Abstract
The proteins of the Dbl family are guanine nucleotide exchange factors (GEFs) of Rho GTPases and are known to be involved in cell growth regulation. Alterations of the normal function of these proteins lead to pathological processes such as developmental disorders, neoplastic transformation, and tumor metastasis. We have previously demonstrated that expression of Dbl oncogene in lens epithelial cells modulates genes encoding proteins involved in epithelial-mesenchymal-transition (EMT) and induces angiogenesis in the lens. Our present study was undertaken to investigate the role of Dbl oncogene in epithelial cells transformation, providing new insights into carcinoma progression.To assess how Dbl oncogene can modulate EMT, cell migration, morphogenesis, and expression of pro-apoptotic and angiogenic factors we utilized bi- and 3-dimensional cultures of MCF-10 A cells. We show that upon Dbl expression MCF-10 A cells undergo EMT. In addition, we found that Dbl overexpression sustains Cdc42 and Rac activation inducing morphological alterations, characterized by the presence of lamellipodia and conferring a high migratory capacity to the cells. Moreover, Dbl expressing MCF-10 A cells form altered 3D structures and can induce angiogenesis by producing proangiogenic factors such as CCL2. These results support a role for Dbl oncogene in epithelial cell differentiation and transformation and suggest the relevance of GEF deregulation in tumor onset and progression.
Abbreviations
| EMT | = | Epithelial-mesenchymal-transition |
| α-SMA | = | α-smooth muscle actin |
Introduction
Epithelial-mesenchymal-transition (EMT) is a fundamental biological process by which epithelial cells are transdifferentiated to a mesenchymal state, and is a normal process during embryonic development and wound healing. EMT has also been implicated in cancer progression, metastasis and invasion.Citation1,2 It begins with the loss of apico-basal polarity, degradation of basement membrane, decrease in epithelial-specific gene expression, including E-cadherin, and acquisition of mesenchymal characteristics, including expression of α-smooth muscle actin (α-SMA) and metalloproteinases. Moreover, cells undergo changes in their cytoskeleton, which enable them to acquire the ability to migrate and invade.Citation3-5 EMT modulates the tumor microenvironment by inducing the upregulation and the secretion of several soluble factors, including angiogenic factors and cytokines, that may induce proliferation of endothelial cells, resulting in neovascularization.Citation6
The Dbl protein is the prototype member of a large family of guanine nucleotide exchange factors (GEFs) for Rho GTPases,Citation7-9 which are known to regulate various physiological processes including actin cytoskeleton organization, cell movement, cell proliferation, cytokinesis, and apoptosis.Citation10-12 The Dbl oncogene was shown to have a strong transforming activity in fibroblasts,Citation13 to affect dendrite elongation in a mouse knock-out model Citation14 and to induce B-cell lymphomas in a mouse knock-in model.Citation15 Moreover, expression of the Dbl oncogene under the metallothionein promoter in transgenic mice affects proliferation, migration, and differentiation of lens epithelial cells.Citation16 We have defined the cluster of genes that are induced or inhibited in the lenses of Dbl transgenic mice at several time points after birth.Citation17 Results obtained indicate modulation of genes encoding proteins involved in EMT, apoptosis, and vasculogenesis, suggesting that Dbl oncogene expression in lens epithelial cells can induce events involved in the early stages of cancer, such as aberrant proliferation, block of differentiation and apoptosis, as well as late events such as EMT, disruption of the basal membrane, and neovascularization.
To further characterize the role of Dbl oncogene in epithelial cancer onset and progression we have analyzed the effects of the Dbl oncogene expression in MCF-10 A epithelial cell line. MCF-10 A are immortalized, non-transformed epithelial cells derived from human fibrocystic mammary tissue. MCF-10 A cells retain the intrinsic ability of mammary epithelial cells to undergo acinar morphogenesis in 3D matrigel cultures, a process that relies on growth factor-dependent proliferation, induction of luminal programmed cell death, establishment of an apico-basal polarity axis, and the deposition of a basal lamina.Citation18 Various oncogenes can have distinct perturbing effects on acinar morphogenesis and can induce supra-cellular effects in the cell colonies.Citation19
We report here that Dbl expressing MCF-10 A cells cultured in matrigel form altered hollow, 3D structures characterized by reduced lumen size, almost complete lack of cleaved-caspase 3 expression and high expression of Ki-67, denoting active proliferation. Moreover, we show that the MCF-10 A cells expressing Dbl oncogene stimulates the invasion and the tube formation potencies of Human Umbilical Vein Endothelial Cells (HUVECs) and that this proangiogenic activity is mediated by CCL2 secretion. In conclusion, our data support a role for Dbl oncogene in epithelial cell differentiation and transformation and in the induction of neovascularisation by stimulating the production of proangiogenic factors.
Results
Dbl oncogene induces EMT in mammary epithelial MCF-10 A cells
EMT leads to aggressive cancer progression and implies the loss of epithelial markers, such as E-cadherin, increased cell motility and expression of mesenchymal genes, such as α-SMA and MMP.Citation20
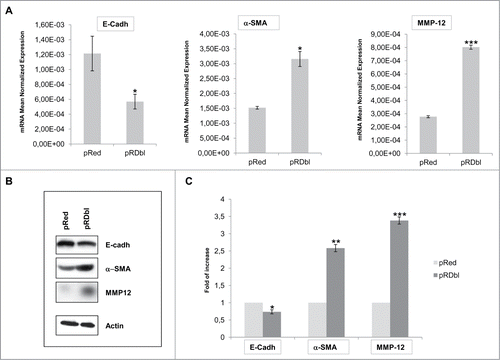
To determine the role of Dbl in governing EMT in human mammary epithelial cells, MCF-10 A cells were infected with a lentiviral vector expressing Dbl oncogene fused to the fluorescence Red epitope (pRDbl) or with the control lentivirus (pRed) and cultured for 2 weeks under puromycin selection. pRDbl and control pRed cells were lysed and E-cadherin, α−SMA and MMP-12 transcripts were quantified by RT-qPCR. shows the relative mRNA expression levels of selected genes in pRDbl respect to pRed cells. A considerable downregulation of E-cadherin and upregulation of α-SMA and MMP-12 transcripts was triggered by the expression of Dbl oncogene in MCF-10 A cells. Cell lysates from pRDbl and pRed MCF-10 A cells were analyzed by Western blotting with anti-E-cadherin, anti-α−SMA, and anti-MMP-12 antibodies. The RT-qPCR results were confirmed by protein expression analysis ()
Figure 1. EMT induction by Dbl oncogene. (A) qRT-PCR analysis of genes involved in EMT. 2 μg of total RNA were reverse-transcribed and qRT-PCR was conducted in triplicate for each target transcript. Expression changes of genes were evaluated in relation to the values obtained in parallel for 2 reference genes. The average results from 3 independent experiments were calculated and represented. The mean values (± SD) are indicated. *P < 0.05; ***P < 0.001. (B) Confirmation of qRT-PCR analysis by Western Blot. Total cell lysates from pRed and pRDbl cells were blotted with anti-E-cadherin, anti-α-SMA and anti-MMP12 antibodies. Actin was used as a loading control. Representative photomicrographs are shown. (C) The optical density of the films was scanned and measured and the average results from 3 independent experiments were calculated and represented; *P < 0.05; **P < 0.005; ***P < 0.001.
