Abstract
TAp73 is a tumor suppressor transcriptional factor, belonging to p53 family. Alteration of TAp73 in tumors might lead to reduced DNA damage response, cell cycle arrest and apoptosis. Carcinogen-induced TAp73−/− tumors display also increased angiogenesis, associated to hyperactivition of hypoxia inducible factor signaling. Here, we show that TAp73 suppresses BNIP3 expression, directly binding its gene promoter. BNIP3 is a hypoxia responsive protein, involved in a variety of cellular processes, such as autophagy, mitophagy, apoptosis and necrotic-like cell death. Therefore, through different cellular process altered expression of BNIP3 may differently contribute to cancer development and progression. We found a significant upregulation of BNIP3 in human lung cancer datasets, and we identified a direct association between BNIP3 expression and survival rate of lung cancer patients. Our data therefore provide a novel transcriptional target of TAp73, associated to its antagonistic role on HIF signaling in cancer, which might play a role in tumor suppression.
Keywords:
Introduction
p73 is a transcriptional factor, belonging to p53 family. The presence of 2 promoters in the TP73 gene gives rise to 2 sets of isoforms: transactivational (TA) domain-containing isoforms, TAp73, regulated by the first promoter (P1), and the N-truncated isoforms, lacking TA domain (from promoter P2), ΔNp73. Alternative splicing can also take place at 3′-end, leading to 7 isoforms varying in activity and specificity α, β, γ, δ, ε, ζ, η.Citation1-5 p53 family is one of the most powerful families of genesCitation6,7; it plays fundamental roles in protection of genome integrityCitation8-13 in germline and somatic cells impacting fertilityCitation14-21 and cancer.Citation22-36 In cancer cells p73 is rarely mutated, but its expression is often deregulated. There is increasing evidence, that TAp73/ΔNp73 expression ratio affects tumor development and progression.Citation37-39 TAp73 is considered a bona fide tumor suppressor, largely mimicking p53 function. It controls cell cycle arrest, apoptosis as well as DNA damage repair.Citation40 Tumor suppressor function of TAp73 has also been recently associated to repression of tumor angiogenesis, through regulation of hypoxia inducible factor (HIF) signaling. TAp73 indeed directly binds HIF-1a protein, promoting its oxygen-independent degradation.Citation41,42 Conversely, ΔNp73 antagonizes TAp73 and it is considered an oncogenic protein.Citation43-46 It can form inactive complexes with TAp73, and also bind common promoters with p53 and TAp73, thus inhibiting their transcriptional activity.Citation47-50 Besides its cancer related function, TAp73 also plays a role in neurogenesis, and its dysregulation is linked with developmental defect and neurodegenerative diseases. In fact, TAp73 is necessary for neuronal differentiation and maintenance of neuronal stem cells.Citation51-54
Hypoxia inducible factors (HIFs) mediate the physiological response to hypoxiaCitation55 regulating processes, such as angiogenesis,Citation56-58 proliferationCitation59-68 and metabolism.Citation64,69-73 The wide transcriptional reprogramming operated by HIF-1, includes the direct transcriptional induction of the Bcl-2 Nineteen kilodalton Interacting Protein (BNIP3).Citation74,75 BNIP3 is a Bcl2-family BH3-only protein, which contributes to cellular processes, such as apoptosis, autophagy, mitophagy and mitochondrial metabolism.Citation76 BNIP3-deficient mice do not display significant physical abnormalities and altered lifespan, however they show decreased postischemic myocardial apoptosis,Citation77-82 suggesting an involvement in hypoxic-dependent cell death. BNIP3 was first shown to localize in mitochondria,Citation83 although later in glial cells was also observed a nuclear localization.Citation84,85 BNIP3 activation causes mitochondrial dysfunction through mitochondrial apoptosis, reduced oxidative phosphorylation and induction of autophagy and mitophagy.Citation86-89
Here, we show a direct regulation by TAp73 on BNIP3 transcription, and we report a possible clinical relevance of this axis for lung cancer patients. Consistently with reduced TAp73 activity, high BNIP3 expression correlates with bad prognosis in patients with lung cancer.
