 expression studies, plasmids were introduced into undifferentiated cells by means of a Nucleofector system (Amaxa Biosystems) according to the manufacturer's instructions. Briefly, 2 × 106 cells were electroporated with 2 μg of pcDNA3-Sohlh1 or pcDNA3-Sohlh2 plasmid vectors or 1μg of each in the case of co-transfection. 200 ng of pEGFP-C1 (Takara) was added in all samples as a control. 24 hr after electroporation, total RNA and cell lysates were prepared. HEK293T cells, cultured in D-MEM-10% FBS, were transiently transfected with the different plasmids by using the jetPei TM Polyplus transfection reagent (Polyplus–TransfectionSA, Società Italiana Chimici, Rome, Italy) according to the manufacturer's protocol. The total amount of DNA for each transfection was kept constant using an empty expression vector.
Quantitative real time RT-PCR
Total RNA was extracted from the cell samples using the RNeasy minikit (#74104, Qiagen). Starting from 1 μg of RNA, first-strand cDNA synthesis was performed with quantiTect reverse transcription kit (#205311, Qiagen). 25 ng of cDNA was amplified with the KAPA SYBR FAST qPCR kit (KK4600, Kapa Biosystem) in accordance to the manufacturer's instructions on an ABI PRISM 7300 Sequence Detection System (Life Technologies). Cycling was performed using the default conditions of the ABI 7300 SDS Software 1.3: 2 min at 95°C, followed by 38 cycles of 15 sec at 95°C, 30 sec at 60°C and 30 sec a 70°C. The relative expression of Stra8 was normalized against Gapdh. The primers used (n. 14–17) are indicated in .
Reporter assay
1 × 105 HEK293T cells or P19EC cells were seeded in 24 wells and co-transfected with 200 ng of reporter plasmids and with 800 ng of pcDNA3-Sohlh1 or pcDNA3-Sohlh2 and 400 ng of each in the case of co-transfection. Each well received also 10 ng of a pRL-TK Vector (Promega) to normalize for transfection efficiency. After 24 hr, 1 μM RA was added and at 48 hr after transfection, cells were washed 3 times with PBS and scraped in 100 μl of reporter lysis buffer (Promega). Luciferase activity in 20 μl of the cell extracts was quantified using the Dual-Luciferase Reporter assay system (Promega). Each extract was assayed 3 times with a Hidex luminometer (RadTech, Italy). The Firefly luciferase activity was divided by Renilla luciferase activity and results expressed as the mean ±SEM of 3 experiments.
Spermatogonia isolation
Enriched spermatogonia cell population was obtained from testes of 7 dpp CD1 mice as previously reported by Rossi et al.Citation48 The cells were cultured for 4 hr in MEM (Life Technologies) with 1 mM lactic acid, 100 U/ml penicillin/streptomycin, 1 mM sodium pyruvate, 1× nonessential amino acids (all from Sigma-Aldrich), and supplemented with 10% FBS to promote adhesion of somatic cells. Spermatogonia were recovered after centrifugation. Separation of KIT+ from KIT− spermatogonia was performed by magnetic-activated cell sorting (MACS) with CD117 conjugated microbeads (Miltenyi Biotech) as previously described.Citation41
Chromatin immunoprecipitation (ChIP)
Proteins obtained from at least 107 spermatogonia or KIT+ and KIT− spermatogonia isolated from testes of 7 dpp mice, were cross-linked to DNA by direct addition to the culture medium of formaldehyde at 1% final concentration for 10 min at 37°C. After sonication, a ChIP assay was performed according to the Abcam protocol. Protein-DNA complexes were immunoprecipitated overnight in the presence of the specific anti-SOHLH1 antibody (Abcam) or rabbit IgGs (Sigma-Aldrich). DNA was purified by phenol/chloroform/isoamylic alcohol extraction and ethanol precipitation, and resuspended in 20 μl of water and used directly for PCR. Primers used were indicated in .