Dbl oncogene alters MCF-10 A monolayer morphology and motility
Cells that undergo EMT reorganize their cortical actin cytoskeleton into one that enables dynamic cell elongation and directional motility.Citation21 Thus, we investigated whether the expression of pRDbl causes changes in MCF-10 A cell morphology (). Phase contrast images showed that cells infected with pRed vector adopt a cobblestone morphology (a′), typical of mammary epithelial cells, with some lamellipodia at the edges of the clusters (c′, e′ and g′). On the contrary, pRDbl cells are characterized by an enlarged and polygonal cell shape (b′), and increase in membrane ruffling and lamellipodia formation (d′, f′ and h′). About 20% of MCF-10 A cell population expressing Dbl oncogene is constituted by giant multinucleated cells, a morphology originally described for Dbl-transformed NIH3T3 cells.Citation22,23 These cells seem to originate as a result of several cycles of nuclear division with no apparent cytokinesis.Citation23
Figure 2. Dbl oncogene expression alters MCF-10 As monolayer morphology. (A) Phase-contrast images of pRed (a′, c′, e′ and g′) and pRDbl (b′, d′, f′ and h′) cells cultured to confluence or sub-confluence in assay media. Magnification: 4X (a′, b′, c′, d′); 20X (e′, f′); 40X (g′, h′). (B) Immunofluorecence analysis of the distribution of pRDbl oncoprotein. pRed and pRDbl cells grown on glass coverslips, were fixed, permeabilized and stained for actin filaments, using FITC-conjugated phalloidin (green), and for nuclei, using DAPI (blue). pRDbl cells show a polygonal shape, and the red fluorescence signal mostly diffused in the cytoplasm and partially localized on the plasma membrane. In contrast, pRed cells appear elongated, and the red signal diffused in the cytoplasm, with no localization along the plasma membrane. The actin cytoskeleton (green) is organized in well-evident short stress fibers and some ruffling and lamellipodia in cells expressing pRDbl, and in thin, long stress fibers in pRed expressing cells. Arrowheads indicate the ruffling and lamellipodia areas of the plasma membrane where the DbI protein localizes, arrows indicate stress fibers. Scale bar: 10 μm.
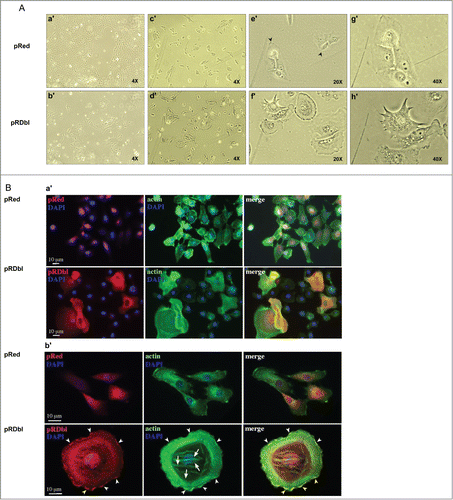
To better visualize the differences observed in phase contrast images, pRDbl and the control cells were plated on glass coverslips and treated with anti-Red to detect Dbl and vector alone, and with FITC-labeled phalloidin to detect actin. Nuclei were visualized with DAPI (). The expression of pRDbl in MCF-10 A cells induces evident cell body enlargement and an increase in the extension of the ruffles and lamellipodia compared to pRed infected cells, which maintain their epithelial cell morphology. Dbl protein was mostly localized on the membrane at the ruffling sites (, arrowheads), in agreement with the knowledge that activated Dbl protein translocates to the plasma membrane where it exerts its GEF activity.Citation24
The regulation of cytoskeletal changes associated with migratory behavior of cells is dependent on Rho GTPase activity.Citation25 In fact, Rho GTPases play a crucial role in the actin cytoskeleton rearrangements, cell motility, and cell–cell dissociation associated with EMT.Citation26,27 Therefore, we investigated whether the morphological changes observed in pRDbl MCF-10 A cells could be correlated with activation of small G proteins by Dbl oncogene. Lysates obtained from cells infected with pRDbl or the control vector were subjected to a pulldown assay. Endogenous, activated Cdc42 and Rac were collected on GST-PAK-CRIB domain fusion protein and analyzed by Western blot. As shown in , the levels of the GTP-bound forms of Cdc42 and Rac increase in pRDbl cells in comparison with pRed cells. We also evaluated active RhoA-GTP by an absorbance based method and found that expression of pRDbl induces RhoA activation ().
Figure 3. Dbl oncogene mediates cell motility. (A) In vivo GEF activity of Dbl oncogene on small G proteins Cdc42 and Rac. pRed and pRDbl cells, serum-starved for 18 hr, were lysed and subjected to the Cdc42 and Rac activation assays. Eluates from GST-PAK precipitates or total cell lysates were subjected to immunoblotting with anti-Cdc42 and anti-Rac antibodies. Specific Dbl product was detected using anti Dbl antibody. Results from one of 3 independent experiments is shown as representative. (B) Cell lysates were subjected to an absorbance based activation assay for RhoA. Results shown represent the mean values ± SD from 3 different experiments. Protein expression level was evaluated by Western blot analysis with specific antibodies using total cell lysates. (C) pRed and pRDbl cells were subjected to a transwell migration assay as described in "Materials and methods." Briefly, cells resuspended in serum-free medium were seeded on the upper chamber. Inserts were incubated for 1, 2, and 4 h and cells on the underside of the insert were fixed and stained with May–Grunwald/Giemsa. Representative photomicrographs are shown (magnification: 20X for 1 and 2 hours, 4X for 4 hours). (D) Cell migration was assayed on pRed and pRDbl MCF-10 A cells cultured overnight in the absence or presence of 100 μM of NSC23766. The average results from 3 independent experiments were calculated and represented. ** P < 0.01. (E) pRed and pRDbl cells cultured overnight in the absence or presence of 100 μM of NSC23766 were lysed and subjected to the Rac activation assays.
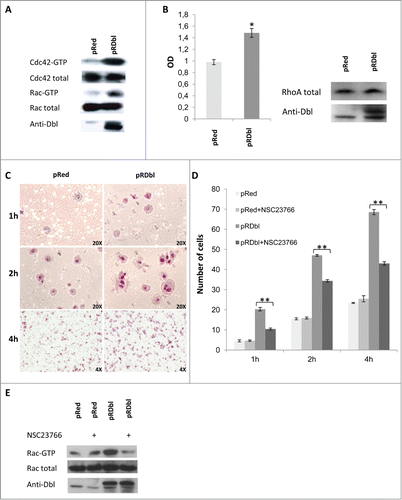
EMT is characterized by an increased cell capacity for migration and invasion.Citation4 Activation of Cdc42 and Rac results in the formation of filopodia and lamellipodia, respectively, and the formation and turnover of these structures are important in cell migration. We performed a transwell assay at different time points to evaluate whether the activation of Rac and Cdc42 in pRDbl cells correlated with cell migration.