Results
TAp73 represses HIF1α and its target BNIP3
To investigate the influence of TAp73 on BNIP3 expression we used SaOS-2 cells with Tet-On system. SaOs-2 is a p53/p63/p73 deficient human osteosarcoma cell line. Expression of TAp73 in these cells can be induced by doxycycline treatment. As shown in and Fig. S1A, 2 µg/ml of doxycycline induced TAp73 expression in a time-dependent manner. Along with TAp73 accumulation we detected decrease in BNIP3 protein levels and as previously described reduced HIF1α ().Citation41,43 p21 was used as positive control of p73 transcriptional activation (). To evaluate whether BNIP3 downregulation was associated to altered transcription of the BNIP3 gene, we performed real-time qPCR in SaOs-2 Tet-On cell line. qPCR also highlighted decrease in BNIP3 mRNA level (). Taken together these data indicate that consistently with TAp73-dependent downregulation of HIF1α BNIP3 is downregulated.
Figure 1. TAp73 overexpression inhibits BNIP3 expression in ostesarcoma cell line. HATAp73 was overexpressed in SAOS2-HATAp73 cell line for 4h, 8h, 16h, 24h. (A) Protein levels of HIF1α, HATAp73, p21, BNIP3 and β-tubulin were analyzed by WB. Figure shows a representative replicate of 3 independent experiments. (B) mRNA levels of p21 and BNIP3 were analyzed by qPCR at different time points after TAp73 induction. Relative expression of genes was normalized against TBP and calculated as fold induction on the time point 0h. Data is reported as mean ± s.d. of two experiments for p21, 3 experiments for BNIP3. *P < 0.05 (Student's T-test).
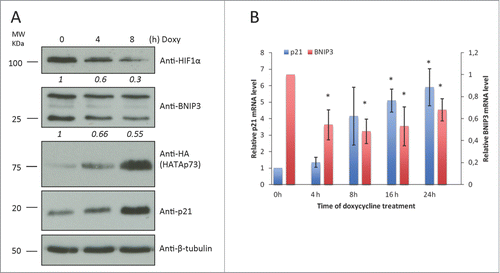
Next we employed H1299, p53-null human non-small cell lung carcinoma (NSCLC) cell line, expressing endogenous TAp73. First, we overexpressed HA-tagged TAp73 for 24 h and exposed the cells to hypoxia during the last 8h of transfection (1% O2) (). Protein and RNA levels of TAp73 and its transcriptional target p21 confirmed TAp73 transcriptional activation (, Fig. S1B, C). Increased levels of BNIP3 mRNA and protein were observed in cells, under hypoxia, as BNIP3 is a hypoxia response gene. TAp73 overexpression in normoxia and hypoxia confirmed the BNIP3 repression observed in SaOs-2 Tet-On mRNA ().
Figure 2. TAp73 inhibits BNIP3 expression in non-small cell lung carcinoma cell line. (A, B) H1299 cells were transfected with HATAp73-alfa for 24h. Cells were subjected to hypoxia for 8h before lysis. (A) BNIP3 mRNA level was analyzed by qPCR. Data is reported as mean ± s.d., n = 3 independent experiments for hypoxia, n = 5 for normoxia. (B) Protein level of BNIP3, HATAp73 and p21 was analyzed by WB. (C) BNIP3 mRNA level was analyzed by qPCR after TAp73 knockdown in H1299 for 48h or 72h. n = 3. (D) Protein level of BNIP3 and p73 after 48h of TAp73 knockdown was analyzed by WB. (A, C) Relative expression of genes was normalized against TBP and calculated as fold induction. Data is reported as mean ± s.d. *P < 0.05 (Student's T-test). (B, D). Figure shows a representative replicate of 3 independent experiments.
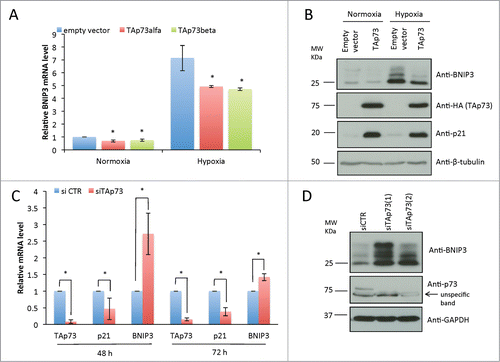
Then we performed knockdown experiment in H1299 cells by transfecting selective siRNA, for TAp73 isoforms. mRNA levels of BNIP3 resulted upregulated after TAp73 silencing, stronger effect was seen after 48h (). Similarly western blot (WB) analysis showed BNIP3 protein accumulation after TAp73 depletion (). Together with the data reported in , these data proved a TAp73-dependent inhibition of BNIP3 expression in an oxygen-independent manner.
