Abstract
Heterogeneity demonstrates that stem cells are constituted by several sub-clones in various differentiation states. The heterogeneous state is maintained by cross-talk among sub-clones, thereby ensuring stem cell adaption. In this study, we investigated the roles of heterogeneity on genetic stability. Three sub-clones (DF2, DF8 and DF18) were isolated from heterogeneous dental stem cells (DSCs), and were proved to be chromosome instability (CIN) after long term expansion. Cell apoptosis were not detected in sub-clones, which exhibited strong tumorigenesis tendency, coupled with weak expression of p53 and aberrant ultra-structure. However, 3 sub-clones did not overexpress tumor related markers or induce tumorigenesis in vivo. The mixed-culture study suggested that 3-clone-mixed culturing cells (DF1) presented apparent decrease in the ratio of aneuploidy. The screening experiment further proved that 3 sub-clones functioned separately in this modification procedure but only mixed culturing all 3 sub-clones, simulated heterogeneous microenvironment, could achieve complete modification. Additionally, osteogenesis capability of 3 sub-clones was partially influenced by CIN while DSCs still kept stronger osteogenesis than sub-clones. These results suggested aberrant sub-clones isolated from heterogeneous DSCs were not tumorigenesis and could modify CIN by cross-talk among themselves, indicating that the heterogeneity played a key role in maintaining genetic stability and differentiation capability in dental stem cells.
Abbreviations
| CIN | = | Chromosome instability |
| DSCs | = | Dental stem cells |
| DFSCs | = | Dental follicle stem cells |
| TEM | = | Transmission Electron Microscope |
| qRT-PCR | = | Quantitative Real time-PCR |
| FCM | = | Flow cytometry |
| TC | = | Tumor Cell |
Introduction
Heterogeneity is the distinct functional and epigenetic states in differentiation.Citation1,2 With most stem cells being heterogeneous,Citation3,4 it might have evolved as a mechanism that enables stem cells to respond to differentiation-inducing signals while retaining their self-renewal potential.Citation5 So heterogeneity could be regarded as the basis for the homeostasis of stem cell niche, just as the role stem cells play in maintaining tissue homeostasis.Citation6,7 The stem cell niche would be broken when sub-clones were separated from heterogeneous cells, thus activating sub-clones from a quiescent state. Homeostasis of stem cell niche is essential for keeping cells away from tumor formation.Citation8 Tumor stem cells and stem cells shared many featuresCitation9 and tumor growth could be caused by transformation of stem cells.Citation10 Stable genome and controllable biological behavior distinguish stem cells from tumor stem cells.
In cell-based therapy, freeze-thaw cycle or long-term expansion of seeding cells may cause unbalanced microenvironment resulting in cell apoptosis, loss of differentiation potential, chromosome instability or even tumor formation.Citation11 Chromosomal aneuploidies are widely recognized as genetic disorders in humans that often lead to spontaneous abortion.Citation12 After massive amplification, CIN or aneuploidy may occur in cell lines isolated from embryonic stem cells (ESCs) or mesenchymal stem cells (MSCs),Citation13,14 which often indicate the tendency of tumor transition.Citation15–17 However, in order to maintain the true properties of stem cells and avoid adverse effects post-transplantation, the preservation of normal chromosomes is necessary.Citation18 Qualified seeding cells should meet at least 2 requirements: (i) the occurrence of a normal genetic karyotype and (ii) the maintenance of chromosomal integrity during long term-culturing and after cryopreservation procedures.Citation19
Previous studies showed sub-clones can be isolated from heterogeneous MSCs and were still pluripotent although with variable differential potentials in vitro.Citation20-22 Both heterogeneous MSCs and most of its sub-clones were found to be promising seeding cells for cell-based therapy. The efficiency and prognosis after in vivo transplantation of heterogeneous stem cells cannot be guaranteed due to the unstable cell population,Citation23,24 although the cell collection was convenient and the regenerated tissue was various.Citation20,25,26 However, controversy still exists on whether the heterogeneous stem cells or one of their sub-clones should be chosen for future clinical applications, because it is still unclear whether they meet above requirements.
DSCs are heterogeneous MSCs.Citation20,27,28 Dental Follicle Stem Cells (DFSCs) were also proved to retain embryonic characteristics.Citation29-31 Heterogeneous DSCs have been utilized in regenerative therapies,Citation32,33 however whether they are safe for clinical application is also unclear. In this study, 3 sub-clones isolated from heterogeneous DFSCs were CIN, but not tumorigenesis. Tumor suppressors, such as p53, p21or E2F1, were the main force to maintain genetic stability and keep cells stable under pressure. In tumor cells, p53 often mutant or partially lost function. However, stem cells could tolerate the loss of p53 by up-regulating p21 and E2F1 when p53 was inhibited.Citation34,35 This study showed that unusual MAD1 and MAD2 expression caused CINCitation36-38 and p21 and E2F1 might keep sub-clones away from tumor formation under p53 loss condition. Moreover, aneuploidy ratio of DF1 which was constituted by 3 sub-clones was obviously decreased but 2-clones-mixed culturing cells were not. In mixing culture cells, expression of MAD1 and MAD2 were also restored. Taking together, our results suggested that heterogeneity played a positive role on maintaining genetic stability of stem cells and further kept them away from transformation. Furthermore, we also found sub-clones lost parts of differentiation capability. Finally, heterogeneous DFSCs were proved to be better seeding cell for cell-based therapy compared with its sub-clones.
Results
Three sub-clones, named DF2, DF8 and DF18 respectively, were isolated from heterogeneous DFSCs. The shapes of them were polygon, spindle and satellite, respectively, while the shapes of DFSCs were varied consisting of all the 3 types (). Proliferation capability of 3 sub-clones was significantly different. To be specific, DF18 showed the highest proliferation activity while DF2 stayed at a relatively quiescent level on the contrary; DF8 and DFSCs showed similar proliferation level (). Additionally, all 3 sub-clones showed low apoptosis ratio (). However, DNA contents analysis () showed aneuploidy was observed in 3 sub-clones. The aneuploidy ratios in DF2, DF8 and DF18 were 33.46%, 53.60% and 13.28% respectively.
Figure 1. (A) The isolation procedure for single sub-clone. (B) Three sub-clones (DF2, DF8 and DF18) were isolated from heterogeneous DFSCs (Scale bar: 100 μm). (C-E) hclust, correlation and heat map analysis of Gene Expression Array for sub-clones and DFSCs. (s1:DF2, s2:DF12, s3:DF8, s4:DF18, s5: DFSCs).