Table 2. Sequences of the primers used in ChIP experiments
Western blot analysis
Protein extraction was performed in Lysis buffer [(50 mM HEPES, pH 7.9, 15 mM MgCl2, 150 mM NaCl, 10% glycerol, 1% TritonX-100, 0.1% SDS, 0.5 mM dithiothreitol, 10 μg/ml phenylmethylsulfonyl fluoride, and protease inhibitor mix (Sigma-Aldrich)] for 30 min on ice. Protein extracts (about 30 μg) were subjected to 10% SDS/PAGE electrophoresis and transferred to PVDF Transfer Membrane Hybond Tm (GE Healthcare Europe GmbH). Membranes were saturated with 5% non-fat dry milk in PBS containing 0.1% Tween20 (PBST) for 1 hr at RT. Incubation with primary antibodies was carried out at 4°C overnight in PBST with 5% BSA. Horseradish peroxidase-conjugated secondary antibody (GE Healthcare Europe GmbH) was used at 1:5000 dilution in PBST for 1 hr at RT. All proteins were detected with an ECL kit (GE Healthcare Europe GmbH) and visualized by chemiluminescence. Primary antibodies were: anti-EGFP mouse (Santa Cruz Technology, 1:1000); anti-Myc mouse (Santa Cruz Technology; 1:1000); anti-SOHLH1 rabbit (Abcam, 1:1000) anti-SOHLH2 guinea pig [gently provided by J.Miyazaki (Osaka University, Japan), 1:500], anti-tubulin rabbit polyclonal (Sigma-Aldrich, 1:1000) and anti-KIT rabbit (see ref. 49).
Statistical analysis
All experiments were replicates at least 3 times. The means tested for homogeneity of variance, and analyzed by ANOVA. The level of significance was set at P < 0.05 and P < 0.01.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
1007721_Supplementary_Materials.zip
Download Zip (127.9 KB)Acknowledgments
We thank Dr. C. Sette and Dr. I. Passacantilli for her helpful suggestions; prof. JL Tilly (Department of Biology, Laboratory of Aging and Infertility Research, Northeastern University, Boston, Massachusetts 02115, USA) for -1,400-Stra8-Egfp plasmid; Prof J.Miyazaki (Division of Stem Cell Regulation Research, Osaka University Graduate School of Medicine, Osaka, Japan) for providing rabbit anti-SOHLH2 antibodies and Mr. G. Bonelli for his expert technical assistance.
Funding
This work was supported by grants from the Ministero dell’Istruzione, dell’Università e della Ricerca Scientifica (PRIN 2010, D.S: 2010C8ERKX_003, Italy).
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- Bouillet P, Oulad-Abdelghani M, Vicaire S, Garnier JM, Schuhbaur B. Efficient cloning of cDNAs of retinoic acid-responsive genes in P19 embryonal carcinoma cells and characterization of a novel mouse gene, Stra1 (mouse LERK-2/Eplg2). Dev Biol 1995; 170:420-33; PMID:7649373; http://dx.doi.org/10.1006/dbio.1995.1226
- Oulad-Abdelghani M, Bouillet P, Décimo D, Gansmuller A, Heyberger S, Dollé P, Bronner S, Lutz Y, Chambon P. Characterization of a premeiotic germ cell-specific cytoplasmic protein encoded by Stra8, a novel retinoic acid-responsive gene. J Cell Biol 1996; 135:469-77; PMID:8896602; http://dx.doi.org/10.1083/jcb.135.2.469
- Baltus AE, Menke DB, Hu YC, Goodheart ML, Carpenter AE, de Rooij DG Page DC. In germ cells of mouse embryonic ovaries, the decision to enter into meiosis precedes premeiotic DNA replication. Nat Genet 2006; 38:1430-4; PMID:17115059; http://dx.doi.org/10.1038/ng1919
- Dokshin GA, Baltus AE, Eppig JJ, Page DC. Oocyte differentiation is genetically dissociable from meiosis in mice. Nat Gen 2013; 45:877-83; PMID:23770609; http://dx.doi.org/10.1038/ng.2672
- Mark M, Jacobs H, Oulad-Abdelghani M, Dennefeld C, Féret B, Vernet N, Codreanu CA, Chambon P, Ghyselinck NB. STRA8-deficient spermatocytes initiate, but fail to complete meiosis and undergo premature chromosome condensation. J Cell Sci 2008; 121:3233-42; PMID:18799790; http://dx.doi.org/10.1242/jcs.035071