TAp73 binds BNIP3 promoter
The ability of TAp73 to inhibit the expression of BNIP3 in an oxygen-independent manner indicated the possibility of an additional HIF-independent regulation of BNIP3 by TAp73. We therefore investigated the hypothesis that TAp73 acts also as a transcriptional factor directly regulating BNIP3 promoter. In support of this hypothesis BNIP3 has been shown as a direct p53 transcriptional target.Citation90 We therefore assessed whether the previous validated p53RE in BNIP3 promoter could also be regulated by TAp73 (). The p53RE is located between −987 and −1021 bp upstream of the transcription start site (TSS) and comprises 2 closely located p53 binding sites. To experimentally validate our hypothesis we used a reporter gene vector, containing the region of BNIP3 promoter showed in (between −1638 and +186 bp from the TSS) upstream of the luciferase reporter gene. We co-transfected H1299 cells with this construct, HA-TAp73-expressing plasmid and control Renilla vector for 20 h. Transfection efficiency was confirmed by WB (). Consistently with our hypothesis luciferase assay showed significant decrease in luciferase activity of approximately 40% after HATAp73 transfection ().
Figure 3. TAp73 directly transactivates p53 response element in the BNIP3 promoter. (A) Schematic image of the BNIP3 promoter region. HRE1, HRE2 – Hypoxia Response Elements. The insert shows p53 responsive element (p53RE), identified by Xi Feng et al.Citation90 located between −1021 and −987 bp upstream of the transcription-start site (TSS). Core p53 binding elements are highlighted in red. (B) BNIP3 promoter activity is repressed by TAp73. H1299 cells were cotransfected with BNIP3 reporter vector and pcDNA or TAp73 as a transactivator. The luciferase assay was performed after 20 h, and normalized by Renilla luciferase activity. Experiment was performed 2 times, mean value ± SD is shown. *P < 0.05 (Student's T-test) (C) Western Blot analysis performed with the same lysates which were used for Luciferase assay was used as a control of the TAp73 expression. (D) Chromatin extracted from SAOS2-HA-TAp73 was incubated with anti-HA or IgG antibodies. Immunoprecipitated DNA was tested by PCR for p53-Response Element in BNIP3 promoter. NG: PCR negative control. Figure shows a representative replicate of 3 independent experiments
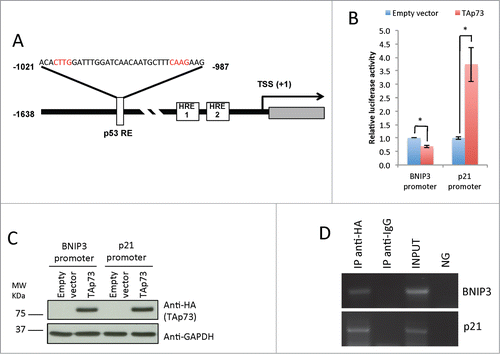
Next, we performed chromatin immunoprecipitation (ChIP) assay for the indicated p53RE in the BNIP3 promoter, in TAp73 SaOs-2 Tet-On cells, after 16 h of doxycycline induction. The specific amplification in anti-HA immunoprecipitated chromatin confirmed a direct binding of TAp73 on BNIP3 human promoter (). Data obtained from luciferase gene reporter assay and ChIP demonstrated that TAp73 suppresses BNIP3 gene expression directly binding its promoter.
The BNIP3 regulation is of clinical importance for lung cancer patients
Trp73−/− and TAp73−/− mice spontaneously develop lung carcinomas, and altered ratio TAp73/ΔNp73 is frequently reported in human lung cancer.Citation91,92 TAp73 is therefore considered a bona fide tumor suppressor, particular relevant in lung tumorigenesis. We wanted therefore to verify whether downstream to TAp73 alteration, BNIP3 upregulation might play a role in lung carcinoma. We employed a bioinformatic approach to assess BNIP3 expression in human lung cancer patient specimens. We used publicly available Hou Lung patient data set to analyze BNIP3 expression in 156 patient samples. Dataset includes 4 groups of patient samples: derived from normal lung, large cell lung carcinoma, lung adenocarcinoma or squamous cell lung carcinoma. Median BNIP3 expression was significantly higher in all lung carcinomas compared to normal lung tissue (). These data suggest that failure of TAp73/BNIP3 axis in lungs may lead to BNIP3 upregulation and may contribute to tumorigenicity.