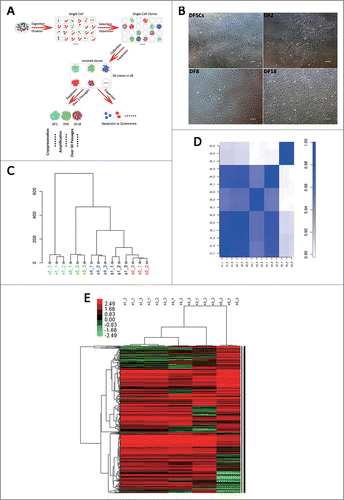
Figure 2. (A) Cell proliferation assay. CCK-8 assay were used to evaluate the proliferation capability of 3 sub-clones and DFSCs. Error bar indicated SEM (n = 3) (B) Cell apoptosis evaluation of 3 sub-clones and DFSCs, using Annexin V-FITC Apoptosis Detection Kit. The apoptosis ratio of DFSCs, DF2, DF8 and DF18 were 2.9%, 0.6%, 4.2% and 4.8% respectively. (C) DNA contents analysis and aneuploidy ratio of 3 sub-clones and DFSCs. Percentage of cells in the S+G2M phases of DFSCs, DF2, DF8 and DF18 were 44.09%, 15.98%, 28.68% and 40.68% respectively. (D) Chromosome number of 3 sub-clones and DFSCs were counted by Giemsa staining assay. (E) Ultrastructures of 3 sub-clones and DFSCs were observed through transmission electron microscope (TEM) (Red quadrangle presented the electronic dense granule). (F) p53 mRNA and protein levels were measured by qRT-PCR and Western blot analysis in DFSCs and sub-clones. (G) mRNA levels of puma were measured by qRT-PCR. (H) Tumor related gene: Tert and K-ras were evaluated by qRT-PCR. Error bar indicated SEM (n = 3). Statistical significance used in this figure: *P < 0.05, **P < 0.01 and ***P < 0.001; ns represented no statistically significant.
Then karyotype analysis was performed to observe the state of a single cell. The chromosome number of DF2, DF8 and DF18 was disorder and even structural aberration was observed in DF8 (). But specific chromosomes lost or gained cannot be identified because of the random alteration of chromosome number.
To evaluate the status of sub-clones, ultra-structures of DFSCs and 3 sub-clones were observed by Transmission Electron Microscope (TEM) (). The electronic dense granule which was the specific marker for DFSCs was observed in all sub-clones and DFSCs. The nucleus of DFSCs, DF8 and DF18 were light-colored euchromatin which indicated cells were at an early stage of development. Nucleus heteromorphy, high nuclear slurry ratios and rough endoplasmic reticulum (RER) expansion, which always occurred in tumor cells, were also observed in 3 sub-clones. DF18 contained rich cell organelles, especially abundant in secondary lysosomes, which indicated that the cells were undergoing active metabolism.
To further verify whether transformation of 3 sub-clones occurred, expression of the key tumor suppressor p53 and 2 oncogenes K-ras and Tert were detected. If aneuploidy occurred, p53 would induce aberrant cells apoptosis. However, the expression of p53 was inhibited in 3 sub-clones in this study () and related apoptosis gene: puma was not up-regulated compared with DFSCs (). At the meantime, K-ras and Tert did not overexpress either in 3 sub-clones compared with DFSCs ().
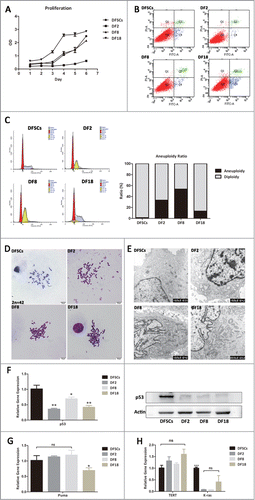
Three sub-clones did not overexpress oncogenes, however it was still unknown whether sub-clones with CIN could transform into tumor cells in the far more complicated in vivo environment. After 4 weeks of transplantation, xenograft tumor formation was found in positive group, but not in sub-clone groups and single-matrigel group (). HE staining showed the xenograft tumor formed in subcutaneous tissue in the positive group and even invaded the muscle layer (). On the contrary, in sub-clone groups, the subcutaneous layer was as normal as the negative group and there was no xenograft neoplasm formation (). Immunofluorescence labeling illustrated the tumor in positive group derived from the transplanted tumor cells (). Interestingly, DF2 was observed scattering in muscular layer however DF8 and DF18 cannot be traced in the subcutaneous tissue ().To sum up, the 3 sub-clones were proved not tumorigenic.
Figure 3. (A) Green fluorescence protein was transfected in 3 sub-clones and tumor cells by lentivirus transfection (Scale bar: 100 μm). (B) Macroscopic appearance of tumor growth 4 weeks after injection of 3 sub-clones and tumor cells. (C and D) HE and Immunofluorescence stain for injection tissues. (White arrows showed the GFP-labeled cells).
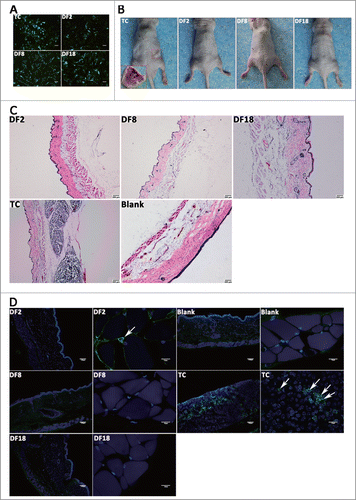
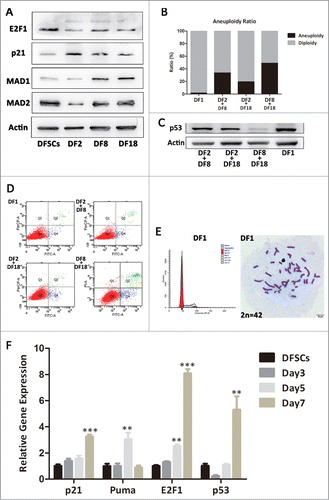
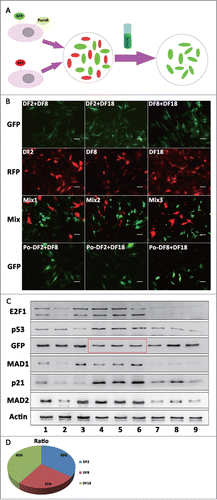
Figure 4. Three sub-clones were mixing cultured by every 2 sub-clones and DF1 was mixed culturing with 3 sub-clones. (A) Protein levels of p21, E2F1, MAD1 and MAD2 were measured by Western blot analysis in 3 sub-clones and DFSCs. (B) Aneuploidy ratio of sub-clones and mixed culturing cells, counted by DNA content analysis. (C) Protein levels of p53 were measured by Western blot analysis in mixed culturing cells. (D) Cell apoptosis evaluation of mixed cells, using Annexin V-FITC Apoptosis Detection Kit. (E) DNA contents and chromosome number analysis for DF1. (F) p53, p21, E2F1 and Puma RNA levels of DF1 were measured by qRT-PCR at the day3,5 and 7 after mixing. Statistical significance used in this figure: *P < 0.05, **P < 0.01 and ***P < 0.001; ns represented no statistically significant.
Since sub-clones with CIN showed no tumorigenicity, the causes of CIN were deserved exploring. Protein expression of E2F1 and p21 were selectively evaluated, trying to find out whether they functioned in CIN (). According to the Western blot results, 3 sub-clones all expressed p21 and E2F1. Sub-clones expressed higher p21 than DFSCs. Specifically, DF2 expressed a higher level of p21 than DF8 and DF18. Expression of p21 were similar in DF8 and DF18. Unlike p21, DF2 expressed the lowest level of E2F1 while DF8 and DF18 were just slightly lower than DFSCs. Protein levels of MAD1 and MAD2, as the key regulators of the spindle checkpoint, were detected by Western blot. DF8 and DF18 expressed higher level of MAD1 than DFSCs; DF2 expressed lower MAD2 than DFSCs (). Hence, disturbing expression of MAD1 and MAD2 may be one of the reasons for CIN in this study.
