Abstract
The Suv39h1 and Suv39h2 H3K9 histone methyltransferases (HMTs) have a conserved role in the formation of constitutive heterochromatin and gene silencing. Using a transgenic mouse model system we demonstrate that elevated expression of Suv39h1 increases global H3K9me3 levels in vivo. More specifically, Suv39h1 overexpression enhances the imposition of H3K9me3 levels at constitutive heterochromatin at telomeric and major satellite repeats in primary mouse embryonic fibroblasts. Chromatin compaction is paralleled by telomere shortening, indicating that telomere length is controlled by H3K9me3 density at telomeres. We further show that increased Suv39h1 levels result in an impaired clonogenic potential of transgenic epidermal stem cells and Ras/E1A transduced transgenic primary mouse embryonic fibroblasts. Importantly, Suv39h1 overexpression in mice confers resistance to a DMBA/TPA induced skin carcinogenesis protocol that is characterized by the accumulation of activating H-ras mutations. Our results provide genetic evidence that Suv39h1 controls telomere homeostasis and mediates resistance to oncogenic stress in vivo. This identifies Suv39h1 as an interesting target to improve oncogene induced senescence in premalignant lesions.
Introduction
The H3K9 specific histone methyltransferases (HMTs) Suv39h1 and Suv39h2 play a central role in the establishment of constitutive heterochromatin and gene silencing.Citation1-3 Pioneering work demonstrated that the Suv39h1 and Suv39h2 histone methyltransferases impose a tri-methylation mark on lysine-9 of histone H3 (H3K9me3) which recruits members of the family of Heterochromatin protein 1 (HP1) to establish the core structure of constitutive heterochromatin at pericentric and telomeric repeats.Citation2,3 Lack of Suv39h HMTases in mice results in a complete loss of H3K9me3 and HP1 at pericentric, telomeric and subtelomeric repeats, submendelian birth-rates and impaired chromosome segregation as well as an increased frequency of spontaneous B cell lymphomas.Citation4 Suv39h HMTases were also demonstrated to silence intact LINE repeats in mouse embryonic stem cells.Citation5 Importantly, Suv39h HMTases were demonstrated to act as central regulators of telomere length, underlining the relevance of H3K9 methylation for the maintenance of genomic stability.Citation6-9 Telomeres are heterochromatic, nucleoprotein structures that ensure genome integrity by protecting chromosomes from end-to-end fusions.Citation10 Loss of telomere function results in de-protection of chromosome ends that triggers a DNA damage response leading to cell cycle arrest and senescence, hallmarks of organismal aging.Citation11 On the other hand, telomere dysfunction increases cancer formation as a consequence of increased genomic instability (for review Citation6,9). Several studies demonstrated that constitutive heterochromatin acts as potent regulator of telomere length in vertebrates, suggesting that telomere chromatin structure impacts on human aging and cancer.Citation7-9 In particular, lack of H3K9me3 at telomeres and subtelomeres drives recombination between telomeric sister chromatids and leads to massive telomere elongation without affecting chromosome end protection.Citation7,8 Moreover, progressive telomere shortening is linked with the loss of telomeric heterochromatin, including H3K9me3.Citation8 This indicates that telomeric heterochromatin is not required for chromosome end protection but has an important role in telomere length regulation.Citation7,9 Suv39h HMTases are also implicated in de novo gene silencing. Retinoblastoma protein 1 was demonstrated to recruit Suv39h1 in order to impose repressive H3K9 methylation at promoter regions of growth promoting E2F target genes, thereby contributing to the establishment of cellular senescence.Citation12 Remarkably, Suv39h HMTases play a central role in the establishment of foci enriched for H3K9me3 and HP1 in the periphery of senescent but not quiescent cells, underlining a link between Suv39h1 and senescence.Citation13 Interestingly, induction of lesions by the oncogenes Ras or BRAF is associated with increased H3K9me3 and HP1 nuclear staining intensity and the expression of senescence markers. In line with this, loss of Suv39h HMTases accelerates Ras or Myc driven tumorigenesis, indicating a central role of Suv39h HMTases in oncogene induced senescence.Citation14,15 Altogether, this indicates an intimate connection of chromatin regulation with oncogene induced senescence in pre-malignant lesions Citation14,16-20. The role of Suv39h HMTases in cancer and telomere regulation has been extensively studied in loss of function model systems.Citation4 However, the effect of elevated Suv39h HMTase expression on telomere regulation and tumorsuppression during oncogenic stress has not yet been addressed in gain of function models in vivo.
Here, we use a conditional Suv39h1 transgenic mouse model system to study the role of Suv39h1 in telomere regulation, and tumor induction using a DMBA/TPA (7,12-Dimethylbenz[α]anthracene/12-O-tetradecanoylphorbol-13-acetate) skin carcinogenesis protocol that is characterized by the accumulation of activating H-ras mutations.Citation4 We demonstrate that elevated Suv39h1 expression significantly increases global H3K9me3 in the skin and primary mouse embryonic fibroblasts of Suv39h1 transgenic mice. Importantly, increased H3K9me3 at telomeres of Suv39h1 transgenic pMEFs is associated with a significant telomere shortening demonstrating that Suv39h1 is a negative regulator of telomere length in embryo derived cells. Ectopic expression of Suv39h1 increases the expression of p53 and its target genes Bax and Puma and limits Ras/E1A induced immortalization of Suv39h1 transgenic primary mouse embryonic fibroblasts (pMEFs). In line with this, Suv39h1 transgenic mice are highly resistant to DMBA/TPA driven skin carcinogenesis. DMBA/TPA driven skin carcinogenesis is a process that involves the accumulation of oncogenic H-Ras mutations, that are reported to trigger oncogene-induced senescence.Citation14,19 Our results show that telomere length regulation is Suv39h1 dosage sensitive in embryo derived cells and that increased Suv39h1 levels modulate p53 tumorsuppressor pathway activity, thereby limiting transformation in the context of oncogenic stress or carcinogenesis. Our results underline the relevance of telomeric heterochromatin for the regulation of telomere length and suggest that Suv39h1 represents an interesting target to improve oncogene-mediated senescence in pre-malignant lesions.