Figure 4. BNIP3 expression is increased in lung carcinomas and correlates with worse patient survival. (A) BNIP3 expression in normal lung and large cell carcinoma, Hou Lung dataset. (B) BNIP3 expression in normal lung and squamous cell lung carcinoma, Hou Lung data set. (C) BNIP3 expression in normal lung and lung adenocarcinoma, Hou Lung dataset. (A–C) n = 156 samples in th Hou Lung data set. (D) Survival analysis of GSE 4573 dataset (patients with lung cancer). Patients were divided in 2 groups: patients with low expression of the BNIP3 gene and with high expression. n = 46 patients with low BNIP3 expression, n = 77 patients with high BNIP3 expression.
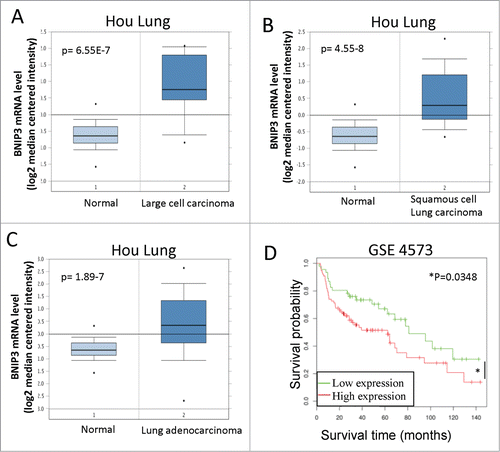
We next used publicly available data set to assess BNIP3 expression impact on patients' survival. Survival rate appeared significantly higher in patients with low BNIP3 expression (). Our data suggest that BNIP3 may have a role in tumorigenesis and progression of lung cancers.
Discussion
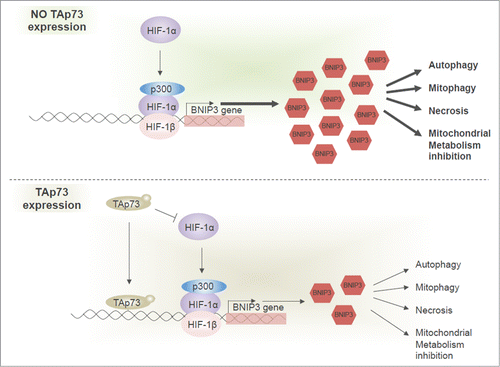
We identified BNIP3 as a novel TAp73 target gene. BNIP3 expression can be regulated by TAp73 via 2 mechanisms (). Here we show that BNIP3 expression is inhibited by TAp73, through its direct binding on BNIP3 promoter. We demonstrated that the p53-like responsive element in the BNIP3 promoter, previously experimentally validated for p53 can also be recognized by TAp73. As BNIP3 is a HIF1α target gene,Citation90 and HIF1α is repressed by TAp73,Citation41 relationship between TAp73 and BNIP3 can also depend on an indirect regulation via HIF1.
Figure 5. TAp73 regulates the BNIP3 expression via 2 mechanisms. TAp73 can directly bind the BNIP3 promoter and inhibit its expression. BNIP3 is upregulated by HIF1 in hypoxia. TAp73 drives HIF1α degradation and, subsequently, can prevent BNIP3 upregulation. No expression of TAp73 enables expression of BNIP3 (upper panel). TAp73 expression leads to lower BNIP3 level impacting different processes, including autophagy, mitophagy, mitochondrial metabolism and necrotic cell death.