Cells were diploid in heterogeneous DFSCs, but CIN occurred when sub-clones were isolated from DFSCs and passaged. Therefore we assumed that mixed culturing sub-clones, simulating heterogeneous microenvironment, should help aberrant cells flee from CIN. To verify this hypothesis, 3 sub-clones were mixed cultured in pairs or in triplet (DF1) for 7 d Aneuploidy ratio of DF1 nearly dropped to normal level, and 2-mixed culturing groups also reduced slightly (). Moreover, p53 expressions in DF1, DF2+DF8 group and DF2+DF18 group recovered as well (). Interestingly, mixed culturing groups by no means showed large amount of apoptosis ().
DNA contents analysis and chromosome number count suggested DF1 was completely modified (). Puma, p53, p21 and E2F1 mRNA levels of DF1 at day3, 5 and 7 suggested the time line of recovering. The mRNA expression of apoptosis gene puma increased at day5 then fell back to normal at day7 (). Therefore DF1did not show high apoptosis ratio at day7 (). Meanwhile, modification related gene such as p53, p21 and E2F1 presented up-regulation over time ().
According to the above results, mixed culturing aberrant sub-clones could modify their CIN. To further explore whether the ratio of aneuploidy changing or the cross reaction among sub-clones functioned in the modification procedure, a screening experiment was designed (). The Western blot results () demonstrated that protein expression level of p53 in DF8+DF18 group was re-activated after addition of DF2 and the re-activated p53 can keep active even when DF2 were screened out. Expression of p21 in DF8+DF18 group was increased after addition of DF2 while other markers were barely changed; However after DF2 were screened out from the reconstituted DF1, expressions of all the proteins decreased. Expressions of E2F1, MAD1, MAD2 and p21 significantly increased after DF8 being added into DF2+DF18 group; When DF8 were screened out from DF1, all of the proteins except p21 decreased back to lower levels. When DF18 was added into DF2+DF8 group, only p21 expression raised and there's no obvious change for other markers; but p21 and MAD2 decreased slightly while MAD1 and E2F1 reduced to a relatively lower level after the screening out of DF18. Alteration of MAD1 and MAD2 showed that cross reaction among sub-clones was the fundamental cause of modification instead of ratio changes. Taken together, only DF1 expressed completely normal levels of cell cycle related proteins and expression of p21 was closely related with the existence of DF2.
Figure 5. (A) The illustration on the Screening flow. (B) Fluorescence labeling on mixed culturing cells and 3 sub-clones. Green fluorescence protein and PuroR were transfected on 2 sub-clones mixed culturing cells by lentivirus transfection, and red fluorescence protein were transfected on 3 sub-clones by lentivirus transfection. Post-mixed (Po-mix) cells were 2-clone-mixed culturing cells experienced screening. (Scale bar: 100 μm) (C) Protein levels of E2F1, GFP, p21, p53, MAD1 and MAD2 were measured by Western blot analysis. No.1, 2 and 3 cells were DF2 +DF8, DF2+DF18, DF8+DF18 which were transfected with GFP separately; No.4, 5 and 6 cells were Mix2, Mix2 and Mix3 in panel B; No.7, 8 and 9 cells were No.4, 5 and 6 cells screened by PuroR. (D) The pie chart showed the ratio of 3 sub-clones in mixing cells according to the intensity of GFP in No. 4, 5 and 6 cells (red frame in panel C).
To investigate the effects of CIN on the differentiation of stem cells, the embryonic and osteogenic characteristics of stem cells were detected using osteoblast (OB) as control group. In this study, DFSCs and 3 sub-clones expressed strongly positive pluripotent markers, like CD29, CD146 and CD90 (). However, only DFSCs and DF2 weakly expressed SSEA1 which was also an embryonic marker (). And only DFSCs positively expressed SOX2, OCT4 and Nanog (). Three sub-clones lost embryonic characteristics after long-term culture.
Figure 6. (A and B) Embryonic properties of 3 sub-clones were evaluated by FCM and western blot. (C) Calcium nodules were visualized using alizarin red after osteogenesis induction of DFSCs, 3 sub-clones and OB. (Scale bar: 100 μm) (D) Osteogenesis related protein levels were measured by Western blot analysis in DFSCs, 3 sub-clones and OB.
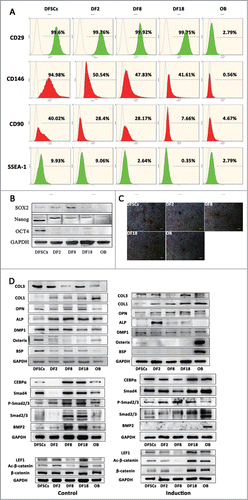
All of 3 sub-clones expressed high level of osteogenic markers () and could be induced mineralization (), indicating that their differentiation capability did not lose. DFSCs expressed the highest osteogenesis related markers except for Col-I; it also expressed high levels of BSP and osterix, which were even higher than OB. Three sub-clones had the similar level of osteogenesis capability and also were positive for most of osteogenesis markers except for osterix which was positively expressed in DFSCs. In summary, the osteogenesis capability was maintained in aberrant sub-clones, though changed to some extent.
To further evaluate specific changes of osteogenesis induced by CIN, WNT and BMP signaling pathway related key proteins were detected (). Before induction, 3 sub-clones expressed similar level of β-catenin as DFSCs and OB while active-β-catenin and LEF1 expressed lower in DF8; and other than DF2, both DF8 and DF18 expressed similar levels of BMP signaling pathway proteins. After induction, WNT signaling pathway proteins of DF8 significantly decreased while those of OB significantly increased; and all of the BMP signaling pathway proteins were upregulated except for BMP2 which only highly expressed in OB. These findings implied the osteogenesis related signaling pathways were indeed changed by the CIN, but the changes might be compensated by the unaffected signaling pathways.
Discussion
In this study, applicable sub-populations were isolated from heterogeneous DFSCs in order to study the heterogeneity of dental stem cells. Although various methods, including the surface marker screening,Citation39-41 single clone isolationCitation19,20,42,43 and any other methods,Citation44,45 could be adopted to isolate different sub-populations, there were no standard methods for isolation to be proposed. Through clones' isolation, single cells from heterogeneous cells were isolated, ensuring the purity of sub-populations. The purity of sub-populations was necessary for heterogeneity analysis avoiding cross-reaction with other populations.
After volumes of amplification, 3 sub-clones isolated from DFSCs were found with disordered chromosome numbers (), aberrant ultrastructure () and weak expression of guard gene p53 (). High expression of MDM2 could result in the inactivity of p53 thus leading to aneuploidy.Citation46,47 However, MDM2 did not overexpress in the present study (Data not shown). Numerical aneuploidy, as a symptom of the CIN, was considered to be a predominant hallmark of cancer, because most of the genetic heterogeneity in tumors was due to CIN.Citation48 Moreover, p53 not only was the tumor suppressorCitation49 but also influenced cell cycle and prevented aneuploidy.Citation50 Weak expression of p53 could cause CIN of sub-clones and even transformation. However, upregulation of p21 was found in all sub-clones. In adult stem cells, p21 not only acted as a target gene for p53 to prevent the activation of p53, induce apoptosis and lead to cell cycle entry symmetric self-renewing divisions but also activated DNA repair, limiting DNA damage accumulation and self-renewal exhaustion independently.Citation34,51 DF2 expressed the strongest p21 than other sub-clones and also exhibited the lowest level of aneuploidy (). Additionally, sub-clones expressing higher levels of p21 than DFSCs also did not form tumor in vivo (). Therefore, p21 might have restricted DNA damage accumulation of sub-clones thereby keeping aberrant sub-clones away from transformation when p53 was inhibited. Expression of E2F1, another protein functioned in DNA repair, was also detected. E2F1 turnover was the key switch that allowed cells to be prepared for another round of re-replication.Citation52 It not only played a direct role in DNA repair, but also regulated the expression of DNA repair genes involved in the upstream DNA damage response.Citation53-57 Although expression was slightly lower than DFSCs, E2F1 still kept active in sub-clones (). The gradient expression of E2F1 in different sub-clones also coincided with their severities of CIN/aneuploidy. To be specific, the highest ratio of aneuploidy occurred in DF8 among 3 sub-clones with the highest expression level of E2F1; while DF2 with the lowest ratio of aneuploidy expressed lowest E2F1. Thus, E2F1 might be able to assist p21 in keeping sub-clones from aneuploidy.