As shown in , pRDbl cells migrate better than control pRed cells. Dbl regulation of cell motility through activation of Rac was confirmed by treatment of pRed and pRDbl MCF-10 A cells with the Rac GTPase-specific inhibitor NSC23766.Citation28 As shown in the presence of NSC23766 inhibits pRDbl cell migration but does not affect the motility of control cells. We performed a pulldown assay to evaluate the inhibitory effect of NSC23766 on Dbl-stimulated Rac activity in vivo. Treatment of pRDbl MCF-10 A cells with 100 μM NSC23766 resulted in inhibition of Rac-GTP compared to untreated cells ().
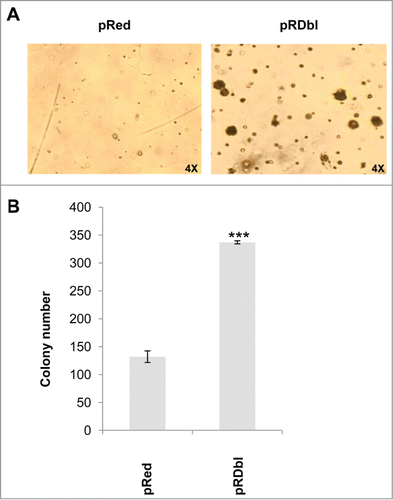
To evaluate whether the induction of EMT correlates with the transforming capacity of Dbl oncogene we performed a soft agar colony formation assay. showed a significant increase in anchorage-indipendent growth in comparison with pRed MCF-10 A cells. Altogether, these results indicate that Dbl oncogene-induced activation of Rho GTPases in MCF-10 A cells stimulates cell migration and promotes transformation.
Figure 4. Dbl oncogene induces anchorage independent growth of MCF-10 A cells. pRed and pRDbl MCF-10 A cells were cultured as indicated in "Materials and methods." Colonies were scored after 14 days and photographed. (A) Representative photomicrographs are shown. (magnification: 4X) (B) The colony number was quantified from 3 independent soft agar assays. *** P < 0.001.
Dbl oncogene affects epithelial cell morphogenesis
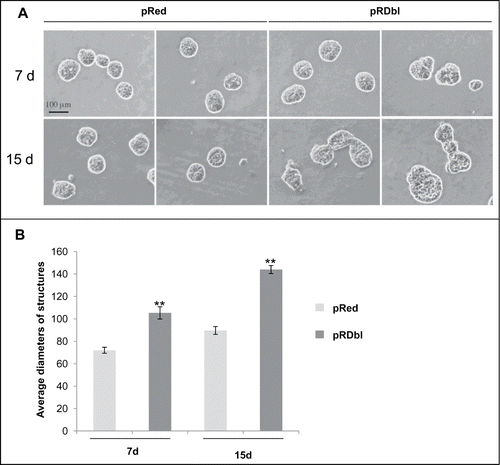
The spontaneously immortalized and non-tumorigenic MCF-10 A epithelial cells possess 3 dimensional (3D) growth ability and form a polarized structure surrounding a hollow lumen. In this process, proapoptotic genes or tumor suppressor genes promote, whereas oncogenes block, acinar differentiation. In fact, oncogenes introduced in MCF-10 A cells disrupt this morphogenetic process, and elicit distinct morphological phenotypes.Citation29-31 We evaluated the effects of Dbl expression on MCF-10 A cell mophogenesis. Stable pools of MCF-10 A cells expressing Dbl oncogene were cultured on a reconstituted basement membrane gel (Matrigel) for 7–15 days. This assay was carried on for different lengths of time to evaluate differences in proliferation capacity between Dbl-expressing and parental cell lines. As shown in , Dbl structures displayed varying degrees of morphological disruption and changes in overall structure size. The average diameter of structures from Dbl expressing cultures was 140 ± 17 μm (mean ± SD) versus 90 ± 13.8 μm (mean ± SD) in controls (). Larger size and morphological alteration of structures were apparent as early as day 7 in 3D culture and progressively increased thereafter.
Figure 5. Dbl oncoprotein affects acinar structures. (A) Phase-contrast images illustrate the development of acinar structures in pRed and pRDbl cells at 7 (7 d) and 15 (15 d) days. Scale bar: 100 μm. (B) The mean (± SD) diameters of pRed and pRDbl acini after the indicated days in basement membrane culture are shown. **P < 0.01.
We then analyzed the expression of the proteins involved in EMT such as E-cadherin and α−SMA during morphogenesis. pRed acini exhibited well-defined cell-cell junctions, illustrated by membrane localization of E-cadherin, concentrated toward the basal pole of the lateral membrane (). Although in pRDbl acini E-cadherin initially displayed a membrane localization similar to that exhibited in the control acini, (data not shown) during the second decade of morphogenesis immunostaining was diminished and became diffusely localized, indicating loss of E-cadherin at adherens junctions. Mislocalization and loss of E-cadherin at cell-cell junctions is an early event that is essential for EMT. On this basis, we investigated whether Dbl expression promotes EMT in 3D culture system by immunostaining the acini with an antibody anti the mesenchymal marker α−SMA. As shown in Dbl expression causes a considerable increase in α-SMA expression vs. control cells.
Figure 6. Dbl oncogene induces loss of proliferative suppression and luminal apoptosis. Acini cultured for 10 days in Matrigel were fixed and immunostained with antibodies against Red epitope (red), E-cadherin (green) and α-SMA (green). Nuclei were counterstained with DAPI (blue). Scale bar: 20 μm.
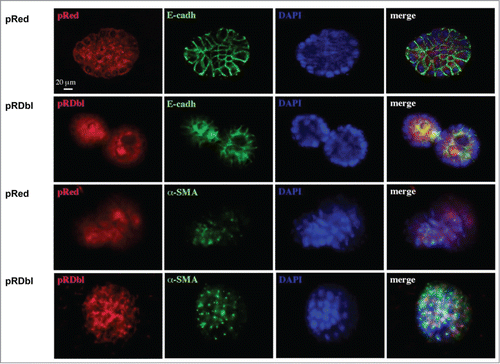
Collectively, these data indicate that Dbl expression alters acinar morphogenesis, leading to large, misshapen structures and multiacinar formations, and induces downregulation of epithelial markers and upregulation of fibroblast markers suggesting that Dbl activation promotes EMT.
Dbl expression prevents luminal apoptosis during morphogenesis
Apoptosis is involved in the elimination of centrally located cells during lumen formation in MCF-10 A acini and in the maintenance of the hollow lumen thereafter. When oncoproteins such as ErbB2 are expressed in these structures, the lumen is filled because of induction of cell proliferation and inhibition of apoptosis.Citation29,32 To study the effect of Dbl expression on apoptosis in the luminal space of mammary structures during morphogenesis, stable pools of MCF-10 A cells expressing Dbl oncogene were cultured on a reconstituted basement membrane gel (Matrigel) for 10 days. 3D structures were then examined for filled lumen by immunostaining with an antibody against the cleaved, activated form of caspase 3. shows a significant filling of the luminal space and reduced apoptosis, as demonstrated by the almost complete lack of cleaved-caspase 3 expression in the lumen of pRDbl-expressing MCF-10 A acini compared with control structures.