TAp73 has tumor-suppressor function, we therefore also investigated a possible involvement of BNIP3 in tumourigenesis. BNIP3 contributes to several processes in cell, which potentially can affect tumor development. The ability to activate apoptosis would indicate a tumor suppressor function for BNIP3, however its pro-necrotic role may lead to pro-tumorigenic effects, as necrosis can promote tumor growth and associates with poor prognosis for patients.Citation93 BNIP3 is also known to lead to autophagy, which may promote both tumor suppression and tumor growth, and the implication of autophagy in cancer progression can be different.Citation94-99 BNIP3 has also been shown to act as a transcriptional factor: if it translocates to nucleus, it suppresses Apoptosis-Inducing Factor expression, preventing cell death, thus showing tumorigenic function.Citation85 Our bioinformatics analysis would suggest an oncogenic function for BNIP3. BNIP3 expression is upregulated in lung carcinomas, and correlates with bad prognosis for patients with lung cancer. Therefore our current data, although still preliminary, might indicate TAp73/BNIP3 negative axis as a novel pathway for TAp73 tumor suppressor function. Consistently, TAp73 loss results in mitochondrial dysfunction.Citation100,101 BNIP3 upregulation as a consequence of TAp73 loss might therefore contribute to TAp73–dependent mitochondrial phenotype and be associated to the complex involvement of p73Citation102-104 and the other family members in regulation of mitochondrial activity,Citation105-108 cell metabolismCitation109-114 and redox homeostasis.Citation115-118 However, currently it is still unclear whether the complex integration of all the p53 family members, in particular the truncated isoform of p73, ΔNp73, and the cancer-associated mutants of p53, impacts and affects the TAp73-dependent antagonism of BNIP3 expression and more generally of hypoxia response. Future studies are demanded to address these aspects.
Overall, we described a novel transcriptional target of TAp73, also involved in hypoxia response, confirming the antagonistic role of TAp73 on HIF signaling and tumourigenesis.
Materials and Methods
Cell cultures
H1299 and SaOS2-Tet-On cell lines were used. Cells were grown in humidified incubator, at 37°C, in atmosphere of 5% CO2 in air. Cells were cultivated in RPMI medium, containing L-glutamine, 4,5 g/L of D-glucose, 2,383 g/L of HEPES Buffer, 1,5 g/L of Sodium Bicarbonate, 110 mg/L of Sodium Pyruvate (Gibco, Life Technologies), supplemented with Penicillin Streptomycin (Gibco, Life Technologies) and 10% (vol/vol) of FBS (Labtech). To generate SaOS2 cell line with inducible expression of HA-TAp73(SaOS2-Tet-On), we used Tet-responsive transcriptional activator rtTA. To induce HA-TAp73 expression in that cell line we treated the cells with 2 ug/ml of doxycycline for indicated period of time.
RNA extraction and quantitative PCR
RNA was extracted from cells by means of RNEasy Mini Kit (Qiagen), according to the Qiagen company protocol. The RNA obtained was quantified by spectrophotometric analysis, and 1 ug of total RNA was used to prepare cDNA with RevertAid H minus First Strand cDNA Synthesis kit (ThermoScientific), using Random primers and protocol from the kit. qPCR was carried out with 1/10 of prepared cDNA and Power SYBR Green PCR Master Mix (Applied Biosystems). Relative gene expression was analyzed in accordance to 7500 Software version 2.0.6 of Applied Biosystems, normalized to housekeeping gene TBP. Sequences of the primers used for the qPCR are: human TAp73: Fw CAGACAGCACCTACTTCGACCTT, Rev CCGCCCACCACCTCATTA; P21: Fw cctgtcactgtcttgtaccct; Rev gcgtttggagtggtagaaatct; TBP: Fw TCAAACCCAGAATTGTTCTCCTTAT; Rev CCTGAATCCCTTTAGAATAGGGTAGA; BNIP3: Fw cctgtcgcagttgggttc; Rev gaagtgcagttctacccaggag.
Western blot analysis
For the protein extraction cells were lysed in RIPA buffer with protease inhibitor cocktail tablets Complete, EDTA-free (Roche) and phosphatase inhibitor cocktail tablets PhosSTOP (Roche). Lysate was measured for protein concentration by using Bio-RAD Protein Assay (Bio-RAD), then mixed with Laemmly loading buffer, and 100 ug of proteins were loaded on 10% SDS-PAGE, and then transferred to polyvinylidene difluoride blotting membranes (Amersham, GE Healthcare). Membranes were blocked for 1 hour in 5% (m/vol) dry milk dissolved in PBS with 1% (vol/vol) Tween-20 (PBST); incubated with primary antibodies overnight and with secondary ones, conjugated with horseradish peroxidase, for 1 hour. Antibodies were diluted in 5% dry milk in PBST: anti-HIF1α 1:250 (Novus Biologicals), anti-HA 1:1000 (Covance), anti-GAPDH 1:40000 (Sigma), anti-p21 1:1000 (Santa Cruz Biotechnology), anti-BNIP3 1:600 (Abcam), anti-β-tubulin 1:3000 (Santa Cruz Biotechnology), anti-P73 1:2000 (Bethyl). SuperSignal West Dura Chemiluminescenr Substrate (Thermo Scientific) was used to detect signal on membranes.