Mutations and/or reduced levels of mitotic checkpoint proteins can cause checkpoint malfunction and CIN.Citation38 Thus the expression of mitotic checkpoint proteins were detected to disclose the cause of CIN. Damage to the checkpoint, which was a partial loss or gain of checkpoint function, would result in aneuploidy during tumorigenesis; One of those types of damage was the change in levels of MAD1 and MAD2, or in the MAD1:MAD2 ratio.Citation58 Three sub-clones expressed different MAD1: MAD2 ratio (). MAD1 were overexpressed in DF8 and DF18, while MAD2 was low-expressed in DF2. Thus, disordered MAD1 and MAD2 ratio might be the cause of CIN in this study.
Interestingly, CIN was modified when sub-clones were mixed culturing. In mixed culture cells, their guard gene p53 was back to strong expression ( and ); DNA repair gene p21 and E2F1 expressions were strengthened and mitotic related genes (MAD1 and MAD2) were also restored (). Stressful microenvironment may render stem cells changing their normal properties to adapt pressure by guard gene, such as p53.Citation49 However, guard gene lost or long–term pressure could cause mitotic disorders, followed by CIN or aneuploidy. Additionally, p53 also played other important roles on stem cells, beside as the guardian of the genome. p53 not only controlled the proliferation and differentiation of stem cells but also provided an effective barrier for the generation of pluripotent stem cell-like cells from terminally differentiated cells.Citation59 Specially, p53 activation may hinder stem cells expansion by several emerging mechanisms including the restriction of self-renewing divisions, inhibition of symmetric division and blocking of reprograming of somatic/progenitor cells into stem cells.Citation59 In the present study, sub-clones weakly expressed p53 thus resulting in symmetric division and maintenance of strong proliferation capability. Our study suggested that the disorder was reversible. Mixed culturing created a similar heterogeneous microenvironment. When aberrant sub-clones were put back to their original microenvironment, abnormity was modified. In this microenvironment, p53, MAD1 and MAD2 expressions were recovered and CIN was modified (). Moreover, the complete modification of DF1 demonstrated that this modification procedure relied on integrality of original microenvironment instead of any particular clone. The more similar between the simulating environment and original environment, the better effects of modification will be achieved.
In addition, CIN of stem cells could also influence their differentiation capability.Citation18 However, in this study, 3 sub-clones still kept part of the embryonic properties () and differentiation capability although they were in a CIN state. Even DF2 remained some embryonic characteristics which might help it survive in vivo. Osterix played an essential role in OB differentiation and bone formation.Citation36 Low expressions of osterix in 3 sub-clones () indicated they were not better cells than DFSCs for osteogenesis differentiation. After osteogenesis induction, only DFSCs expressed osterix, which further proved above conclusion. WNT and BMP signaling pathway were involved in many biological processes,Citation60-64 such as osteogenesis, tumor formation and so on. In this study, WNT signaling pathway was evidently changed in DF8 and BMP signaling pathway were changed in DF2 (). The down-regulation of β-catenin in DF8 implied the WNT signaling pathway was mainly involved in other processes instead of osteogenesis (). Moreover, our study suggested that mutual complementation among these signaling pathways might exist to maintain osteogenesis capability.
Both of the genetic stability and differentiation capability of stem cells were essential for cell-based therapy. Our results have unearthed an unexpected yet crucial role of heterogeneity in maintaining genetic stability and differentiation capability and provided some basis for choosing seeding cells for cell-based therapy. Moreover, heterogeneity was widespread in stem cells suggesting that this might be a general phenomenon in the development and regeneration of many tissues or organs.
Materials and Methods
The isolation procedure for single clone
To isolate sub-clones from rat-DFSCs, we adopted a limited dilution protocol to isolate single cell and amplify the clones. The 1st passage of rat-DFSCs in a logarithmic growth phase was re-suspended at a density of 10 cells/ml in α-MEM medium with 20% FBS. Subsequently, 100 μl of cell suspension was sampled into wells of a 96-well plate. Following 24 hours of culture, the adherent cells were visualized under a light microscope to determine if only one single cell existed in a well. The well with one single DFSC was regarded as an effective well and marked as a clone using the labeling protocol: DFx, where x = 1, 2, 3, etc. The medium was altered every 2 d.
The amplified cells were digested and transferred to a 48-well plate when the cells reached ˜66% confluence in the 96-well plate. Once fully confluent, DFx were successively passaged into a 24-well, 12-well, or 6-well plate, a T-25 cell culture flask (Costar, MA, USA), and a T-75 cell culture flask. The DFx clone populations were passaged and amplified until their expansion ceased, or beyond 30 passages for perpetually expanding lines ().
Four sub-clones (DF2, DF8, DF12 and DF18) were isolated from heterogeneous DFSCs and amplified successfully for more than 5 passages through culturing for 90-95 d (). Because DF2 and DF12 were proved to be very similar by Cluster Analysis, Correlation Analysis and heat map of Gene Expression Array (S1:DF2; S2:DF12; S3:DF8; S4:DF18; S5: DFSCs) (), DF2 and DF12 were collectively referred to as DF2 for the following research.
RNA extraction and gene expression array
Total RNA was extracted using RNAiso following the manufacturer's instructions and checked for a RIN number to inspect RNA integration by an Agilent Bioanalyzer 2100 (Agilent technologies, Santa Clara, CA, US).Qualified total RNA was further purified by RNeasy mini kit (Cat#74106, QIAGEN, GmBH, Germany) and Rnase-Free Dnase Set (Cat#79254, QIAGEN, GmBH, Germany). Total RNA was amplified and labeled by Low Input Quick Amp Labeling Kit, One-Color (Cat#5190-2305, Agilent technologies, Santa Clara, CA, US), following the manufacturer's instructions. Labeled cRNA were purified by Rneasy mini kit (Cat#74106, QIAGEN, GmBH, Germany). Each Slide was hybridized with 1.65 μg Cy3-labeled cRNA using Gene Expression Hybridization Kit (Cat#5188–5242, Agilent technologies, Santa Clara, CA, US) in Hybridization Oven (Cat#G2545A, Agilent technologies, Santa Clara, CA, US), according to the manufacturer's instructions. After 17 hours hybridization, slides were washed in staining dishes (Cat#121, Thermo Shandon, Waltham, MA, US) with Gene Expression Wash Buffer Kit (Cat#5188–5327, Agilent technologies, Santa Clara, CA, US), followed the manufacturer's instructions. Slides were scanned by Agilent Microarray Scanner (Cat#G2565CA, Agilent technologies, Santa Clara, CA, US) with default settings, Dye channel: Green, Scan resolution = 5 μm, PMT 100%, 10%, 16 bit. Data were extracted with Feature Extraction software 10.7 (Agilent technologies, Santa Clara, CA, US). Raw data were normalized by Quantile algorithm, Gene Spring Software 11.0 (Agilent technologies, Santa Clara, CA, US).