Figure 7. Dbl expression prevents luminal apoptosis during morphogenesis. Acini were cultured in Matrigel for 10 days, fixed and immunostained with antibodies against Red epitope (red), activated caspase-3 (green), and Ki-67 (green). Nuclei were counterstained with DAPI (blue). Scale bar: 20 μm.
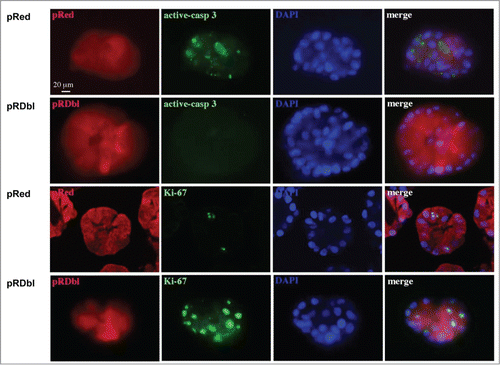
Next, we examined the effects of Dbl expression on cell proliferation during morphogenesis by immunostaining for Ki-67, a marker of cycling cells as well as EMT.Citation33 We observed that pRDbl-expressing MCF-10 A acini exhibit high levels of Ki-67 positive cells during morphogenesis (), indicating that inhibition of cell death and increased cell proliferation account for the increased size and filled lumens of pRDbl MCF-10 A acini.
Dbl oncogene induces endothelial cell migration
Tumor cells undergoing EMT are known to increase the secretion of specific factors, including cytokines, chemokines, and growth factors, which play an important role in inducing angiogenesis and tumor progression.Citation6
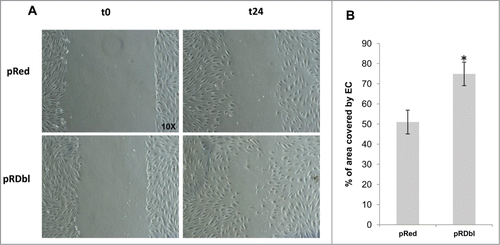
We examined whether pRDbl cells secrete factors that induce endothelial cell migration. HUVECs were co-cultured with pRDbl MCF-10 A cells and migration was evaluated by scratch wound healing assays. As shown in , pRDbl cells co-cultured in conditioned medium increased endothelial cell migration by up to 25% compared to the control.
Figure 8. Dbl expression induces endothelial cell migration. (A) HUVEC migration was evaluated by the scratch assay. Representative images of healing in the presence of MCF-10 pRed or pRDbl at 0 and 24 h (t = 0 and t = 24) are reported (magnification: 10X). (B) Data are reported as a percentage of area covered by endothelial cells (EC) after co-culture with MCF-10 pRed or pRDbl. The experiment was performed 3 times in duplicate (2 wells/experiment). Cells were photographed and cell-free areas were measured by image analysis. *P < 0.05.
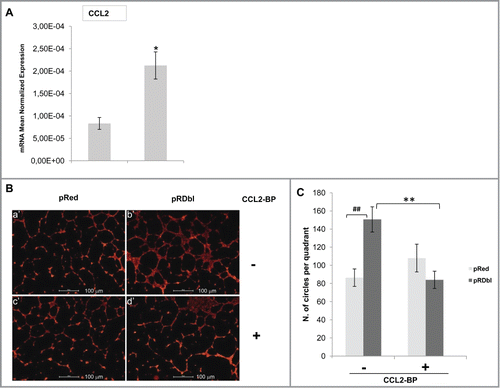
Dbl oncogene can promote neovascularisation in the lens by upregulating proangiogenic factors, such as CCL2/MCP-1.Citation17 On this basis, we evaluated whether MCF-10 A cells expressing Dbl synthesize and secrete the chemokine CCL2/MCP-1. shows a significant upregulation of CCL2 mRNA in MCF-10 A cells expressing Dbl oncogene vs control. The initial phase of angiogenesis involves organization of individual endothelial cells into a 3-dimensional pseudo-capillary-like structure. Therefore, we evaluated whether secreted CCL2 could stimulate formation of capillary-like structures.Citation34 HUVECs, plated in a thin layer of Matrigel, were co-cultured with pRed or pRDbl MCF-10 A cells and the number of intact ring that HUVECs are able to form was determined. HUVECs, cultured with pRDbl MCF-10 A cells, organized into a network of pseudo-capillary tubes that invaded the gel (; Fig. S1, b′). Quantification of the number of circles formed by tubular HUVECs highlights the significant increase of capillary tube formation induced by pRDbl cells versus that induced by co-culture with pRed MCF-10 A cells (). Furthermore, HUVECs co-cultured with pRDbl MCF-10 A cells in the presence of 1 μg/ml CCL2 blocking peptide failed to form capillary tubes (; ; Fig. S1 d′), indicating that Dbl oncogene expression in MCF-10 A cells controls CCL2/MCP-1 output and the angiogenic phenotype.
Figure 9. CCL2 stimulates pseudocapillary tube formation. (A) qRT-PCR analysis of CCL2 gene. 2 μg of total RNA were reverse-transcribed and qRT-PCR was conducted in triplicate. Expression changes of genes were evaluated in relation to the values obtained in parallel for 2 reference genes. The mean values (±SD) are indicated. *P < 0.05. (B) Endothelial cell organization on Matrigel in the co-culture system with pRed or pRDbl MCF-10 A. HUVECs were seeded on Matrigel layers and maintained for 18 h in the presence of pRed (a′,c′) or pRDbl MCF-10 A(b′,d′) cells. Where indicated 1 μg/ml of CCL2 blocking peptide (CCL2-BP) was added (c′,d′). Scale bar: 100 μm. (C) Data in the graph are reported as angiogenic index, calculated as the number of complete circles counted/field at 4X magnification by image analysis. ##P < 0.01 **P < 0.05.
Discussion
The Dbl oncogene was originally isolated from a human B-cell lymphoma and the mammary carcinoma cell line MCF-7 as 2 activated versions of the same proto-oncogene.Citation22,35 Dbl protein is the prototype of the large family of GEFs for Rho GTPases Cdc42, RhoA and Rac. GEFs are involved in cell growth regulation and play an essential role in establishing epithelial cell polarity. Dbl expression in mammary epithelial cells has been previously linked to transformation and stimulation of motility.Citation36-38
In this work we provide evidence that Dbl oncogene causes morphological changes, increases migratory behavior, and promotes transformation of MCF-10 A cells. Cells expressing Dbl also exhibit increase in Cdc42, Rac, and Rho activation in the absence of growth factor stimulation, compatible with the constitutive activation of GTPase exchange activity of Dbl oncogene. Moreover, the expression of Dbl oncogene in MCF-10 A epithelial cells activates downstream effectors to induce EMT-like processes and to stimulate angiogenesis.