- Anderson EL, Baltus AE, Roepers-Gajadien HL, Hssold TJ, de Rooij DG, van Pelt AM, Page DC. Stra8 and its inducer, retinoic acid, regulate meiotic initiation in both spermatogenesis and oogenesis in mice. Proc Natl Acad Sci U S A 2008; 105:14976-980; PMID:18799751; http://dx.doi.org/10.1073/pnas.0807297105
- Tedesco M, La Sala G, Barbagallo F, De Felici M, Farini D. STRA8 shuttles between nucleus and cytoplasm and displays transcriptional activity. J Biol Chem 2009; 284:35781-793; PMID:19805549; http://dx.doi.org/10.1074/jbc.M109.056481
- Choi Y J, Yoon J W, Pyo CW, Kim JA, Bae SH, Park S S. A possible role of STRA8 as a transcription factor. Genes and Genomics 2010; 32:521-26; http://dx.doi.org/10.1007/s13258-010-0059-9
- Al Tanoury Z, Piskunov A, Rochette-Egly, C. Vitamin A and retinoid signaling: genomic and non genomic effects. J Lipid Res 2013; 54:1761-75; PMID:23440512; http://dx.doi.org/10.1194/jlr.R030833
- Rochette-Egly C. Retinoic acid signaling and mouse embryonic stem cell differentiation: Cross talk between genomic and non-genomic effects of RA. Biochim Biophys Acta 2014; 1388-1981(14)00078-X
- Griswold MD, Hogarth CA, Bowles J, Koopman P. Initiating meiosis: the case for retinoic acid. Biol Reprod 2012; 86:1-7; PMID:22075477; http://dx.doi.org/10.1095/biolreprod.111.096610
- Giuili G, Tomljenovic A, Labrecque N, Oulad- Abdelghani M, Rassoulzadegan M, Cuzin F. Murine spermatogonial stem cells: targeted transgene expression and purification in an active state. EMBO Rep 2002; 3:753-59; PMID:12151334; http://dx.doi.org/10.1093/embo-reports/kvf149
- Kumar S, Chatzi C, Brade T, Cunningham TJ, Zhao X, Duester G. Sex-specific timing of meiotic initiation is regulated by Cyp26b1 independent of retinoic acid signalling. Nat Commun 2011; 2:151-59; PMID:21224842; http://dx.doi.org/10.1038/ncomms1136
- Le Bouffant R, Souquet B, Duval N, Duquenne C, Hervé R, Frydman N, Robert B, Habert R, Livera G. Msx1 and Msx2 promote meiosis initiation. Development 2011; 138:5393-402; PMID:22071108; http://dx.doi.org/10.1242/dev.068452
- Matson CK, Murphy MW, Griswold MD, Yoshida S, Bardwell VJ, Zarkower D. The mammalian doublesex homolog DMRT1 is a transcriptional gatekeeper that controls the mitosis versus meiosis decision in male germ cells. Dev Cell. 2010; 19:612-24; PMID:20951351; http://dx.doi.org/10.1016/j.devcel.2010.09.010
- Krentz AD, Murphy MW, Sarver AL, Griswold MD, Bardwell VJ, Zarkower D. DMRT1 promotes oogenesis by transcriptional activation of Stra8 in the mammalian fetal ovary. Dev Biol 2011; 356:63-70; PMID:21621532; http://dx.doi.org/10.1016/j.ydbio.2011.05.658
- Zhang T, Murphy MW, Gearhart MD, Bardwell VJ, Zarkower D. The mammalian Doublesex homolog DMRT6 coordinates the transition between mitotic and meiotic developmental programs during spermatogenesis. Development 2014; 141:3662-71; PMID:25249458; http://dx.doi.org/10.1242/dev.113936
- Murphy MW, Sarver AL, Rice D, Hatzi K, Ye K, Melnick A, Heckert LL, Zarkower D, Bardwell VJ. Genome-wide analysis of DNA binding and transcriptional regulation by the mammalian Doublesex homolog DMRT1 in the juvenile testis. Proc Natl Acad Sci U S A. 2010; 107:13360-5; PMID:20616082; http://dx.doi.org/10.1073/pnas.1006243107
- Feng YM, Liang GJ, Pan B, Qin XS, Zhang XF, Chen CL, Li L, Cheng SF, De Felici M, Shen W. Notch pathway regulates female germ cell meiosis progression and early oogenesis events in fetal mouse. Cell Cycle 2014; 13:782-91; PMID:24398584; http://dx.doi.org/10.4161/cc.27708
- Wang N, Tilly JL. Epigenetic status determines germ cell meiotic commitment in embryonic and postnatal mammalian gonads. Cell Cycle 2010; 9:339-49; PMID:20009537; http://dx.doi.org/10.4161/cc.9.2.10447