Cell proliferation analysis
Cell Counting Kit-8 (CCK-8, Dojindo, Japan) was utilized to quantitatively evaluate the viability of the 3 sub-clones and DFSCs. 2 × 103 cells were cultivated on 96-well plates (Thermo, USA). The original cultivation medium was replaced by 120 μl α-MEM with 10% FBS containing 12 μl CCK-8 for each well of 96-well plate at the same time of consecutive 7 d. After incubation at 37°C for 4 h, 100 μl of the above solution was taken from each sample and added to one well of another new 96-well plate. Three parallel replicates were prepared and the absorbance at 450 nm was detected using a spectrophotometer (Thermo, USA).
DNA contents and cell apoptosis analysis
For the evaluation of DNA contents by Flow cytometry (FCM), when cells reached 70% confluence they were digested, precipitates were washed twice with 0.01 M PBS, and were re-suspended in 1 ml of physiological saline with repeated vibration to ensure a single-cell suspension. 2 ml of cold, dehydrated alcohol was quickly mixed with the cell suspension to fix cells at 4°C for 24–48 hours. Finally, the cells were washed twice with PBS, stained with 100 mg/ml PI (Cell Cycle and Apoptosis Analysis Kit, Beyotime Institute of Biotechnology, China) at 4°C for 30 min and subjected to cell-cycle analysis using Elite ESP flow cytometry (Beckman Coulter, CA, USA.).
For the evaluation of cell apoptosis by FCM, when cells reached 70% confluence they were harvested using 0.25% trypsin and re-suspended in binding buffer. Cells were subsequently incubated with Annexin-V–FITC and PtdIns (Cell Cycle and Apoptosis Analysis Kit, Beyotime Institute of Biotechnology, China) in the dark for 15 min. Apoptosis analysis was performed utilizing Elite ESP flow cytometry (Beckman Coulter, CA, USA.).
Flow cytometric (FCM) analysis
Approximately 5 × 105 cells were incubated with anti-CD90 (ab225, Abcam, MA, USA; 1:100 for Flow Cytometric); CD146 (FAB3250P, R&D system, MN, USA; 1:100 for Flow Cytometric); SSEA1(sc-21702, Santa Cruz, MA, USA; 1:200 for Flow Cytometric) and CD29(555005, BD Bioscience, CA, USA; 1:100 for Flow Cytometric) according to the manufacturers' protocols. FITC-conjugated, isotype-matching immunoglobulins were used to determine non-specific staining. The secondary reagents included goat anti-mouse and goat anti-rat IgG-FITC (Santa Cruz). Cells were analyzed on a FACS Caliber (Becton-Dickinson, CA, USA), and data were analyzed using CXP software.
Transmission electron microscopy (TEM)
A total of 1 × 106 cells were harvested and centrifuged (3000 g, 10 min) to form pellets, then fixed in 2% glutaraldehyde in 0.1 M cacodylate buffer (PH 7.3) for 1 h at room temperature and post-fixed in aq. 2%(V/V) osmium tetroxide for a further 1 h. The cells were then dehydrated in an ethanol series (50%, 70%, 95% and 100%) and embedded in Epon 812 resin. Ultrathin sections were manufactured and stained with uranyl- acetate and lead citrate. The samples were viewed with a JEM 100 SX electron microscope.
Karyotype analysis
1/100 volume colcemid stock solution was added into the medium of dividing cells. After 2–3 hours of procedure and culture in an incubator, the culture medium was aspirated and cells were washed 3 times by PBS. Processed cells were digested by trypsin and transferred into a new centrifuge tube. Cells were washed 3 times in PBS and treated with hypotonic solution (0.075 M KCl) for 30 min at 37°C, prefixed with some drops of ice-cold fixative (methanol:glacial acetic acid, 3:1,v/v), fixed with new cold fixative and then stored at −4°C. About 3 drops of the cell suspension were dropped on cold, dry slides. Slides were stored at room temperature in a dry place and then stained with Gimsa (Sigma-Aldrich, USA) for karyotype analysis. The slides were observed and pictured on microscope (Olympus, Japan).
Quantitative real time-PCR
For gene-detection, total RNA was extracted from cells using RNAiso Plus (Takara, Japan), according to the manufacturer's instructions. The cDNA (cDNA) synthesis was performed using Revert Aid First Stand cDNA Synthesis Kit (Thermo scientific, USA). For quantitative reverse transcription polymerase chain reaction (qRT-PCR), 1 μl of cDNA with the SYBR Premix Ex Taq II (Perfect real time) (Takara, Japan) was performed with an Eco Real-Time PCR System (Illumina, USA). All of the operating procedures are according to the manufacturer's protocol. The primer pairs for p53, puma, K-ras, Tert, p21 and E2F1 were shown in .
Table 1. The primer pairs for p53, puma, K-ras, Tert, p21 and E2F1
Western blot
Proteins were prepared with Total Protein Extraction Kit (KeyGene, China). After standard SDS–polyacrylamide gel electrophoresis and Western blotting procedures, proteins were visualized using an electrochemiluminescence system (GE, USA). Blotting band intensities were quantified densitometrically using Quantity One software.
Antibodies
Anti-p53(ab26, Abcam); Anti-E2F1(#3742, Cell Signaling Technology); Anit-p21(ab18209, Abcam); Anti-MAD1(#4682, Cell Signaling Technology); Anti-MAD2(#4636, Cell Signaling Technology); Anti-actin(ab3280, Abcam); Anti-GFP(AB105-01, TIANGEN); Anti-Sox2(ab97959, Abcam); Anti-Nanog(sc-33760, santa cruz); Anti-OCT4(ab18976, Abcam); Anti- COL-1(ab90395, Abcam); Anti-OPN(ab8448, Abcam); Anti-ALP(ab95462, Abcam); Anti-DMP1(sc-73633, santa cruz); Anti-osterix(ab22552, Abcam); Anti-BSP(ab52128, Abcam); Anti-GAPDH(200306–7E4, Zen); Anti-LEF1(ab85052. Abcam), Anti-active-β-cantenin (#8814, Cell Signaling Technology); Anti-β-cantenin (sc-59737, santa cruz); Anti-CEBPα (ab40761, Abcam); Anti-Smad4 (ab40759, Abcam); Anti-phospho Smad2/3(ab63399, Abcam); Smad2/3(sc-8332, santa cruz); BMP2(sc-6895, santa cruz).
Lentivirus transfection labeling cells
A total of 5 × 104 cells were seeded into each well of a 6-well plate respectively. At 50% confluence, 1 ml α-MEM without FBS but containing 1.5 μl GFP or RFP labeled lentivirus (NeuronBiotech, China) and 0.5 μl polybrene were added into each well. After cultured for 8 h, the supernatant and lentivirus which didn't infect the stem cells were replaced by 2 ml fresh α-MEM supplemented with 10% FBS. After 3 d later, the infected cells were observed and photographed under a fluorescence microscope (Leica Optical, Germany).