Results
Increased levels of global H3K9me3 in Suv39h1 overexpressing mice
In order to study the biological consequences of Suv39h1 overexpression in vivo we have generated conditional transgenic mice. A construct containing the Suv39h1 cDNA downstream of a floxed transcriptional stop-cassette (pA), that terminates transgene-transcription driven from the upstream located CAGGS promoter was used to generate a transgenic mouse line (; see Material and Methods). Transgenic mice were crossed with homozygous CMV-Cre transgenic mice (ref. Citation21) to obtain animals with excised transcriptional stop-cassette (Suv39h1ΔStop/CMV-Cre; in this study: “Suv39h1ΔStop” mice) and heterozygous CMV-Cre mice (in this study: “control”). To verify Cre-recombinase mediated deletion of the transcriptional stop-cassette, we have performed southern blotting analysis using a specific probe that discriminates between transgene insertions before and after deletion of the stop-cassette, but also detects the endogenous Suv39h1 (Figure S1A). The excision of the stop-cassette in pMEFs obtained from Suv39h1 and Cre double transgenic mice is highly variable, with a maximum frequency of 98% (Figure S1A). Importantly, the efficiency of Cre-mediated deletion of the stop-cassette correlates with transgene expression, reaching a maximum 45-fold overexpression of Suv39h1 mRNA in primary mouse embryonic fibroblasts (pMEFs) derived from Suv39h1ΔStop embryos (Figure S1A, B). Consistent with the role of Suv39h1 as histone H3 specific HMTase, pMEFs derived from Suv39h1ΔStop embryos display significantly increased levels of tri-methylated histone lysine-9 of histone H3 (H3K9me3; ; Figure S1C). This indicates that Suv39h1 HMTase expressed from the transgenic construct is enzymatically active.
Figure 1. Suv39h1 transgenic mice. (A) Schematic representation of the Suv39h1 transgenic vector. Restriction sites for transgene-cassette excision and DNA gel blotting are indicated. (B) Increased H3K9me3 expression levels in pMEFs obtained from control and Suv39h1ΔStop transgenic mice as determined by western blotting. Antibodies against global histone H3 were used to control for equal loading. (C) Quantification of H3K9me3 levels shown in (B). H3K9me3 levels were normalized to global histone H3 levels. (D) Suv39h1 expression levels in pMEFs derived from control and Suv39h1ΔStop transgenic mice as determined by western blotting using nuclear extracts (pMEFs). Whole cell extracts prepared from H1299 transiently transfected with a control vector (pcDNA) or HA-tagged Suv39h1 (HA-Suv39h1) were loaded to verify specificity of the anti-Suv39h1 antibody. Weak Suv39h1 signals in Suv39h1ΔStop pMEFs are due to low antibody specificity. Arrows indicated specific Suv39h1 band; L.E.: long exposure, SE: short exposure. N = number of different pMEF pool used for each genotype; standard deviations are indicated; a Student's t-test was used to calculate statistical significance.
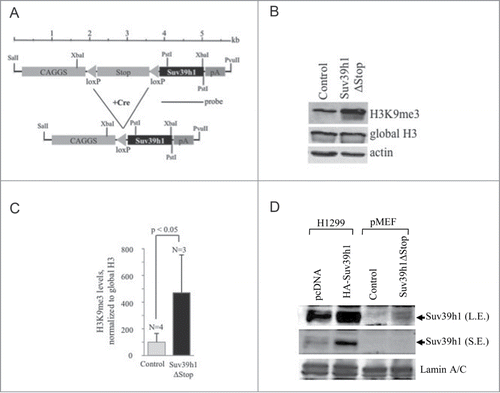
Finally, western blotting using specific anti-Suv39h1 antibodies confirmed ectopic expression of Suv39h1 in pMEFs obtained from Suv39h1ΔStop embryos (). Of note, Suv39h1ΔStop animals were phenotypically normal and did not display weight differences when compared to control mice (Figure S2A, B).
We conclude that deletion of the floxed transcriptional stop-cassette results in Suv39h1 transgene expression, and consequently increased global levels of H3K9me3 in Suv39h1ΔStop pMEFs.
Suv39h1 overexpression impacts on HP1β and increases H3K9me3 in transgenic animals
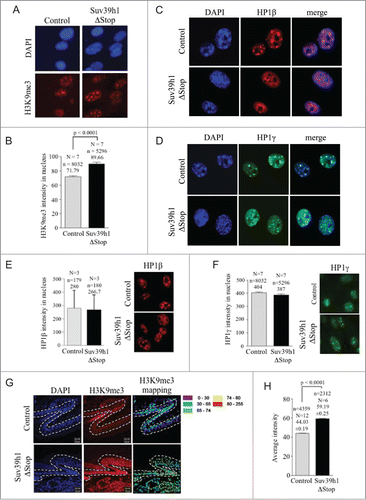
We next wished to investigate the impact of Suv39h1 overexpression on chromatin structure in transgenic animals. In line with western blotting experiments we found that Suv39h1 overexpression correlates with a significant increase of H3K9me3 staining intensity in the nuclei and at chromocenters of Suv39h1ΔStop pMEFs (). High-affinity binding of HP1 proteins to H3K9me3 represents a key-step in the formation of constitutive heterochromatin at pericentric and telomeric repeats.Citation2,7 To test whether ectopic Suv39h1 impacts on the macroscopic structure of constitutive heterochromatin we performed immunostainings for HP1β and HP1γ on Suv39h1ΔStop pMEFs. We found that Suv39h1 overexpression changes dense HP1β staining at DAPI rich chromocenters to a more dispersed staining pattern; however increased Suv39h1 expression does not have an obvious impact on HP1γ staining pattern (). Remarkably, although H3K9me3 levels are efficiently increased in Suv39h1ΔStop pMEFs, we did not find evidence for a biologically relevant increase of HP1β or HP1γ levels in the nuclei of pMEFs from transgenic animals, as detected by quantitative immunofluorescence (). We next wished to study the effect of Suv39h1 expression on H3K9me3 levels in vivo. To validate the impact of Suv39h1 transgene expression on chromatin structure we measured global H3K9me3 levels on tail sections prepared from adult Suv39h1ΔStop mice using confocal microscopy. Consistent with data from pMEFs, we found significantly increased levels of H3K9me3 in the nuclei of skin cells from Suv39h1ΔStop transgenic mice (). Together, this indicates that Cre-mediated deletion of the stop cassette results in Suv39h1 overexpression that increases global H3K9me3 abundance and modulates the distribution of nuclear HP1β, without affecting global HP1β and HP1γ levels.