- Chen W, Jia W, Wang K, Si X, Zhu S, Duan T, Kang J. Distinct roles for CBP and p300 on the RA-mediated expression of the meiosis commitment gene Stra8 in mouse embryonic stem cells. PLoS One 2013; 8(6):e66076; PMID:23785470; http://dx.doi.org/10.1371/journal.pone.0066076
- Sleutels F, Soochit W, Bartkuhn M, Heath H, Dienstbach S, Bergmaier P, Franke V, Rosa-Garrido M, van de Nobelen S, Caesar L. et al. The male germ cell gene regulator CTCFL is functionally different from CTCF and binds CTCF-like consensus sites in a nucleosome composition-dependent manner. Epigenetics Chromatin 2012; 5:8; PMID:22709888; http://dx.doi.org/10.1186/1756-8935-5-8
- Yokobayashi S, Liang CY, Kohler H, Nestorov P, Liu Z, Vidal M, van Lohuizen M, Roloff TC, Peters AH. PRC1 coordinates timing of sexual differentiation of female primordial germ cells. Nature 2013; 495:236-40; PMID:23486062; http://dx.doi.org/10.1038/nature11918
- Ballow D, Meistrich M L, Matzuk M, Rajkovic A. Sohlh1 is essential for spermatogonial differentiation. Dev Biol 2006a; 294:161-7; PMID:16564520; http://dx.doi.org/10.1016/j.ydbio.2006.02.027
- Hao J, Yamamoto M, Richardson TE, Chapman KM, Denard BS, Hammer RE, Zhao GQ, Hamra FK. Sohlh2knockout mice are male-sterile because of degeneration of differentiating type A spermatogonia. Stem Cells 2008; 26:1587-97; PMID:18339773; http://dx.doi.org/10.1634/stemcells.2007-0502
- Toyoda S, Miyazaki T, Miyazaki S, Yoshimura T, Yamamoto M, Tashiro F, Yamato E, Miyazaki J. Sohlh2affects differentiation of KIT positive oocytes and spermatogonia. Dev Biol 2009; 325:238-48; PMID:19014927; http://dx.doi.org/10.1016/j.ydbio.2008.10.019
- Suzuki H, Ahn HW, Chu T, Bowden W, Gassei K, Orwig K, Rajkovic A. SOHLH1 and SOHLH2 coordinate spermatogonial differentiation. Dev Biol 2011; 361:301-12; PMID:22056784; http://dx.doi.org/10.1016/j.ydbio.2011.10.027
- Pangas SA, Choi Y, Ballow DJ, Zhao Y, Westphal H, Matzuk MM, Rajkovic A. Oogenesis requires germ cell-specific transcriptional regulators Sohlh1and Lhx8. Proc Natl Acad Sci USA 2006; 103:8090-95; PMID:16690745; http://dx.doi.org/10.1073/pnas.0601083103
- Choi Y, Yuan D, Rajkovic A. Germ cell-specific transcriptional regulator Sohlh2 is essential for early mouse folliculogenesis and oocyte-specific gene expression. Biol Reprod 2008; 79:1176-82; PMID:18753606; http://dx.doi.org/10.1095/biolreprod.108.071217
- Barrios F, Filipponi D, Campolo F, Gori M, Bramucci F, Pellegrini M, Ottolenghi, S, Rossi P, Jannini EA, Dolci S. SOHLH1 and SOHLH2 control Kit expression during postnatal male germ cell development. J Cell Sci 2012; 125:1455-64; PMID:22328502; http://dx.doi.org/10.1242/jcs.092593
- Rossi P. Transcriptional control of KIT gene expression during germ cell development. Int J Dev Biol 2013; 57:179-84; PMID:23784828; http://dx.doi.org/10.1387/ijdb.130014pr
- Zhou Q, Nie R, Li Y, Friel P, Mitchell D, Hess R A, Small C, Griswold MD. Expression of stimulated by retinoic acid gene 8 (STRA8) in spermatogenic cells induced by retinoic acid: an in vivo study in vitamin A-sufficient postnatal murine testes. Biol Reprod 2008; 79:35-42; PMID:18322276; http://dx.doi.org/10.1095/biolreprod.107.066795
- Nayernia K, Li M, Jaroszynski L, Khusainov R, Wulf G. Stem cell based therapeutical approach of male infertility by teratocarcinoma derived germ cells. Hum Mol Genet 2004; 13:1451-60; PMID:15163638; http://dx.doi.org/10.1093/hmg/ddh166
- Sadate-Ngatchou PI, Payne CJ, Dearth AT, Braun RE. Cre recombinase activity specific to postnatal, premeiotic male germ cells in transgenic mice. Genesis 2008; 46:738-42; PMID:18850594; http://dx.doi.org/10.1002/dvg.20437