Screening experiment
Green fluorescence protein with puromycin-resistant was transfected on 2 sub-clones mixed cells by lentivirus and red fluorescence protein were transfected on 3 sub-clones by lentivirus. Single sub-clones with RFP were added into 2 sub-clones mixed cells with GFP to constitute DF1. Mixed DF1 was screened by puromycin after 7 d mixing culture. The puromycin-resistant cells (2 sub-clones mixed culturing cells) were isolated and enriched by applying puromycin in culture medium. Proteins of pre-screening cells, 3 sub-clones mixed culturing cells and post-screening cells were extracted respectively. No.1, 2 and 3 group stood for GFP-(DF2+DF8), GFP-(DF2+DF18) and GFP-(DF8+DF18); No.4, 5 and 6 group stood for GFP-(DF2+DF8) +RFP-DF18, GFP-(DF2+DF18)+RFP-DF8 and GFP-(DF8+DF18)+RFP-DF2; 7, 8 and 9 stood for post-screening GFP-(DF2+DF8), GFP-(DF2+DF18) and GFP-(DF8+DF18).
In vivo transplantation experiment
Adult NOD SCID mice were injected at the subcutaneous site with 1 × 105 cells combined with matrigel (BD Biosciences, USA), while those injected with matrigel only were regarded as negative controls. The SCC9 tumor cells were injected as a positive control. Animals were maintained in good conditions with access ad libitum to food and fresh water, and were monitored for tumor formation. Mice were sacrificed after 4 weeks culture and transplanted parts were obtained for further study.
Immunofluorescence (IF)
Transplanted parts were collected and fixed in paraformaldehyde for 12 hours, 4°C. The fixed tissues were dehydrated in 20% and 30% sucrose solutions for 2 times respectively. After dehydration finished, the samples were embedded and frozen sliced. Frozen slices were hydrated 5 min, 3 times before staining. And then slices were incubated for overnight at 4°C by GFP antibodies and were then incubated with Fluorescein (FITC)-conjugated Affinipure goat anti-mouse immunoglobulin G (Beijing Zhongshan Biotech Co., Ltd.) for 2 h at 37 C. Finally, slices were incubated with DAPI for 3 minutes. The samples were viewed under a confocal laser scanning microscopy (Olympus, Japan).
Osteogenic differentiation
Cells were induced with osteogenic inducing medium containing 10% FBS, 10 mM β-glycerophosphate (Sigma), 100 nM dexamethasone (Sigma), 50 μg/ml ascorbic acid and 0.01 μM 1,25-dihydroxy-vitamin D3 (Sigma) for 14 d. Medium was changed every 2 d. After 14 d of culture, induced DFCs were washed 3 times in PBS after being fixed in 4% paraformaldehyde for 10 minutes and then incubated in 0.1% alizarin red solution (Sigma) in Tris-HCl (pH 8.3) at 37°C for 30 minutes. Cells were washed and observed using a phase-contrast inverted microscope (Nikon, Japan).
Statistical analysis
Quantitative data were presented as mean SEM. Statistical analysis was conducted using Graph-Pad Prism software Version 5.0 (Graph-Pad software, Inc., La Jolla, CA, USA). A Student-Newman-Keuls test was performed to determine the statistical significance between experimental groups. A value of P < 0.05 was considered statistically significant.
Disclosure of Potential Conflicts of Interest
The authors declare no conflict of interest.
Acknowledgments
We appreciate the colleagues in National Engineering Laboratory for Oral Regenerative Medicine in Sichuan University, and particularly Dr. Gang Chen and Dr. Chao Yang for their excellent work, excellent assistance, and stimulating discussions.
Funding
This study was supported by National Basic Research Program (China, 2010CB944800), Nature Science Foundation (China, 81271095, 81271119, 81200792 and 81300848), International Cooperation Program of China (China, 2013DFG32770 and 2011DFA51970), Trans—Century Training Pro-gramme Foundation for the Talents of Humanities and Social Science by the State Eduation Commission (China, NCET-13-0385), Doctoral Foundation of Ministry of Education (China, 20110181120067, 20110181110089 and 20120181120013), China postdoctoral science foundation (China, 2012M511934), Key Technology R&D Program of Sichuan Province (2010FZ0055, 2012SZ0013, 12ZC0493, 13ZC0971, 2013GZX0158, 2013SZ0015 and 13ZC0979) and Basic Research Program of Sichuan Province (2011JY0125, 12JC0212, 2012JY0077 and 2013JY0019).
References
- Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, Vrana J, Jones K, Grotewold L, Smith A. Nanog safeguards pluripotency and mediates germline development. Nature 2007; 450:1230-4; PMID:18097409; http://dx.doi.org/10.1038/nature06403
- Hayashi K, Lopes SM, Tang F, Surani MA. Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Sell 2008; 3:391-401; PMID:18940731; http://dx.doi.org/10.1016/j.stem.2008.07.027
- Singh AM, Hamazaki T, Hankowski KE, Terada N. A heterogeneous expression pattern for Nanog in embryonic stem cells. Stem cells 2007; 25:2534-42; PMID:17615266; http://dx.doi.org/10.1634/stemcells.2007-0126
- Bianco P, Riminucci M, Gronthos S, Robey PG. Bone marrow stromal stem cells: nature, biology, and potential applications. Stem Cells 2001; 19:180-92; PMID:11359943; http://dx.doi.org/10.1634/stemcells.19-3-180
- Graf T, Stadtfeld M. Heterogeneity of embryonic and adult stem cells. Cell Stem Cell 2008; 3:480-3; PMID:18983963; http://dx.doi.org/10.1016/j.stem.2008.10.007
- Solozobova V, Blattner C. p53 in stem cells. World J Biol Chem 2011; 2:202-14; PMID:21949570; http://dx.doi.org/10.4331/wjbc.v2.i9.202
- Valtieri M, Sorrentino A. The mesenchymal stromal cell contribution to homeostasis. J Cell Physiol 2008; 217:296-300; PMID:18615579; http://dx.doi.org/10.1002/jcp.21521
- Chery L, Lam HM, Coleman I, Lakely B, Coleman R, Larson S, Aguirre-Ghiso JA, Xia J, Gulati R, Nelson PS, et al. Characterization of single disseminated prostate cancer cells reveals tumor cell heterogeneity and identifies dormancy associated pathways. Oncotarget 2014; 5:9939-51; PMID:25301725
- Boesch M, Zeimet AG, Reimer D, Schmidt S, Gastl G, Parson W, Spoeck F, Hatina J, Wolf D, Sopper S. The side population of ovarian cancer cells defines a heterogeneous compartment exhibiting stem cell characteristics. Oncotarget 2014; 5:7027-39; PMID:25216521
- Ben-David U, Benvenisty N. The tumorigenicity of human embryonic and induced pluripotent stem cells. Nat Rev Cancer 2011; 11:268-77; PMID:21390058; http://dx.doi.org/10.1038/nrc3034