Figure 2. Suv39h1 overexpression increases H3K9me3 in transgenic pMEFs and animals: (A) Increased nuclear H3K9me3 levels in pMEFs obtained from control and Suv39h1ΔStop transgenic mice as determined by automated confocal microscopy. (B) Quantification of confocal H3K9me3 immunofluorescence of samples shown in (A) Suv39h1 overexpression results in a significant increase in H3K9me levels. (C) Suv39h1 overexpression results in a re-distribution of HP1β in pMEFs, as determined by immunofluorescence staining. (D) Suv39h1 does not impact on the localization of HP1γ, as determined by immunofluorescence staining. (E) Suv39h1 overexpression does not impact on global HP1β levels in Suv39h1ΔStop pMEFs, as determined by classic quantitative immunofluorescence. Left panel: quantification; right panel, representative images. (F) Suv39h1 overexpression does not impact on global HP1γ levels in Suv39h1ΔStop pMEFs, as determined by automated confocal quantitative immunofluorescence. Left panel: quantification; right panel, representative images. (G) Immuno-histochemistry on adult skin sections using H3K9me3 specific antibodies. Suv39h1ΔStop mice display increase H3K9me3 levels as demonstrated by confocal microscopy and signal intensity mapping (see material methods). H3K9me3 intensity ranges are indicated by a color code that is based on arbitrary telomere fluorescence units. (H) Quantification of average H3K9me3 intensity. Average H3K9me3 levels are significantly increased in tail skin sections of Suv39h1ΔStop mice. N = number of independently prepared pMEF cultures or mice tested; n = number of nuclei analyzed; arbitrary fluorescence intensity values are indicated; standard deviations are indicated; p-values indicate statistical significance.
Ectopic Suv39h1 expression drives telomere shortening in pMEFs
We previously showed that Suv39h1 and Suv39h2 double knock out (Suv39hDN) pMEFs and mouse embryonic stem cells (mESCs) display a loss of chromatin compaction at telomeres resulting in recombination dependent telomere elongation.Citation7 This indicates that telomeric chromatin structure is a major regulator of telomere length.Citation9 To investigate whether increased Suv39h1 expression impacts on telomere regulation we performed telomere specific chromatin immunoprecipitation and telomere length measurements. Consistent with immunofluorescence data, we found that ectopic Suv39h1 expression causes an approximately 50% increase in H3K9me3 at telomeric and major satellite repeats without affecting the abundance of HP1γ or H4K20me3 (). As expected, Suv39h1 does not impact on the abundance of TRF1 at telomeric TTAGGG repeats ().Citation7 Performing quantitative DNA FISH using a fluorescently labeled telomere repeat specific probe we found that Suv39h1 overexpression results a significant overall telomere shortening in Suv39h1ΔStop pMEFs when compared with control pMEFs (P < 0.0001; ). In line with this, we found that Suv39h1 transgenic mice display an increased number of short telomeres (< 30 kb) and reduced number of long telomeres (>70 kb) (). Our data is in line with the reported telomere elongation in Suv39DN cells and underlines the role of Suv39h1 as negative regulator of telomere length.Citation7 Progressive telomere shortening results in decreased subtelomeric DNA methylation in telomerase knock-out pMEFs.Citation8 However, the modest telomere shortening in Suv39h1 overexpressing pMEFs did not have an effect on subtelomeric DNA methylation (Fig. S4). This suggests, that pronounced telomere shortening is required to induce alteration in subtelomeric DNA methylation. In contrast to Suv39h1ΔStop pMEFs that show a 10% reduction of telomere length (), we were not able to detect telomere shortening in total skin or basal layer cells of these mice (Figure S3A-C). We propose that rapid cell proliferation and activation of various pathways involved in cell specification events during adult skin formation can indirectly impact on telomere homeostasis, thereby occulting the negative effect of Suv39h1 on telomere length.
Figure 3. Compaction of telomeric chromatin and telomere shortening in Suv39h1ΔStop pMEFs. (A) Suv39h1ΔStop pMEFs display augmented H3K9me3 levels at telomeric repeats, as determined by ChIP. Representative images are shown. (B) Quantification of immunoprecipitated telomeric DNA using the indicated antibodies; signals were normalized to the input material. (C) Suv39h1ΔStop pMEFs display augmented H3K9me3 levels at pericentric major satellite repeats. Representative images are shown. (D) Quantification of immunoprecipitated major satellite repeats; signals were normalized to the input material. (E) Quantitative telomere Q-FISH on metaphase chromosomes obtained from control and Suv39h1ΔStop pMEFs. Suv39h1ΔStop transgenic pMEFs (right panel) display overall telomere shortening when compared to control littermates (left panel). (F) Average telomere length is significantly shorter in Suv39h1ΔStop transgenic mice when compared to pMEFs with normal Suv39h1 expression. (G) Accumulation of short telomeres (<30 kb) and reduction of long (>70 kb) upon Suv39h1 overexpression in Suv39h1ΔStop pMEFs. N, number of independently prepared pMEF cultures analyzed; standard deviations are indicated; n, number of telomeres analyzed; statistical significance is indicated by p-values
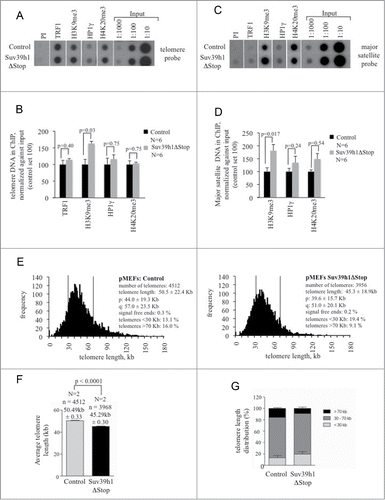
We conclude that Suv39h1 overexpression drives telomere shortening in pMEFs obtained from transgenic mouse embryos, underlining the role of Suv39h HMTases as negative regulator of telomere length.