During EMT, the decrease in epithelial characteristics is indicated by the decreased expression of E-cadherin, which downregulates epithelial cell migration, and is accompanied by the acquisition of mesenchymal features, including the expression of α−SMA and MMPs.Citation39 Our results indicate that Dbl expression not only induces morphological transformation of MCF-10 A but also makes these cell to undergo EMT, leading to the downregulation of epithelial-specific markers, like E-cadherin, upregulation of mesenchymal markers, such as α−SMA, and induction of expression of EMT markers, such as Ki-67 and MMP-12, all concurrently contributing to the cytoskeleton alterations and increased motility.Citation40
Angiogenesis, essential for physiological as well as pathological conditions, including tumor growth and metastasis, is a complex multistep process that includes endothelial cells proliferation, migration, and differentiation of endothelial cells, degradation of extracellular matrix, microtubule formation, and sprouting of new capillary branches.Citation41 Although activated oncogenes increase tumor cell proliferation and decrease their apoptosis, these activities are not sufficient to expand tumor mass beyond a microscopic size. Induction of angiogenesis is a necessary step for expansion of tumor mass. In this respect, EMT not only increases tumor cell migration and invasion, but also promotes tumor angiogenesis by inducing expression of angiogenic factors and cytokines.Citation42
Indeed, we show that the expression of Dbl oncogene in MCF-10 A cells causes upregulation of the pro-angiogenic factor CCL2/MCP-1 that increases endothelial cell migration and ability to form a network of pseudocapillary tubes. CCL2 is a potent angiogenic chemokine that acts directly on endothelial cells to promote angiogenesis.Citation43,44 Moreover, CCL2 is highly expressed by breast tumor cells and has causative roles in breast malignancy and metastasis.Citation45
During normal acinar morphogenesis, lumen formation involves both the block of proliferation and the apoptosis of centrally localized cells that are not in contact with the basement membrane.Citation29 The 3D MCF-10 A cultures provide a model that can be useful to determine the pathways that regulate acini structure formation as well as to identify genes and pathways that induce phenotypic alterations similar to those associated with tumor progression, such as filling of the luminal space, loss of polarization, escape from proliferative suppression, invasive behavior, and loss of cell adhesion.Citation46
Filling of the luminal space occurs when increase in cell proliferation is associated with inhibition of apoptosis, either because of expression of anti-apoptotic genes or because of expression of oncogenes with anti-apoptotic activities. For example, activated ErbB2 impinges on multiple aspects of acini development by disturbing mechanisms that establish cell polarity, inducing a complex multiacinar phenotype, increasing of cell proliferation, filling of the luminal space, and protection from apoptosis, all properties comparable to those of early stage cancer.Citation32,47
Dbl-expressing MCF-10 A cells form altered hollow, multi-acinar 3D structures characterized by filled lumen, an almost complete lack of cleaved-caspase 3 expression and high expression of Ki-67, indicating that Dbl oncogene provides significant protection of cells within the lumen from apoptosis and enhances proliferation, thus overcoming the signals causing proliferative arrest.
In summary, Dbl oncogene expression in MCF-10 A cells induces some of the changes that characterize tumor progression. In fact, we observed downregulation of E-cadherin, the lack of which is implicated in the loss of cell-cell adhesion,Citation48 and upregulation of mesenchimal markers such as α-Sma, characteristics of EMT, with consequent changes in cell morphology and cytoskeleton organization. Moreover, Dbl oncogene induces expression of MMP-12, one of the metalloproteinases that, because of their extracellular matrix degrading activity, exert a pro-invasive action,Citation49 disruption of glandular structure, inhibition of anoikis and stimulation of proliferation,Citation50 hallmarks of epithelial tumors. Finally, Dbl stimulates the expression of the pro-angiogenic factor CCL2, necessary for the outgrowth of new vessels, essential for tumor growth and metastasis.
The identification of a new regulatory loop between angiogenesis and EMT in Dbl-transformed epithelial cells represents a novel step forward in understanding the mechanisms and the relevance of GEF deregulation in tumor onset and progression.
Material and Methods
Antibodies, reagents, inhibitors
Polyclonal antibody anti-Dbl was purchased from Santa Cruz Biotechnology. Polyclonal antibody anti-DsRed was purchased from Clontech. Monoclonal antibody against Rac was purchased from Upstate Biotecnology. Polyclonal antibody against Cdc42 was purchased from BD Transduction Laboratories. Mouse monoclonal antibody against RhoA was purchased from Santa Cruz Biotechnology. Monoclonal antibody against α-SMA was purchased from DAKO. Polyclonal antibody against Cleaved Caspase-3 was purchased from Cell Signaling. Rabbit monoclonal anti MMP-12 was purchased from Novus Biologicals. Mouse monoclonal anti human E-cadherin was kindly provided by Dr. Defilippi (MCB, University of Turin). CCL2 blocking peptide was purchased from Fitzgerald Industry International. Rac GTPase inhibitor NSC23766 was kindly provided by Dr. Y. Zheng.
Cell cultures
MCF-10 A cells, an immortalized, non-transformed epithelial cell line derived from human fibrocystic mammary tissue, were kindly provided by Dr. Defilippi (University of Turin). MCF-10 A cells were maintained in DMEM/F12 (Gibco) supplemented with 5% horse serum, 20 ng/ml EGF, 10 μg/ml insulin, 0.1 μg/ml cholera toxin, 0.5 μg/ml hydrocortisone, 50 U/ml penicillin and 50μg/ml streptomycin. Human Umbilical Cord Vein Endothelial Cells (HUVECs) were from Cambrex. Cells were maintained in basal EGM-2 supplemented with 10% FBS (Hyclone).
Lentivirus Production
The pLVX-DsRed-Monomer-C1 vector (Clontech) was utilized to obtain a lentiviral construct expressing Dbl conjugated with a red fluorescent protein. Recombinant lentiviruses were produced in HEK-293 T cells using the Lenti-X Lentiviral Expression kit (Clontech) as described by the manufacturer. MCF-10 A cells were infected with lentiviruses expressing the red fluorescent protein alone (pRed) or the Dbl product fused to the red fluorescent protein (pRDbl) in the presence of 10 μg/ml of polybrene. pRed and pRDbl infected cells were cultured in DMEM/F12 supplemented with 5% horse serum and 40 μg/ml puromycin.
In vivo Rho GTPases activation assay
Pull-down assays were carried out as previously described.Citation51 Briefly, glutathione-coupled Sepharose 4B beads bound to recombinant GST–PAK CRIB domain fusion protein were incubated with cell extracts at 4°C for 45 min, eluted in Laemmli buffer and analyzed for the presence of Rac1 and Cdc42 by Western blot.
Activated RhoA was detected by using a G‑Lisa RhoA Activation assay (Absorbance based) (Cytoskeleton Inc.) as indicated by the manufacturer. Aliquots of cell lysates were subjected to SDS PAGE and immunoblotting to evaluate pRDbl and RhoA expression levels.
Western Blot analysis
pRed and pRDbl cells were lysed in StaphA buffer containing 1 mM AEBSF and 10 mg/ml each of aprotinin and leupeptin. Lysates were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to PVDF membrane (Merck Millipore). Lysates were probed with specific antibodies to detect E-cadherin, α-SMA, MMP-12 and Dbl expression level. The blots were reprobed with anti-β-actin (Santa Cruz) antibody to evaluate protein loading. Immunocomplexes were visualized by West Dura extended chemioluminescent detection using a horseradish peroxidase (HRP)-conjugated secondary antibody (Thermo Fisher Scientific). The optical density of the film was scanned and measured with Quantity One v. 2–3 Image software (Versa Doc, Biorad).