- Imudia AN, Wang N, Tanaka Y, White YA, Woods DC, Tilly JL. Comparative gene expression profiling of adult mouse ovary-derived oogonial stem cells supports a distinct cellular identity. Fertil Steril. 2013; 100:1451-8; PMID:23876535; http://dx.doi.org/10.1016/j.fertnstert.2013.06.036
- Raverdeau M, Gely-Pernot A, Féret B, Dennefeld C, Benoit G, Davidson I, Chambon P, Mark M, Ghyselinck NB. Retinoic acid induces Sertoli cell paracrine signals for spermatogonia differentiation but cell autonomously drives spermatocyte meiosis. Proc Natl Acad Sci U S A 2012; 109:16582-587; PMID:23012458; http://dx.doi.org/10.1073/pnas.1214936109
- Filipponi D, Hobbs RM, Ottolenghi S, Rossi P, Jannini EA, Pandolfi PP, Dolci S. Repression of kit expression by Plzf in germ cells. Mol Cell Biol. 2007; 27:6770-81; PMID:17664282; http://dx.doi.org/10.1128/MCB.00479-07
- Kumar S, Cunningham TJ, Duester G. Resolving molecular events in the regulation of meiosis in male and female germ cells. Sci Signal 2013; 6:pe25; PMID:23943607
- Rossi P, Dolci S. Paracrine mechanisms involved in the control of early stages of Mammalian spermatogenesis. Front Endocrinol 2013; 4: art 181; PMID:24324457; http://dx.doi.org/10.3389/fendo.2013.00181
- Song HW, Wilkinson MF. Transcriptional control of spermatogonial maintenance and differentiation. Semin Cell Dev Biol 2014; 30:14-26; PMID:24560784; http://dx.doi.org/10.1016/j.semcdb.2014.02.005
- Pellegrini M, Filipponi D, Gori M, Barrios F, Lolicato F, Grimaldi P, Rossi P, Jannini EA, Geremia R, Dolci S. ATRA and KL promote differentiation toward the meiotic program of male germ cells. Cell Cycle 2008; 7:3878-88; PMID:19098446; http://dx.doi.org/10.4161/cc.7.24.7262
- Vernet N, Dennefeld C, Guillou F, Chambon P, Ghyselinck NB, Mark M. Prepubertal testis development relies on retinoic acid but not rexinoid receptors in Sertoli cells. EMBO J 2006; 25:5816-25; PMID:17124491; http://dx.doi.org/10.1038/sj.emboj.7601447
- Toyoda S, Yoshimura T, Mizuta J, Miyazaki J. Auto-regulation of the Sohlh1gene by the SOHLH2/SOHLH1/SP1 complex: implications for early spermatogenesis and oogenesis. PLoS One 2014; 9:e101681; PMID:25003626; http://dx.doi.org/10.1371/journal.pone.0101681
- Blais A, Tsikitis M, Acosta-Alvear D, Sharan R, Kluger Y, Dynlacht BD. An initial blueprint for myogenic differentiation. Genes Dev 2005; 19:553-69; PMID:15706034; http://dx.doi.org/10.1101/gad.1281105
- Sartorelli V, Puri PL, Hamamori Y, Ogryzko V, Chung G, Nakatani Y, Wang JY, Kedes L. Acetylation of MyoD directed by PCAF is necessary for the execution of the muscle program. Mol Cell 1999; 4:725-34; PMID:10619020; http://dx.doi.org/10.1016/S1097-2765(00)80383-4
- Tapscott SJ. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development 2005; 132:2685-95; PMID:15930108; http://dx.doi.org/10.1242/dev.01874
- Barrera LO, Ren B. The transcriptional regulatory code of eukaryotic cells-insights from genome-wide analysis of chromatin organization and transcription factor binding. Curr Opin Cell Biol 2006; 18:291-8; PMID:16647254; http://dx.doi.org/10.1016/j.ceb.2006.04.002
- Rossi P, Dolci S, Albanesi C, Grimaldi P, Ricca R, Geremia R. Follicle-stimulating hormone induction of steel factor (SLF) mRNA in mouse Sertoli cells and stimulation of DNA synthesis in spermatogonia by soluble SLF. Dev Biol 1993; 155:68-74; PMID:7677988; http://dx.doi.org/10.1006/dbio.1993.1007
- Albanesi C, Geremia R, Giorgio M, Dolci S, Sette C, Rossi P. A cell- and developmental stage-specific promoter drives the expression of a truncated c-kit protein during mouse spermatid elongation. Development 1996; 122:1291-302; PMID:8620856