- Bolouri H. Network dynamics in the tumor microenvironment. Semin Cancer Biol 2014; 30:52-9; PMID: 24582766; http://dx.doi.org/10.1016/j.semcancer.2014.02.007
- Biancotti JC, Benvenisty N. Aneuploid human embryonic stem cells: origins and potential for modeling chromosomal disorders. Regenerative Med 2011; 6:493-503; PMID:21749207; http://dx.doi.org/10.2217/rme.11.27
- Mayshar Y, Ben-David U, Lavon N, Biancotti JC, Yakir B, Clark AT, Plath K, Lowry WE, Benvenisty N. Identification and classification of chromosomal aberrations in human induced pluripotent stem cells. Cell Stem Cell 2010; 7:521-31; PMID:20887957; http://dx.doi.org/10.1016/j.stem.2010.07.017
- Miura M, Miura Y, Padilla-Nash HM, Molinolo AA, Fu B, Patel V, Seo BM, Sonoyama W, Zheng JJ, Baker CC, et al. Accumulated chromosomal instability in murine bone marrow mesenchymal stem cells leads to malignant transformation. Stem Cells 2006; 24:1095-103; PMID:16282438; http://dx.doi.org/10.1634/stemcells.2005-0403
- Boveri T. Concerning the origin of malignant tumours by Theodor Boveri. Translated and annotated by Henry Harris. J Cell Sci 2008; 121 Suppl 1:1-84; PMID:18089652; http://dx.doi.org/10.1242/jcs.025742
- A SA, H BC, J SS. Application of UV-spectrophotometric methods for estimation of tenofovir disoproxil fumarate in tablets. Pakistan J Pharm Sci 2009; 22:27-9; PMID:19168416;
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144:646-74; PMID:21376230; http://dx.doi.org/10.1016/j.cell.2011.02.013
- Moon SH, Kim JS, Park SJ, Lim JJ, Lee HJ, Lee SM, Chung HM. Effect of chromosome instability on the maintenance and differentiation of human embryonic stem cells in vitro and in vivo. Stem Cell Res 2011; 6:50-9; PMID:20920899; http://dx.doi.org/10.1016/j.scr.2010.08.006
- Colter DC, Sekiya I, Prockop DJ. Identification of a subpopulation of rapidly self-renewing and multipotential adult stem cells in colonies of human marrow stromal cells. Proc Natl Acad Sci U S A 2001; 98:7841-5; PMID:11427725; http://dx.doi.org/10.1073/pnas.141221698
- Guo W, Gong K, Shi H, Zhu G, He Y, Ding B, Wen L, Jin Y. Dental follicle cells and treated dentin matrix scaffold for tissue engineering the tooth root. Biomaterials 2012; 33:1291-302; PMID:22088889; http://dx.doi.org/10.1016/j.biomaterials.2011.09.068
- Muraglia A, Cancedda R, Quarto R. Clonal mesenchymal progenitors from human bone marrow differentiate in vitro according to a hierarchical model. J Cell Sci 2000; 113 (Pt 7)1161-6; PMID:10704367;
- Lee CC, Christensen JE, Yoder MC, Tarantal AF. Clonal analysis and hierarchy of human bone marrow mesenchymal stem and progenitor cells. Exp Hematol 2010; 38:46-54; PMID:19900502; http://dx.doi.org/10.1016/j.exphem.2009.11.001
- Yoshimura K, Asano Y, Aoi N, Kurita M, Oshima Y, Sato K, Inoue K, Suga H, Eto H, Kato H, et al. Progenitor-enriched adipose tissue transplantation as rescue for breast implant complications. Breast J 2010; 16:169-75; PMID:19912236; http://dx.doi.org/10.1111/j.1524-4741.2009.00873.x
- Collins CA, Olsen I, Zammit PS, Heslop L, Petrie A, Partridge TA, Morgan JE. Stem cell function, self-renewal, and behavioral heterogeneity of cells from the adult muscle satellite cell niche. Cell 2005; 122:289-301; PMID:16051152; http://dx.doi.org/10.1016/j.cell.2005.05.010
- Sherwood RI, Christensen JL, Conboy IM, Conboy MJ, Rando TA, Weissman IL, Wagers AJ. Isolation of adult mouse myogenic progenitors: functional heterogeneity of cells within and engrafting skeletal muscle. Cell 2004; 119:543-54; PMID:15537543; http://dx.doi.org/10.1016/j.cell.2004.10.021
- Yang R, Chen M, Lee CH, Yoon R, Lal S, Mao JJ. Clones of ectopic stem cells in the regeneration of muscle defects in vivo. PloS One 2010; 5:e13547; PMID: 20975999; http://dx.doi.org/10.1371/journal.pone.0013547
- Wagner W, Feldmann RE Jr, Seckinger A, Maurer MH, Wein F, Blake J, Krause U, Kalenka A, Burgers HF, Saffrich R, et al. The heterogeneity of human mesenchymal stem cell preparations-evidence from simultaneous analysis of proteomes and transcriptomes. Exp Hematol 2006; 34:536-48; PMID:16569600; http://dx.doi.org/10.1016/j.exphem.2006.01.002
- Guo W, Chen L, Gong K, Ding B, Duan Y, Jin Y. Heterogeneous dental follicle cells and the regeneration of complex periodontal tissues. Tissue Eng Part A 2012; 18:459-70; PMID:21919800; http://dx.doi.org/10.1089/ten.TEA.2011.0261
- Hou LT, Liu CM, Chen YJ, Wong MY, Chen KC, Chen J, Thomas HF. Characterization of dental follicle cells in developing mouse molar. Arch Oral Biol 1999; 44:759-70; PMID:10471160;
- Wu J, Jin F, Tang L, Yu J, Xu L, Yang Z, Wu G, Duan Y, Jin Y. Dentin non-collagenous proteins (dNCPs) can stimulate dental follicle cells to differentiate into cementoblast lineages. Biol Cell/Under Auspices Eur Cell Biol Organ 2008; 100:291-302; PMID:18042041; http://dx.doi.org/10.1042/BC20070092
- Yao S, Pan F, Prpic V, Wise GE. Differentiation of stem cells in the dental follicle. J Dental Res 2008; 87:767-71; PMID:18650550;
- Kabir R, Gupta M, Aggarwal A, Sharma D, Sarin A, Kola MZ. Imperative role of dental pulp stem cells in regenerative therapies: a systematic review. Nigerian J Surg: Off Pub Nigerian Surg Res Soc 2014; 20:1-8; PMID:24665194; http://dx.doi.org/10.4103/1117-6806.127092
- Lymperi S, Ligoudistianou C, Taraslia V, Kontakiotis E, Anastasiadou E. Dental stem cells and their applications in dental tissue engineering. Open Dent J 2013; 7:76-81; PMID:24009647; http://dx.doi.org/10.2174/1874210601307010076
- Insinga A, Cicalese A, Faretta M, Gallo B, Albano L, Ronzoni S, Furia L, Viale A, Pelicci PG. DNA damage in stem cells activates p21, inhibits p53, and induces symmetric self-renewing divisions. Proc Natl Acad Sci U S Am 2013; 110:3931-6; PMID:23417300; http://dx.doi.org/10.1073/pnas.1213394110
- Udayakumar T, Shareef MM, Diaz DA, Ahmed MM, Pollack A. The E2F1/Rb and p53/MDM2 pathways in DNA repair and apoptosis: understanding the crosstalk to develop novel strategies for prostate cancer radiotherapy. Semin Radiat Oncol 2010; 20:258-66; PMID:20832018; http://dx.doi.org/10.1016/j.semradonc.2010.05.007
- Schvartzman JM, Duijf PH, Sotillo R, Coker C, Benezra R. Mad2 is a critical mediator of the chromosome instability observed upon Rb and p53 pathway inhibition. Cancer Cell 2011; 19:701-14; PMID:21665145; http://dx.doi.org/10.1016/j.ccr.2011.04.017