Cell transfection
For TAp73 overexpression in H1299 cell line 1.2 E6 cells were seeded per 10 cm dish 24 h before transfection. Transfection was performed with 10 ug DNA (pcDNA empty or pcDNA with HA-TAp73) per 10 cm dish using Lipofectamine 2000 Reagent (Invitrogen). Cells were collected 24 h after transfection.
For TAp73 knockdown in H1299 cell line 1.2 E6 cells were seeded per 10 cm dish 24 h before transfection. Transfection was performed using 50 nM siRNA (control siRNA (Ambion) or siTAp73 (Ambion)) and Lipofectamine RNAiMAX (Invitrogen). Each dish was split in two 24 h after transfection; cells were collected 48 h and 72 h after transfection.
For luciferase assay H1299 cells were seeded 20 h before transfection in 12-well plates, 1.5 E5 cells per well. Transfection was carried out by means of Lipofectamine 2000 (Invitrogen). Cells were cotransfected with 0.05 ng/well pcDNA with HA-TAp73 plasmid or empty pcDNA plasmid, 1 ug/well pRL-cytomegalovirus vector and 800 ng/well BNIP3 promoter luciferase reporter vector or p21 promoter luciferase reporter vector.
Luciferase assay
Cells were lyzed 20 h after transfection, and Firefly luciferase activity was measured, normalized to Renilla luciferase activity with Dual-Glo Luciferase Assay System (Promega), in accordance with Dual-Glo Luciferase Assay System protocol. Light emission over 1s was measured with luminometer.
Chromatin immunoprecipitation assay
SaOS2-Tet-On cell line was used for ChIP assay. TAp73 overexpression for 24 h was achieved by doxycycline treatment. Then cells were collected, fixed in 37% formaldehyde, and subjected to sonication for DNA shearing. Chromatin was immunoprecipitated with anti-HA antibodies (Covance) or unspecific immunoglobulin G (IgG) antibodies (Invitrogen) with a ChIP assay Kit (Invitrogen), and the promoter region, containing potential p73 response element, was amplified using the designed BNIP3 promoter primers. For positive control p21 promoter primers were used. The sequences of BNIP3-ChIP primers are following: 5′ -AGCGTTTCTGGGGCGCACCTTG- 3′ and 5′ -GGGACTGGGAGGCACTTTTCAGAGGA- 3′.
Bioinformatic analyses
By using Oncomine®database and Oncomine®Research Edition (available via Internet https://www.oncomine.org/resource/main.html) we gained access to Hou Lung dataset, analyzed it for BNIP3 expression and compared BNIP3 expression in normal lung with expression in large cell carcinoma, squamous cell lung carcinoma or lung adenocarcinoma.
Gene expression data set GSE4573 was downloaded. Patients were divided in 2 cohorts, in accordance to level of the BNIP3 expression. Kaplan-Meier curves, demonstrating survival, were built up for both cohorts. P-value is measured by Students t-test.Citation119,120
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
1044178_Fig_S1.pdf
Download PDF (146.8 KB)Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- Rufini A, Agostini M, Grespi F, Tomasini R, Sayan BS, Niklison-Chirou MV, Conforti F, Velletri T, Mastino A, Mak TW, et al. p73 in Cancer. Genes Cancer 2011; 2:491-502; PMID:21779517; http://dx.doi.org/10.1177/1947601911408890
- Engelmann D, Meier C, Alla V, Putzer BM. A balancing act: orchestrating amino-truncated and full-length p73 variants as decisive factors in cancer progression. Oncogene 2014; 0; PMID:25381823.