Suv39h1 transgenic mice are resistant to skin carcinogenesis
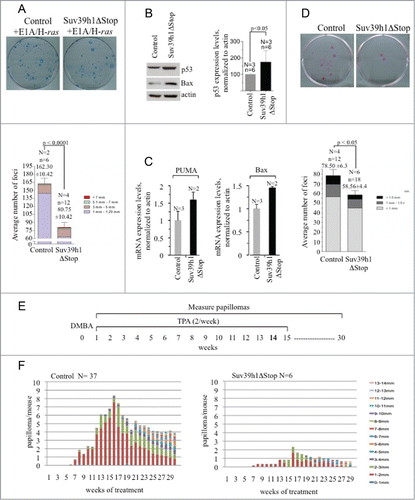
Recent data indicate a role for Suv39h1 in the control of proliferation and maintenance of genomic stability. Suv39hDN mice develop B cell lymphomas at increased frequency and have a reduced capacity to activate a senescence program in response to oncogenic stress.Citation4,14 Moreover, Suv39h1 overexpression induces mitotic defects in human cancer cells and increases the immortalization of primary erythroblasts.Citation22,23 We next set out to test a possible role of Suv39h1 in the control of cell proliferation of primary cells and oncogene-induced transformation. To test this idea we transduced primary fibroblasts derived from control and Suv39h1ΔStop embryos with retroviral expression vectors for E1A and H-ras and performed colony forming assays. Importantly, we found that Suv39h1 transgene expression caused a 50% reduction of colony forming potential of E1A/H-ras transduced pMEFs (). This points toward a role of Suv39h1 in antagonizing immortalization driven by the Ras and E1A pathways. To identify a potential mechanism that contributes to the increased resistance to oncogenic transformation of Suv39h1ΔStop pMEFs we examined the p53 tumorsuppressor pathway in the context of Suv39h1 overexpression. We found that Suv39h1ΔStop pMEFs display a significant upregulation of p53 protein levels that is paralleled by upregulation of p53 target genes Bax and Puma () This suggests that p53 is transcriptional active and that increased tumor-surveillance by the p53 pathway can mediate increased resistance to oncogenic transformation in Suv39h1ΔStop pMEFs. This model is in line with the central role of Suv39h1 and p53 in mediating Ras-oncogene induced senescence.Citation14,24 In line with the data from Suv39h1ΔStop pMEFs we found that primary keratinocytes prepared from the back skin of Suv39h1ΔStop animals showed a significant, 25% reduction in colony forming potential compared to control cells (). We consequently wished to examine a potential tumor suppressive role of Suv39h1 in vivo. We decided to use a chemical skin carcinogenesis protocol that involves the acquisition of oncogenic mutations. The DMBA/TPA protocol is a classic tool to study chemical skin carcinogenesis that is driven by the accumulation of oncogenic H-ras mutations.Citation25 The back skin of experimental mice was treated with the DNA damaging agent DMBA, followed by a repeated treatment with the mitogen TPA for 29 weeks to promote the formation of neoplastic lesions (). As expected, control mice efficiently develop papillomas during the course of the experiment (). Importantly, Suv39h1ΔStop mice exhibit a dramatically reduced frequency of papilloma formation, indicating that ectopic Suv39h1 expression confers resistance to DMBA/TPA induced carcinogenesis (). This indicates that increased Suv39h1 expression levels confers increased tumor protection in transgenic mice. Together our data suggest that ectopic Suv39h1 expression increases p53 protein expression to mediate improved tumorsuppression under conditions involving oncogenic stress.
Figure 4. Suv39h1 overexpression limits immortalization and confers resistance to skin carcinogenesis. (A) Colony forming assay of control and Suv39h1ΔStop transgenic pMEFs after retroviral transduction of H-ras and E1A. Suv39h1 overexpression results in reduced immortalization as indicated by limited colony formation potential. Color code indicates diameter of formed colonies (mm). (B) Left panel: western blotting indicates elevated p53 protein levels in Suv39h1ΔStop pMEFs. Actin was used as a loading control. Right panel: quantification of western blotting results. (C) Quantitative real-time PCR of p53 target genes Bax and Puma. Actin was used to normalize expression levels. (D) Colony forming assays using primary mouse keratinocytes prepared from the back skin of control or Suv39h1ΔStop newborn mice. Suv39h1ΔStop keratinocytes display reduced colony forming potential. Color code indicates diameter of formed colonies (mm). (E) Schematic representation of the DMBA/TPA skin carcinogenesis protocol. After an initial DMBA treatment, the back skin of experimental mice was treated twice per week with TPA for a total period of 15 weeks. Papilloma formation and size was monitored in weekly inspections. (F) Papilloma formation in control and Suv39h1ΔStop mice during DMBA/TPA skin carcinogenesis. Papilloma formation is efficiently impaired in Suv39h1ΔStop mice. Color code indicates diameter of formed papillomas (mm). N, number of mice or independently prepared cell cultures analyzed; n, number of total numbers of experiments or replicas analyzed; standard deviations are indicated; statistical significance is indicated by p-values.