Real-Time quantitative (qRT)-PCR analysis
Total RNA was purified using the RNeasy MiniKit from Qiagen, and controlled for integrity with an Agilent Bioanalyzer 2100 (Agilent Technologies). RNA was quantified by NanoDrop (NanoDrop Technologies) and reverse-transcribed into double-stranded cDNA on a GeneAmp PCR System 2700 thermal cycler (Applied Biosystems) using the SuperScript Double-Stranded cDNA synthesis kit (Invitrogen). qRT-PCR was performed on a 7500 Real Time PCR System (Applied Biosystems) in triplicate for each target transcript using SYBR Green PCR Master Mix (Applied Biosystems) and sense/antisense oligonucleotide primers () synthesized by TIB Molbiol (Genova, Italy) as detailed.Citation52 Expression data were normalized on the values obtained in parallel for 2 reference genes (Actin Related Protein 2/3 Complex subunit 1, ARPC1B; ribosomal protein S19, RPS19) using the Bestkeeper software, and relative expression values were calculated using Q-gene software.Citation52 Data were analyzed with Graph Pad Prism 5 software v. 5.00 (GraphPad Software).
Table 1. Primer pairs used for qRT-PCR
Cell migration
Cell migration was assayed in Boyden chambers (8.0 μm pore size, FALCON cell culture insert). pRed and pRDbl MCF-10 A cells were cultured overnight in the absence or presence of 100 μM of NSC23766. The cells were trypsinized and counted. One × 105 cells were resuspended in serum‑free medium and added to the upper chamber. Inserts, previously coated with fibronectin on the lower side, were incubated for 1, 2 and 4 h at 37°C with 700 μl of complete medium in the lower chamber. Cells on the inside of the inserts were removed with a cotton swab, and cells on the underside of the insert were fixed and stained with May‑Grunwald/Giemsa. The number of cells were counted and the average number of the cells that had transmigrated was determined.
For the scratch assay, pRed or pRDbl (8 × 104) cells were plated on transwell inserts (12 mm diameter, polycarbonate membranes with 0.4 μm pores, Corning, Lowell, MA, USA) and cultured to confluence. HUVECs were plated in 12-well plates at the concentration of 8 × 104 cells/well and cultured to 90% confluence in EGM-2 medium supplemented with 10% FBS. HUVECs were then starved for 24 h in EGM-2 medium containing 0.1% FBS. Cell monolayers were then scored vertically down the center of each well with a sterile tip and each well was washed with PBS to remove detached cells. Medium was replaced with fresh EGM-supplemented with 0.1% FBS. The transwell inserts containing pRed or pRDbl cells were then placed on top of HUVECs and cultured for another 24 h.
Images of the wound in each well were acquired at time 0 (t0) and after 24 h (t24) under phase contrast microscope at the magnification of 10X. The area covered by the cells was measured by using Image J and the ratio between t24 and t0 was performed. Results are expressed as percentage of healing taking as reference the area at time 0.
Anchorage-independent growth assay
For soft agar assay, cells were detached from plates, resuspended in DMEM/F-12 medium with 5% HS and 0.3% agarose (UltraPureTM by Invitrogen), and plated on top of a 0.5% agarose layer at the concentration of 104 and 105 cells per plate. Cells were fed once a week with DMEM/F-12 supplemented with 5% HS and growing colonies were scored after 14 days and photographed.
Three-dimensional morphogenetic assay
3D morphogenetic assays were conducted as previously described in http://muthuswamylab.cshl.edu/ml protocols.html and for a review see refs.Citation30,53,54
Immunofluorescence analysis
pRed and pRDbl cells were plated onto glass coverslips, fixed with 4% paraformaldehyde in PBS for 30 minutes at 25°C, treated with 0.1 M glycine in PBS for 20 minutes at 25°C and with 0.1% Triton X-100 in PBS for additional 5 minutes at 25°C to allow permeabilization. Filamentous actin was visualized by incubating the cells with FITC-labeled phalloidin (Sigma) in PBS for 45 min at 25°C, and nuclei were stained with 10 μg/ml DAPI (Sigma). Coverslips were mounted with Mowiol (Calbiochem) for observation.
To analyze 3D acinar structures, immunostaining of 3D cultured MCF10 A cells was performed as described in http://muthuswamylab.cshl.edu/protocols/IF protocol.pdf
Fluorescence signals were visualized with an AxioObserver inverted microscope (Zeiss), equipped with the ApoTome System (Carl Zeiss Inc.). In the case of active caspase 3 and Ki-67 cells were scanned in a series of 0.5 μm sequential sections with an ApoTome System (Zeiss), connected to an AxioObserver inverted microscope; images were all acquired by the digital camera Axio CAM MRm (Zeiss) and analysis was performed by the Axiovision software (Zeiss). Reconstruction of a selection of 3 central out of the total number of the serial optical sections was shown in each figure. Red epitope was visualized by incubating cells with Anti-Red antibody (Clontech).
Matrigel assay
pRed or pRDbl MCF-10 A cells (8 × 104) were cultured on transwell inserts (12 mm diameter, polycarbonate membranes with 0.4 μM pores, Corning,) to confluence. The inserts were placed on top of HUVECs plated in 12-well plates at 13.5 × 104 cells/well onto a thin layer (300 μl) of basement membrane matrix (Matrigel; Becton Dickinson) in EGM-2 medium supplemented with 0.1% FBS. In some experiments 1 μg/ml of CCL-2 blocking peptide was added to the lower chamber. After 18 h, the transwells and the medium were removed and HUVECs were fixed with 4% paraformaldehyde in PBS for 30 min at 25°C and permeabilized with 0.1% Triton X‑100 for 5 min. Filamentous actin was visualized by incubating the cells with DY554 phalloidin (Thermo Fisher Scientific). Images of fluorescent cells were obtained using a Nikon Eclipse TE 300 inverted microscope and acquired by a Nikon digital camera DS-5 MC and NIS Element software (Nikon). Quantification of the tubular structures was performed by counting the number of complete circles produced by interlinking tubular HUVECs.Citation55
Statistical analysis
Data are the mean ± SD of 3 independent experiments, unless differently specified. The paired Student′s t-test was used to determine the significance of the result (P < 0.05).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
1021516_Figure_S1.pdf
Download PDF (356.9 KB)Acknowledgments
We thank Dr. Y. Zheng for providing Rac inhibitor NSC23766. We are grateful to Sara Barzaghi for secretarial assistance.