- Ryan SD, Britigan EM, Zasadil LM, Witte K, Audhya A, Roopra A, Weaver BA. Up-regulation of the mitotic checkpoint component Mad1 causes chromosomal instability and resistance to microtubule poisons. Proc Natl Acad Sci U S A 2012; 109:E2205-14; PMID:22778409; http://dx.doi.org/10.1073/pnas.1201911109
- Schvartzman JM, Sotillo R, Benezra R. Mitotic chromosomal instability and cancer: mouse modelling of the human disease. Nat Rev Cancer 2010; 10:102-15; PMID:20094045; http://dx.doi.org/10.1038/nrc2781
- El-Sayed KM, Paris S, Graetz C, Kassem N, Mekhemar M, Ungefroren H, Fandrich F, Dorfer C. Isolation and characterisation of human gingival margin-derived STRO-1/MACS and MACS cell populations. Int J Oral Sci 2014; 7:80-8; PMID:25257881; http://dx.doi.org/10.1038/ijos.2014.41
- Xu J, Wang W, Kapila Y, Lotz J, Kapila S. Multiple differentiation capacity of STRO-1+/CD146+ PDL mesenchymal progenitor cells. Stem Cells Dev 2009; 18:487-96; PMID:18593336; http://dx.doi.org/10.1089/scd.2008.0113
- Majore I, Moretti P, Hass R, Kasper C. Identification of subpopulations in mesenchymal stem cell-like cultures from human umbilical cord. Cell Commun Signaling: CCS 2009; 7:6; PMID:19302702; http://dx.doi.org/10.1186/1478-811X-7-6
- Stewart MH, Bosse M, Chadwick K, Menendez P, Bendall SC, Bhatia M. Clonal isolation of hESCs reveals heterogeneity within the pluripotent stem cell compartment. Nat Methods 2006; 3:807-15; PMID:16990813; http://dx.doi.org/10.1038/nmeth939
- Sengers BG, Dawson JI, Oreffo RO. Characterisation of human bone marrow stromal cell heterogeneity for skeletal regeneration strategies using a two-stage colony assay and computational modelling. Bone 2010; 46:496-503; PMID:19818885; http://dx.doi.org/10.1016/j.bone.2009.10.002
- Tormin A, Brune JC, Olsson E, Valcich J, Neuman U, Olofsson T, Jacobsen SE, Scheding S. Characterization of bone marrow-derived mesenchymal stromal cells (MSC) based on gene expression profiling of functionally defined MSC subsets. Cytotherapy 2009; 11:114-28; PMID:19242838; http://dx.doi.org/10.1080/14653240802716590
- Smith JG, Smith AJ, Shelton RM, Cooper PR. Recruitment of dental pulp cells by dentine and pulp extracellular matrix components. Exp Cell Res 2012; 318:2397-406; PMID:22819733; http://dx.doi.org/10.1016/j.yexcr.2012.07.008
- Freedman DA, Wu L, Levine AJ. Functions of the MDM2 oncoprotein. Cell Mol Life Sci: CMLS 1999; 55:96-107; PMID:10065155
- Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, Pellman D. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature 2005; 437:1043-7; PMID:16222300; http://dx.doi.org/10.1038/nature04217
- Heng HH, Bremer SW, Stevens JB, Horne SD, Liu G, Abdallah BY, Ye KJ, Ye CJ. Chromosomal instability (CIN) what it is and why it is crucial to cancer evolution. Cancer Metast Rev 2013; 32:325-40; PMID:23605440; http://dx.doi.org/10.1007/s10555-013-9427-7
- Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol 2007; 8:275-83; PMID:17380161; http://dx.doi.org/10.1038/nrm2147
- Notterman D, Young S, Wainger B, Levine AJ. Prevention of mammalian DNA reduplication, following the release from the mitotic spindle checkpoint, requires p53 protein, but not p53-mediated transcriptional activity. Oncogene 1998; 17:2743-51; PMID:9840938; http://dx.doi.org/10.1038/sj.onc.1202210
- Lee J, Hoi CS, Lilja KC, White BS, Lee SE, Shalloway D, Tumbar T. Runx1 and p21 synergistically limit the extent of hair follicle stem cell quiescence in vivo. Proc Natl Acad Sci U S Am 2013; 110:4634-9; PMID:23487742; http://dx.doi.org/10.1073/pnas.1213015110
- Zielke N, Kim KJ, Tran V, Shibutani ST, Bravo MJ, Nagarajan S, van Straaten M, Woods B, von Dassow G, Rottig C, et al. Control of Drosophila endocycles by E2F and CRL4(CDT2). Nature 2011; 480:123-7; PMID:22037307; http://dx.doi.org/10.1038/nature10579
- Lin WC, Lin FT, Nevins JR. Selective induction of E2F1 in response to DNA damage, mediated by ATM-dependent phosphorylation. Genes Dev 2001; 15:1833-44; PMID:11459832
- Maser RS, Mirzoeva OK, Wells J, Olivares H, Williams BR, Zinkel RA, Farnham PJ, Petrini JH. Mre11 complex and DNA replication: linkage to E2F and sites of DNA synthesis. Mol Cell Biol 2001; 21:6006-16; PMID:11486038
- Liu K, Lin FT, Ruppert JM, Lin WC. Regulation of E2F1 by BRCT domain-containing protein TopBP1. Mol Cell Biol 2003; 23:3287-304; PMID:12697828
- Guo R, Chen J, Zhu F, Biswas AK, Berton TR, Mitchell DL, Johnson DG. E2F1 localizes to sites of UV-induced DNA damage to enhance nucleotide excision repair. J Biol Chem 2010; 285:19308-15; PMID:20413589; http://dx.doi.org/10.1074/jbc.M110.121939
- Chen J, Zhu F, Weaks RL, Biswas AK, Guo R, Li Y, Johnson DG. E2F1 promotes the recruitment of DNA repair factors to sites of DNA double-strand breaks. Cell Cycle 2011; 10:1287-94; PMID:21512314; http://dx.doi.org/10.4161/cc.10.8.15341
- Schuyler SC, Wu YF, Kuan VJ. The Mad1-Mad2 balancing act - a damaged spindle checkpoint in chromosome instability and cancer. J Cell Sci 2012; 125:4197-206; PMID:23093575; http://dx.doi.org/10.1242/jcs.107037
- Bonizzi G, Cicalese A, Insinga A, Pelicci PG. The emerging role of p53 in stem cells. Trends Mol Med 2012; 18:6-12; PMID:21907001; http://dx.doi.org/10.1016/j.molmed.2011.08.002
- Gregory CA, Gunn WG, Reyes E, Smolarz AJ, Munoz J, Spees JL, Prockop DJ. How Wnt signaling affects bone repair by mesenchymal stem cells from the bone marrow. Ann New York Acad Sci 2005; 1049:97-106; PMID:15965110; http://dx.doi.org/10.1196/annals.1334.010
- Rosenbluh J, Wang X, Hahn WC. Genomic insights into WNT/beta-catenin signaling. Trends Pharmacol Sci 2014; 35:103-9; PMID:24365576; http://dx.doi.org/10.1016/j.tips.2013.11.007
- James AW. Review of signaling pathways governing MSC osteogenic and adipogenic differentiation. Scientifica 2013; 2013:684736; PMID:24416618; http://dx.doi.org/10.1155/2013/684736
- Heggebo J, Haasters F, Polzer H, Schwarz C, Saller MM, Mutschler W, Schieker M, Prall WC. Aged human mesenchymal stem cells: the duration of bone morphogenetic protein-2 stimulation determines induction or inhibition of osteogenic differentiation. Orthopedic Rev 2014; 6:5242; PMID:25002931; http://dx.doi.org/10.4081/or.2014.5242
- Kevin A. Maupin CJD, Bart O. Williams. A comprehensive overview of skeletal phenotypes associated with alterations in Wnt/β-catenin signaling in humans and mice. Bone Res 2013; 1:27-71; PMID:http://dx.doi.org/10.4248/BR201301004