Discussion
The histone H3K9 specific histone methyltransferases Suv39h1 and Suv39h2 are essential for the formation of constitutive heterochromatin at pericentric and telomeric repeats but are also critically involved in the silencing of gene expression.Citation3,12 In particular, Suv39h1 has an important role in mediating the irreversible silencing of cell cycle promoting E2F genes to promote senescence in primary cells but also Ras-oncogene mediated senescence in premalignant tumors.Citation13,14 Lack of Suv39h1 and Suv39h2 results in genomic instability and loss of telomeric heterochromatin leading to an uncontrolled telomere elongation.Citation4,6 Here, we use a transgenic mouse model system to investigate the effect of increased Suv39h1 levels in the context of telomere regulation and chemical oncogenesis. In contrast to a previously reported Suv39h1 transgenic mouse model (ref. Citation23) Suv39h1ΔStop mice did not display alterations in body-weight. We suggest that this discrepancy is due to the variable efficiency of Cre-mediated deletion of the transcriptional Stop cassette in our mouse model system (Fig. S1A, B).
Consistent with the telomere elongation phenotype in Suv39h DN pMEFs and mESCs (ref. Citation7), we observed that increased telomeric and global H3K9me3 levels drive telomere shortening in pMEFs obtained from Suv39h1 transgenic mice. Interestingly, increased H3K9me3 levels were not paralleled by increased levels of nuclear HP1γ and HP1β or enrichment of HP1γ at telomeres. We speculate that the moderate increase of H3K9me3 in Suv39h1 transgenic mice is not sufficient to increase the recruitment of HP1γ to constitutive heterochromatin. Our results demonstrate that H3K9me3 abundance is an efficient regulator of telomere length in embryo-derived cells. However, we were not able to link an increased H3K9me3 with telomere shortening in adult tail skin sections. We speculate that cell proliferation coupled with various cell differentiation pathways that control skin formation might also impact on the expression of telomere regulators and thereby over-ride telomere shortening caused by Suv39h1 overexpression in embryo-derived fibroblasts. Suv39h1 overexpression has been reported to de-localize HP1β and to impair proliferation of cancer cells.Citation22 In addition, a previous study show that Suv39h1 overexpression can increase p53 expression.Citation23 Here, we found that ectopic Suv39h1 confers increased resistance to oncogenic stress conditions. In particular we show that pMEFs obtained from Suv39h1ΔStop embryos display significantly increased p53 protein levels. The fact that p53 target gene Bax and Puma are upregualted in Suv39h1ΔStop pMEFs suggest that p53 is transcriptional active, indicative for increased tumor surveillance. Elevated basal p53 tumorsuppressor activity in Suv39h1ΔStop pMEFs offers an attractive model to explain the increased resistance of Suv39h1ΔStop pMEFs to oncogenic transformations mediated via E1A/H-ras. This is in line with a study that demonstrates that loss of Suv39h1 or p53 impairs oncogene induced senescence, a key tumorsuppressive mechanism in premalignant lesions.Citation14,24 In line with increased resistance to E1A/H-ras induced transformation we found that Suv39h1ΔStop transgenic animals display a remarkable resistance to DMBA/TPA mediated skin carcinogenesis, a process that has been demonstrated to involve the acquisition of oncogenic H-ras mutations.Citation25 We propose that Suv39h1 dependent increase of basal expression levels of p53 protein as observed in Suv39h1ΔStop pMEFs provides a model to explain increased resistance to chemical carcinogenesis of Suv39h1 overexpressing animals. This model is supported by the reported loss of oncogene induced senescence in the absence of p53 and Suv39h1, but also by the fact that a modest increase in p53 protein levels in Super-p53 mice is sufficient to mediate increased resistance to 3MC chemical carcinogenesis.Citation14,24,26
Altogether, our transgenic mouse model underlines the role of the Suv39h1 HMTase as negative regulator of telomere length and identifies Suv39h1 as an interesting target modulate p53 tumorsuppressor activity and improving resistance to oncogene induced transformation.
Material and Methods
Generation of transgenic animals and cell culture
Human non-small cell lung carcinoma H1299 cells were obtained from ATCC and have not been cultured for longer than 6 months. pMEFs and H1299 cells were cultured in DMEM high glucose (Life Technologies), 10% heat-inactivated FBS (Gibco). H1299 cells were transiently transfected with pcDNA and pcDNA-HA-Suv39h1 using Lipofectamine 2000 (Life Technologies) according to the manufacturer's suggestions.
To prepare a vector for transgene injection, the coding region of the murine Suv39h1 ORF was amplified by PCR and cloned downstream of a CAGGS promoter, a transcriptional Stop cassette, flanked by loxP sites and upstream of a transcriptional termination cassette (Figure S1A).Citation27 Constructs were verified by DNA sequencing. The CMV-loxP-Stop-loxP-Suv39h1-pA cassette was excised from the vector using PvuII/SalI, purified and injected into the pronuclei of fertilized oocytes obtained from C57BL/6 × CBA F1 mice at the CNIO transgenics unit and founder mice were established. Founder animals were backcrossed for 3 generations to a pure C57BL/6 background to obtain the Suv39h1 transgenic line studied here. Germline transmission of the transgene was verified by Southern blotting using a Suv39h1 cDNA probe that detects the Suv39h1 transgene but also endogenous Suv39h1 locus (). Suv39h1Stop transgenic animals were crossed with homozygous CMV-Cre recombinase transgenic animals (ref. Citation21) to give rise to Suv39h1ΔStop-CMV-Cre mice (in this study: Suv39h1ΔStop) and heterozygous CMV-Cre (in this study “control” mice) offsprings. A PCR strategy was developed that allows the detection of the transgene before and after Cre-recombinase mediated deletion of the loxP-Stop-loxP cassette (primer sequences available upon request). In pMEFs deletion of the loxP-Stop-loxP cassette was verified by Southern blotting and PCR (Figure S1A). Transgene expression was confirmed by quantitative real-time PCR. Mice were generated and maintained at the CNIO under specific-pathogen-free conditions in accordance with the recommendations of the Federation of European Laboratory Animal Science Associations. All efforts were made to minimize suffering.
Skin carcinogenesis
Age-matched (8- to 12-wk-old) mice of each genotype (37 control; 6 Suv39h1ΔStop) were shaved and treated with a single dose of DMBA (7,12-Dimethylbenz[α]anthracene; 0.1 μg/μL in acetone; Sigma). Mice were subsequently treated twice weekly with TPA (12-O-tetradecanoylphorbol-13-acetate; 12.5 μg in 200 μL of acetone each treatment; Sigma) for 15 wk. Papilloma formation and growth was followed in weekly examinations during additional 15 weeks () Citation25,28.