Funding
This work was supported by grants from Italian Association for Cancer Research (AIRC-IG10272), from the Compagnia di San Paolo, from the Italian Health Ministry.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- Hay ED. The mesenchymal cell, its role in the embryo, and the remarkable signaling mechanisms that create it. Dev Dyn 2005; 233:706-20; PMID:15937929; http://dx.doi.org/10.1002/dvdy.20345
- Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol 2006; 7:131-42; PMID:16493418; http://dx.doi.org/10.1038/nrm1835
- Huber MA, Kraut N, Beug H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr Opin Cell Biol 2005; 17:548-58; PMID:16098727; http://dx.doi.org/10.1016/j.ceb.2005.08.001
- Lee JM, Dedhar S, Kalluri R, Thompson EW. The epithelial-mesenchymal transition: new insights in signaling, development, and disease. J Cell Biol 2006; 172:973-81; PMID:16567498; http://dx.doi.org/10.1083/jcb.200601018
- Micalizzi DS, Farabaugh SM, Ford HL. Epithelial-mesenchymal transition in cancer: parallels between normal development and tumor progression. J Mammary Gland Biol Neoplasia 2010; 15:117-34; PMID:20490631; http://dx.doi.org/10.1007/s10911-010-9178-9
- Palena C, Hamilton DH, Fernando RI. Influence of IL-8 on the epithelial-mesenchymal transition and the tumor microenvironment. Future Oncol 2012; 8:713-22; PMID:22764769; http://dx.doi.org/10.2217/fon.12.59
- Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol 2005; 6:167-80; PMID:15688002; http://dx.doi.org/10.1038/nrm1587
- Erickson JW, Cerione RA. Structural elements, mechanism, and evolutionary convergence of Rho protein-guanine nucleotide exchange factor complexes. Biochemistry 2004; 43:837-42; PMID:14744125; http://dx.doi.org/10.1021/bi036026v
- Schmidt A, Hall A. Guanine nucleotide exchange factors for Rho GTPases: turning on the switch. Genes Dev 2002; 16:1587-609; PMID:12101119; http://dx.doi.org/10.1101/gad.1003302
- Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol 2005; 21:247-69; PMID:16212495; http://dx.doi.org/10.1146/annurev.cellbio.21.020604.150721
- Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science 2003; 302:1704-9; PMID:14657486; http://dx.doi.org/10.1126/science.1092053
- Burridge K, Wennerberg K. Rho and Rac take center stage. Cell 2004; 116:167-79; PMID:14744429; http://dx.doi.org/10.1016/S0092-8674(04)00003-0
- Ron D, Tronick SR, Aaronson SA, Eva A. Molecular cloning and characterization of the human dbl proto-oncogene: evidence that its overexpression is sufficient to transform NIH/3T3 cells. EMBO J 1988; 7:2465-73; PMID:3056717
- Hirsch E, Pozzato M, Vercelli A, Barberis L, Azzolino O, Russo C, Vanni C, Silengo L, Eva A, Altruda F. Defective dendrite elongation but normal fertility in mice lacking the Rho-like GTPase activator Dbl. Mol Cell Biol 2002; 22:3140-8; PMID:11940671; http://dx.doi.org/10.1128/MCB.22.9.3140-3148.2002
- Ognibene M, Barbieri O, Vanni C, Mastracci L, Astigiano S, Emionite L, Salani B, Fedele M, Resaz R, Tenca C, et al. High frequency of development of B cell lymphoproliferation and diffuse large B cell lymphoma in Dbl knock-in mice. J Mol Med (Berl) 2011; 89:493-504; PMID:21221514; http://dx.doi.org/10.1007/s00109-010-0712-4
- Eva A, Graziani G, Zannini M, Merin LM, Khillan JS, Overbeek PA. Dominant dysplasia of the lens in transgenic mice expressing the dbl oncogene. New Biol 1991; 3:158-68; PMID:2065011
- Fardin P, Ognibene M, Vanni C, De SA, Varesio L, Eva A. Induction of epithelial mesenchimal transition and vasculogenesis in the lenses of Dbl oncogene transgenic mice. PLoS One 2009; 4:e7058; PMID:19759912; http://dx.doi.org/10.1371/journal.pone.0007058
- Herr R, Wohrle FU, Danke C, Berens C, Brummer T. A novel MCF-10 A line allowing conditional oncogene expression in 3D culture. Cell Commun Signal 2011; 9:17; PMID:21752278; http://dx.doi.org/10.1186/1478-811X-9-17
- Debnath J, Brugge JS. Modelling glandular epithelial cancers in three-dimensional cultures. Nat Rev Cancer 2005; 5:675-88; PMID:16148884; http://dx.doi.org/10.1038/nrc1695
- Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer 2002; 2:442-54; PMID:12189386; http://dx.doi.org/10.1038/nrc822
- Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol 2014; 15:178-96; PMID:24556840; http://dx.doi.org/10.1038/nrm3758
- Eva A, Aaronson SA. Isolation of a new human oncogene from a diffuse B-cell lymphoma. Nature 1985; 316:273-5; PMID:3875039; http://dx.doi.org/10.1038/316273a0
- Ron D, Zannini M, Lewis M, Wickner RB, Hunt LT, Graziani G, Tronick SR, Aaronson SA, Eva A. A region of proto-dbl essential for its transforming activity shows sequence similarity to a yeast cell cycle gene, CDC24, and the human breakpoint cluster gene, bcr. New Biol 1991; 3:372-9; PMID:2065022
- Vanni C, Mancini P, Gao Y, Ottaviano C, Guo F, Salani B, Torrisi MR, Zheng Y, Eva A. Regulation of proto-Dbl by intracellular membrane targeting and protein stability. J Biol Chem 2002; 277:19745-53; PMID:11907027; http://dx.doi.org/10.1074/jbc.M111025200
- Nobes CD, Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell 1995; 81:53-62; PMID:7536630; http://dx.doi.org/10.1016/0092-8674(95)90370-4
- Edme N, Downward J, Thiery JP, Boyer B. Ras induces NBT-II epithelial cell scattering through the coordinate activities of Rac and MAPK pathways. J Cell Sci 2002; 115:2591-601; PMID:12045229
- Keely PJ, Westwick JK, Whitehead IP, Der CJ, Parise LV. Cdc42 and Rac1 induce integrin-mediated cell motility and invasiveness through PI(3)K. Nature 1997; 390:632-6; PMID:9403696; http://dx.doi.org/10.1038/37656
- Gao Y, Dickerson JB, Guo F, Zheng J, Zheng Y. Rational design and characterization of a Rac GTPase-specific small molecule inhibitor. Proc Natl Acad Sci U S A 2004; 101:7618-23; PMID:15128949; http://dx.doi.org/10.1073/pnas.0307512101
- Muthuswamy SK, Li D, Lelievre S, Bissell MJ, Brugge JS. ErbB2, but not ErbB1, reinitiates proliferation and induces luminal repopulation in epithelial acini. Nat Cell Biol 2001; 3:785-92; PMID:11533657; http://dx.doi.org/10.1038/ncb0901-785
- Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10 A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 2003; 30:256-68; PMID:12798140; http://dx.doi.org/10.1016/S1046-2023(03)00032-X
- Reginato MJ, Mills KR, Becker EB, Lynch DK, Bonni A, Muthuswamy SK, Brugge JS. Bim regulation of lumen formation in cultured mammary epithelial acini is targeted by oncogenes. Mol Cell Biol 2005; 25:4591-601; PMID:15899862; http://dx.doi.org/10.1128/MCB.25.11.4591-4601.2005
- Debnath J, Mills KR, Collins NL, Reginato MJ, Muthuswamy SK, Brugge JS. The role of apoptosis in creating and maintaining luminal space within normal and oncogene-expressing mammary acini. Cell 2002; 111:29-40; PMID:12372298; http://dx.doi.org/10.1016/S0092-8674(02)01001-2
- Scholzen T, Gerdes J. The Ki-67 protein: from the known and the unknown. J Cell Physiol 2000; 182:311-22; PMID:10653597; http://dx.doi.org/10.1002/(SICI)1097-4652(200003)182:3%3c311::AID-JCP1%3e3.0.CO;2-9
- Arnaoutova I, George J, Kleinman HK, Benton G. The endothelial cell tube formation assay on basement membrane turns 20: state of the science and the art. Angiogenesis 2009; 12:267-74; PMID:19399631; http://dx.doi.org/10.1007/s10456-009-9146-4
- Noguchi T, Galland F, Batoz M, Mattei MG, Birnbaum D. Activation of a mcf.2 oncogene by deletion of amino-terminal coding sequences. Oncogene 1988; 3:709-15; PMID:2577874
- Liu XF, Ohno S, Miki T. Nucleotide exchange factor ECT2 regulates epithelial cell polarity. Cell Signal 2006; 18:1604-15; PMID:16495035; http://dx.doi.org/10.1016/j.cellsig.2006.01.007
- Prag S, Parsons M, Keppler MD, Ameer-Beg SM, Barber P, Hunt J, Beavil AJ, Calvert R, Arpin M, Vojnovic B, et al. Activated ezrin promotes cell migration through recruitment of the GEF Dbl to lipid rafts and preferential downstream activation of Cdc42. Mol Biol Cell 2007; 18:2935-48; PMID:17538024; http://dx.doi.org/10.1091/mbc.E06-11-1031
- Murakami M, Meneses PI, Knight JS, Lan K, Kaul R, Verma SC, Robertson ES. Nm23-H1 modulates the activity of the guanine exchange factor Dbl-1. Int J Cancer 2008; 123:500-10; PMID:18470881; http://dx.doi.org/10.1002/ijc.23568
- Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest 2009; 119:1420-8; PMID:19487818; http://dx.doi.org/10.1172/JCI39104
- Navaraj A, Finnberg N, Dicker DT, Yang W, Matthew EM, El-Deiry WS. Reduced cell death, invasive and angiogenic features conferred by BRCA1-deficiency in mammary epithelial cells transformed with H-Ras. Cancer Biol Ther 2009; 8:2417-44; PMID:20038817; http://dx.doi.org/10.4161/cbt.8.24.10850
- Potente M, Gerhardt H, Carmeliet P. Basic and therapeutic aspects of angiogenesis. Cell 2011; 146:873-87; PMID:21925313; http://dx.doi.org/10.1016/j.cell.2011.08.039
- Fantozzi A, Gruber DC, Pisarsky L, Heck C, Kunita A, Yilmaz M, Meyer-Schaller N, Cornille K, Hopfer U, Bentires-Alj M, et al. VEGF-mediated angiogenesis links EMT-induced cancer stemness to tumor initiation. Cancer Res 2014; 74:1566-75; PMID:24413534; http://dx.doi.org/10.1158/0008-5472.CAN-13-1641
- Salcedo R, Ponce ML, Young HA, Wasserman K, Ward JM, Kleinman HK, Oppenheim JJ, Murphy WJ. Human endothelial cells express CCR2 and respond to MCP-1: direct role of MCP-1 in angiogenesis and tumor progression. Blood 2000; 96:34-40; PMID:10891427
- Keeley EC, Mehrad B, Strieter RM. Chemokines as mediators of neovascularization. Arterioscler Thromb Vasc Biol 2008; 28:1928-36; PMID:18757292; http://dx.doi.org/10.1161/ATVBAHA.108.162925
- Soria G, Ben-Baruch A. The inflammatory chemokines CCL2 and CCL5 in breast cancer. Cancer Lett 2008; 267:271-85; PMID:18439751; http://dx.doi.org/10.1016/j.canlet.2008.03.018
- Wang CC, Jamal L, Janes KA. Normal morphogenesis of epithelial tissues and progression of epithelial tumors. Wiley Interdiscip Rev Syst Biol Med 2012; 4:51-78; PMID:21898857; http://dx.doi.org/10.1002/wsbm.159
- Aranda V, Haire T, Nolan ME, Calarco JP, Rosenberg AZ, Fawcett JP, Pawson T, Muthuswamy SK. Par6-aPKC uncouples ErbB2 induced disruption of polarized epithelial organization from proliferation control. Nat Cell Biol 2006; 8:1235-45; PMID:17060907; http://dx.doi.org/10.1038/ncb1485
- Derksen PW, Liu X, Saridin F, van der Gulden H, Zevenhoven J, Evers B, van Beijnum JR, Griffioen AW, Vink J, Krimpenfort P, et al. Somatic inactivation of E-cadherin and p53 in mice leads to metastatic lobular mammary carcinoma through induction of anoikis resistance and angiogenesis. Cancer Cell 2006; 10:437-49; PMID:17097565; http://dx.doi.org/10.1016/j.ccr.2006.09.013
- Jechlinger M, Grunert S, Tamir IH, Janda E, Ludemann S, Waerner T, Seither P, Weith A, Beug H, Kraut N. Expression profiling of epithelial plasticity in tumor progression. Oncogene 2003; 22:7155-69; PMID:14562044; http://dx.doi.org/10.1038/sj.onc.1206887
- Kim YN, Koo KH, Sung JY, Yun UJ, Kim H. Anoikis resistance: an essential prerequisite for tumor metastasis. Int J Cell Biol 2012; 2012:306879; PMID:22505926; http://dx.doi.org/10.1155/2012/306879
- Vanni C, Visco V, Mancini P, Parodi A, Ottaviano C, Ognibene M, Manazza AD, Retta SF, Varesio L, Torrisi MR, et al. Inhibition of PI3 K induces Rac activation and membrane ruffling in proto-Dbl expressing cells. Cell Cycle 2006; 5:2657-65; PMID:17172840; http://dx.doi.org/10.4161/cc.5.22.3445
- Raggi F, Blengio F, Eva A, Pende D, Varesio L, Bosco MC. Identification of CD300 a as a new hypoxia-inducible gene and a regulator of CCL20 and VEGF production by human monocytes and macrophages. Innate Immun 2013; PMID:24131792
- Seton-Rogers SE, Lu Y, Hines LM, Koundinya M, LaBaer J, Muthuswamy SK, Brugge JS. Cooperation of the ErbB2 receptor and transforming growth factor beta in induction of migration and invasion in mammary epithelial cells. Proc Natl Acad Sci U S A 2004; 101:1257-62; PMID:14739340; http://dx.doi.org/10.1073/pnas.0308090100
- Seton-Rogers SE, Brugge JS. ErbB2 and TGF-beta: a cooperative role in mammary tumor progression? Cell Cycle 2004; 3:597-600; PMID:15107620; http://dx.doi.org/10.4161/cc.3.5.886
- Hull MA, Thomson JL, Hawkey CJ. Expression of cyclooxygenase 1 and 2 by human gastric endothelial cells. Gut 1999; 45:529-36; PMID:10486360; http://dx.doi.org/10.1136/gut.45.4.529