Telomere length analyses on tail skin sections and pMEF metaphase spreads
Paraffin-embedded skin sections were de-paraffinated and hybridized with a PNA-telomeric probe, and telomere fluorescence was determined by confocal microscopy and telomapping. Telomapping is based on confocal telomere quantitative fluorescence in situ hybridization that allows to detect gradients of telomere length within tissues or cell populations.Citation29 This technique can be adopted to immunostainings. For telomere Q-FISH on metaphase spreads exponentially growing primary mouse embryonic fibroblasts (pMEFs) were incubated with 0.1 μg/mL colcemid (GIBCO) for 5 h at 37°C and then fixed in methanol:acetic acid (3:1). Q-FISH was performed as described.Citation30 Images were captured in a linear acquisition mode using a COHU CCD camera on a Leica DMRB microscope.
Immunohistochemistry
Skin samples were fixed in 10% buffered formalin, embedded in paraffin wax and sectioned at 4 μm. After de-paraffination, sections were processed with 10 mM sodium citrate (pH 6.0) for 20 min at 95°C. After blocking in 5% goat serum cells were incubate with respective primary and secondary antibodies. Immnofluorescence stainings on pMEFs were carried out as previously described.Citation31 Following primary antibodies were used: a rabbit anti-H3K9me3 antibody (Upstate 07–442); a mouse monoclonal anti-HP1γ specific antibody (Upstate 05–690), or a rabbit polyclonal anti-HP1β specific antibody (abcam 10478). Primary antibodies were incubated for 1 hour at room temperature. Secondary Cy3-goat anti-rabbit antibody (1:400; Jackson Immuno Research Laboratories, Inc.) and a FITC-goat anti-mouse antibody (1:400; Jackson ImmunoResearch Laboratories, Inc.) for 60 min at room temperature; slides were mounted in Vectashield with 4′,6-diamino-2 phenylindole. Quantitative image analysis of immunostainings (4',6-diamidino-2-phenylindole) of pMEFs and mouse tail skin sections was performed by confocal microscopy (H3K9me3 and HP1γ mapping) using a TCS-SP2-A-OBS-UV Leica microscope with Metamorph software (version 6.3r6) platform or by classic quantitative immunofluorescence (Zeiss Axiovert 200 M microscope, equipped with a Zeiss AxioCam MRm digital camera and AxioVision Rel. 4.8 imaging software followed by ImageJ picture analysis), as previously described.Citation29,31,32
Isolation, culture and colony forming assays using primary mouse keratinocytes
Skin keratinocyes were isolated from the back skin of newborn mice as described. For colony forming assays 10Citation4 or 5 × 105 keratinocytes were seeded on a mitomycin C treated J2-3T3 feeder layer and cultivated in CnT-O2 medium with supplement and antibiotics (CELLnTEC, Advanced Cell Systems AG) for 11 d Subsequently cells were fixed with 18,5% formaldehyde and stained in with 1% Rhodamine B. Colony size and numbers were assessed.
Western blots
Whole cell and nuclear extract was prepared as previously described.Citation31 Extracts were separated on SDS–polyacrylamide gels. After transfer, the membranes were incubated with an anti-H3K9me3 polyclonal antibody (Upstate, 07–442), anti-Suv39h1 monoclonal antibody “…(Abcam ab1791), a polyclonal rabbit anti-Bax antibody (Cell signaling 2772), mouse monoclonal anti-p53 antibody (abcam [PAb 240] (ab26)), goat polyclonal anti Lamin A/C antibody (Santa Cruz (N18): sc-6215) or polyclonal anti-global histone H4 antibodies (Abcam, ab10158). Antibody binding was detected after incubation with a secondary antibody coupled to horseradish peroxidase.
Chromatin Immunoprecipitation (ChIP)
Chromatin immunoprecipitation of telomeric and subtelomeric chromatin was performed as previously described.Citation7 The following antibodies were used for immunoprecipitation: polyclonal rabbit anti-mouse TRF1 (produced in house), a monoclonal anti-HP1γ antibody (Upstate, 05–690), a polyclonal rabbit anti-H3K9me3 antibody (Upstate 07–442) and a polyclonal rabbit anti-H4K20me3 antibody (Upstate 07–749). DNA was purified from immunoprecipitated chromatin, transferred to a nitrocellulose membrane and probed with a radio labeled telomeric or major satellite probe. Signal intensities were quantified using ImageJ and normalized to the ChiP Input.
Quantitative real-time PCR
Total RNA was prepared using QIAzol Lysis Reagent (Qiagen) and reverse transcribed with QuantiTect Reverse Transcription Kit (Qiagen) according to the manufacturer's instructions. Quantitative PCR was performed using the QuantiFast SYBR Green PCR Kit (Qiagen) and analyzed with the CFX96 Real-Time PCR Detection System (BIO-RAD). mRNA levels were normalized to actin. PCR primers used for quantitative Real-Time PCR (ref.Citation33): Bax FW: 5′ GGAGCAGCTTGGGAGCG 3′, Bax REV: 5′ AAAAGGCCCCTGTCTTCA 3′; Puma FW: 5′ ACCTCAACGCGCAGTAC 3′, Puma REV: 5′ TGAGG GTCGGTGTCGAT 3′; β-actin FW: 5′ CACACCCGCCACCAGTTC 3′, β-actin REV: 5′ CCCATTCCCACCATCACACC 3′.
Bisulfite sequencing
Bisulfite sequencing of subtelomeric regions in Chr1q and Chr2q and B1SINE repeats was carried out as described previously.Citation7
Colony formation assays using pMEFs
Low passage of pMEFs of control and Suv39h1ΔStop transgenic mice were infected with a recombinant, replication deficient retroviral vector expressing E1A and H-ras as previously described.Citation34 Puromycin selection for 3 d was used to eliminate non-transduced cells. After 19 d colonies were stained with Giemsa and colony number and size was determined. pMEFs were cultured in DMEM, high glucose; 10% fetal bovine serum; L-glutamine and Pen/Strep (all Invitrogen).
Statistical analysis
The Wilcoxon–Mann–Whitney rank sum test was used for statistical comparisons of the mean telomere length in MEFs. For all other experiments a Student's t-test was used to calculate statistical significance.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Author contributions
EP, FJ, RD and VB carried out experiments and analyzed obtained data; SS and RB analyzed obtained data; SS, RB and MAB, conception and design of work. SS, and MAB wrote the manuscript; SS and MAB, study supervision.
KCCY_A_1021517_supplemental_files.zip
Download Zip (410.7 KB)Acknowledgments
We thank R. Serrano for mouse care and genotyping and Orlando Dominguez for bisulfite sequencing.
Funding
This work was supported by the European Union (TELOSENS FIGH-CT-2002-00217, INTACT LSHC-CT-2003-506803) to MAB, the Josef Steiner Cancer Research Award, 2003 to MAB and by the AIRC Start up grant (10299) to SS. EP, VB and RD are participants of the SDBM PhD school of Molecular Medicine at the University of Trieste, Italy.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Additional information
Notes on contributors
Eleonora Petti
EP, FJ and VB carried out experiments and analyzed obtained data; SS and RB analyzed obtained data; SS, RB and MAB, conception and design of work. SS, and MAB wrote the manuscript; SS and MAB, study supervision.
Fabian Jordi
EP, FJ and VB carried out experiments and analyzed obtained data; SS and RB analyzed obtained data; SS, RB and MAB, conception and design of work. SS, and MAB wrote the manuscript; SS and MAB, study supervision.
Valentina Buemi
EP, FJ and VB carried out experiments and analyzed obtained data; SS and RB analyzed obtained data; SS, RB and MAB, conception and design of work. SS, and MAB wrote the manuscript; SS and MAB, study supervision.
Roberta Benetti
EP, FJ and VB carried out experiments and analyzed obtained data; SS and RB analyzed obtained data; SS, RB and MAB, conception and design of work. SS, and MAB wrote the manuscript; SS and MAB, study supervision.
Maria A Blasco
EP, FJ and VB carried out experiments and analyzed obtained data; SS and RB analyzed obtained data; SS, RB and MAB, conception and design of work. SS, and MAB wrote the manuscript; SS and MAB, study supervision.
Stefan Schoeftner
EP, FJ and VB carried out experiments and analyzed obtained data; SS and RB analyzed obtained data; SS, RB and MAB, conception and design of work. SS, and MAB wrote the manuscript; SS and MAB, study supervision.
References
- Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 2000; 406:593-9; PMID:10949293; http://dx.doi.org/10.1038/35020506
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 2001; 410:116-20; PMID:11242053; http://dx.doi.org/10.1038/35065132
- Fodor BD, Shukeir N, Reuter G, Jenuwein T. Mammalian Su(var) genes in chromatin control. Annu Rev Cell Dev Biol 2010; 26:471-501; PMID:19575672; http://dx.doi.org/10.1146/annurev.cellbio.042308.113225
- Peters AH, O'Carroll D, Scherthan H, Mechtler K, Sauer S, Schöfer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 2001; 107:323-37; PMID:11701123; http://dx.doi.org/10.1016/S0092-8674(01)00542-6
- Bulut-Karslioglu A, De La Rosa-Velázquez IA, Ramirez F, Barenboim M, Onishi-Seebacher M, Arand J, Galán C, Winter GE, Engist B, Gerle B, et al. Suv39h-Dependent H3K9me3 Marks Intact Retrotransposons and Silences LINE Elements in Mouse Embryonic Stem Cells. Mol Cell 2014; 55:277-90; PMID:24981170; http://dx.doi.org/10.1016/j.molcel.2014.05.029
- Blasco MA. Telomeres and human disease: ageing, cancer and beyond. Nat Rev Genet 2005; 6:611-22; PMID:16136653; http://dx.doi.org/10.1038/nrg1656
- García-Cao M, O'Sullivan R, Peters AHFM, Jenuwein T, Blasco MA. Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat Genet 2004; 36:94-9; PMID:14702045; http://dx.doi.org/10.1038/ng1278
- Benetti R, García-Cao M, Blasco MA. Telomere length regulates the epigenetic status of mammalian telomeres and subtelomeres. Nat Genet 2007; 39:243-50; PMID:17237781; http://dx.doi.org/10.1038/ng1952
- Blasco MA. The epigenetic regulation of mammalian telomeres. Nat Rev Genet 2007; 8:299-309; PMID:17363977; http://dx.doi.org/10.1038/nrg2047
- Chan SRWL, Blackburn EH. Telomeres and telomerase. Philos Trans R Soc Lond B Biol Sci 2004; 359:109-21; PMID:15065663; http://dx.doi.org/10.1098/rstb.2003.1370
- d'Adda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T, Saretzki G, Carter NP, Jackson SP. A DNA damage checkpoint response in telomere-initiated senescence. Nature 2003; 426:194-8; PMID:14608368; http://dx.doi.org/10.1038/nature02118
- Nielsen SJ, Schneider R, Bauer UM, Bannister AJ, Morrison A, O'Carroll D, Firestein R, Cleary M, Jenuwein T, Herrera RE, et al. Rb targets histone H3 methylation and HP1 to promoters. Nature 2001; 412:561-5; PMID:11484059; http://dx.doi.org/10.1038/35087620
- Narita M, Nũnez S, Heard E, Narita M, Lin AW, Hearn SA, Spector DL, Hannon GJ, Lowe SW. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 2003; 113:703-16; PMID:12809602; http://dx.doi.org/10.1016/S0092-8674(03)00401-X
- Braig M, Lee S, Loddenkemper C, Rudolph C, Peters AHFM, Schlegelberger B, Stein H, Dörken B, Jenuwein T, Schmitt CA. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature 2005; 436:660-5; PMID:16079837; http://dx.doi.org/10.1038/nature03841
- Reimann M, Lee S, Loddenkemper C, Dörr JR, Tabor V, Aichele P, Stein H, Dörken B, Jenuwein T, Schmitt CA. Tumor stroma-derived TGF-beta limits myc-driven lymphomagenesis via Suv39h1-dependent senescence. Cancer Cell 2010; 17:262-72; PMID:20227040; http://dx.doi.org/10.1016/j.ccr.2009.12.043
- Lee AC, Fenster BE, Ito H, Takeda K, Bae NS, Hirai T, Yu ZX, Ferrans VJ, Howard BH, Finkel T. Ras proteins induce senescence by altering the intracellular levels of reactive oxygen species. J Biol Chem 1999; 274:7936-40; PMID:10075689; http://dx.doi.org/10.1074/jbc.274.12.7936
- Collado M, Gil J, Efeyan A, Guerra C, Schuhmacher AJ, Barradas M, Benguría A, Zaballos A, Flores JM, Barbacid M, et al. Tumour biology: senescence in premalignant tumours. Nature 2005; 436:642; PMID:16079833; http://dx.doi.org/10.1038/436642a
- Michaloglou C, Vredeveld LCW, Soengas MS, Denoyelle C, Kuilman T, van der Horst CMAM, Majoor DM, Shay JW, Mooi WJ, Peeper DS. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature 2005; 436:720-4; PMID:16079850; http://dx.doi.org/10.1038/nature03890
- Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, Schurra C, Garre’ M, Nuciforo PG, Bensimon A, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 2006; 444:638-42; PMID:17136094; http://dx.doi.org/10.1038/nature05327
- Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou L-VF, Kolettas E, Niforou K, Zoumpourlis VC, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 2006; 444:633-7; PMID:17136093; http://dx.doi.org/10.1038/nature05268
- Zinyk DL, Mercer EH, Harris E, Anderson DJ, Joyner AL. Fate mapping of the mouse midbrain-hindbrain constriction using a site-specific recombination system. Curr Biol 1998; 8:665-8; PMID:9635195; http://dx.doi.org/10.1016/S0960-9822(98)70255-6
- Melcher M, Schmid M, Aagaard L, Selenko P, Laible G, Jenuwein T. Structure-function analysis of SUV39H1 reveals a dominant role in heterochromatin organization, chromosome segregation, and mitotic progression. Mol Cell Biol 2000; 20:3728-41; PMID:10779362; http://dx.doi.org/10.1128/MCB.20.10.3728-3741.2000
- Czvitkovich S, Sauer S, Peters AH, Deiner E, Wolf A, Laible G, Opravil S, Beug H, Jenuwein T. Over-expression of the SUV39H1 histone methyltransferase induces altered proliferation and differentiation in transgenic mice. Mech Dev 2001; 107:141-53; PMID:11520670; http://dx.doi.org/10.1016/S0925-4773(01)00464-6
- Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 1997; 88:593-602; PMID:9054499; http://dx.doi.org/10.1016/S0092-8674(00)81902-9
- Balmain A, Ramsden M, Bowden GT, Smith J. Activation of the mouse cellular Harvey-ras gene in chemically induced benign skin papillomas. Nature 307:658-60; PMID:6694757; http://dx.doi.org/10.1038/307658a0
- García-Cao I, García-Cao M, Martín-Caballero J, Criado LM, Klatt P, Flores JM, Weill J-C, Blasco MA, Serrano M. “Super p53” mice exhibit enhanced DNA damage response, are tumor resistant and age normally. EMBO J 2002; 21:6225-35; PMID:12426394; http://dx.doi.org/10.1093/emboj/cdf595
- Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 1991; 108:193-9; PMID:1660837; http://dx.doi.org/10.1016/0378-1119(91)90434-D
- Muñoz P, Blanco R, Flores JM, Blasco MA. XPF nuclease-dependent telomere loss and increased DNA damage in mice overexpressing TRF2 result in premature aging and cancer. Nat Genet 2005; 37:1063-71; PMID:16142233; http://dx.doi.org/10.1038/ng1633
- Flores I, Canela A, Vera E, Tejera A, Cotsarelis G, Blasco MA. The longest telomeres: a general signature of adult stem cell compartments. Genes Dev 2008; 22:654-67; PMID:18283121; http://dx.doi.org/10.1101/gad.451008
- Samper E, Goytisolo FA, Slijepcevic P, van Buul PP, Blasco MA. Mammalian Ku86 protein prevents telomeric fusions independently of the length of TTAGGG repeats and the G-strand overhang. EMBO Rep 2000; 1:244-52; PMID:11256607; http://dx.doi.org/10.1093/embo-reports/kvd051
- Dinami R, Ercolani C, Petti E, Piazza S, Ciani Y, Sestito R, Sacconi A, Biagioni F, le Sage C, Agami R, et al. miR-155 drives telomere fragility in human breast cancer by targeting TRF1. Cancer Res 2014;; PMID:24876105
- Blanco R, Muñoz P, Flores JM, Klatt P, Blasco MA. Telomerase abrogation dramatically accelerates TRF2-induced epithelial carcinogenesis. Genes Dev 2007; 21:206-20; PMID:17234886; http://dx.doi.org/10.1101/gad.406207
- Schuler M, Maurer U, Goldstein JC, Breitenbücher F, Hoffarth S, Waterhouse NJ, Green DR. p53 triggers apoptosis in oncogene-expressing fibroblasts by the induction of Noxa and mitochondrial Bax translocation. Cell Death Differ 2003; 10:451-60; PMID:12719722; http://dx.doi.org/10.1038/sj.cdd.4401180
- Efeyan A, Murga M, Martinez-Pastor B, Ortega-Molina A, Soria R, Collado M, Fernandez-Capetillo O, Serrano M. Limited role of murine ATM in oncogene-induced senescence and p53-dependent tumor suppression. PLoS One 2009; 4:e5475; PMID:19421407; http://dx.doi.org/10.1371/journal.pone.0005475