- Grespi F, Amelio I, Tucci P, Annicchiarico-Petruzzelli M, Melino G. Tissue-specific expression of p73 C-terminal isoforms in mice. Cell Cycle 2012; 11:4474-83; PMID:23159862; http://dx.doi.org/10.4161/cc.22787
- Conforti F, Yang AL, Agostini M, Rufini A, Tucci P, Nicklison-Chirou MV, Grespi F, Velletri T, Knight RA, Melino G, et al. Relative expression of TAp73 and DeltaNp73 isoforms. Aging 2012; 4:202-5; PMID:22388545
- Luh LM, Kehrloesser S, Deutsch GB, Gebel J, Coutandin D, Schafer B, Agostini M, Melino G, Dotsch V. Analysis of the oligomeric state and transactivation potential of TAp73alpha. Cell Death Differ 2013; 20:1008-16; PMID:23538419; http://dx.doi.org/10.1038/cdd.2013.23
- Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nat Rev Cancer 2009; 9:749-58; PMID:19776744; http://dx.doi.org/10.1038/nrc2723
- Belyi VA, Levine AJ. One billion years of p53/p63/p73 evolution. Proc Natl Acad Sci U S A 2009; 106:17609-10; PMID:19826090; http://dx.doi.org/10.1073/pnas.0910634106
- Levine AJ, Tomasini R, McKeon FD, Mak TW, Melino G. The p53 family: guardians of maternal reproduction. Nat Rev Mol Cell Biol 2011; 12:259-65; PMID:21427767; http://dx.doi.org/10.1038/nrm3086
- Nair BC, Krishnan SR, Sareddy GR, Mann M, Xu B, Natarajan M, Hasty P, Brann D, Tekmal RR, Vadlamudi RK. Proline, glutamic acid and leucine-rich protein-1 is essential for optimal p53-mediated DNA damage response. Cell Death Differ 2014; 21:1409-18; PMID:24786831; http://dx.doi.org/10.1038/cdd.2014.55
- Dashzeveg N, Taira N, Lu ZG, Kimura J, Yoshida K. Palmdelphin, a novel target of p53 with Ser46 phosphorylation, controls cell death in response to DNA damage. Cell Death Dis 2014; 5:e1221; PMID:24810057; http://dx.doi.org/10.1038/cddis.2014.176
- Zambetti GP. Expanding the reach of the p53 tumor suppressor network. Cell Death Differ 2014; 21:505-6; PMID:24608846; http://dx.doi.org/10.1038/cdd.2014.13
- Zaccara S, Tebaldi T, Pederiva C, Ciribilli Y, Bisio A, Inga A. p53-directed translational control can shape and expand the universe of p53 target genes. Cell Death Differ 2014; 21:1522-34; PMID:24926617; http://dx.doi.org/10.1038/cdd.2014.79
- Bisio A, Zamborszky J, Zaccara S, Lion M, Tebaldi T, Sharma V, Raimondi I, Alessandrini F, Ciribilli Y, Inga A. Cooperative interactions between p53 and NFkappaB enhance cell plasticity. Oncotarget 2014; 5:12111-25; PMID:25401416
- Hu W, Feng Z, Teresky AK, Levine AJ. p53 regulates maternal reproduction through LIF. Nature 2007; 450:721-4; PMID:18046411; http://dx.doi.org/10.1038/nature05993
- Inoue S, Tomasini R, Rufini A, Elia AJ, Agostini M, Amelio I, Cescon D, Dinsdale D, Zhou L, Harris IS, et al. TAp73 is required for spermatogenesis and the maintenance of male fertility. Proc Natl Acad Sci U S A 2014; 111:1843-8; PMID:24449892; http://dx.doi.org/10.1073/pnas.1323416111
- Suh EK, Yang A, Kettenbach A, Bamberger C, Michaelis AH, Zhu Z, Elvin JA, Bronson RT, Crum CP, McKeon F. p63 protects the female germ line during meiotic arrest. Nature 2006; 444:624-8; PMID:17122775; http://dx.doi.org/10.1038/nature05337
- Van Nostrand JL, Brady CA, Jung H, Fuentes DR, Kozak MM, Johnson TM, Lin CY, Lin CJ, Swiderski DL, Vogel H, et al. Inappropriate p53 activation during development induces features of CHARGE syndrome. Nature 2014; 514:228-32; PMID:25119037
- Kang HJ, Feng Z, Sun Y, Atwal G, Murphy ME, Rebbeck TR, Rosenwaks Z, Levine AJ, Hu W. Single-nucleotide polymorphisms in the p53 pathway regulate fertility in humans. Proc Natl Acad Sci U S A 2009; 106:9761-6; PMID:19470478; http://dx.doi.org/10.1073/pnas.0904280106
- Hu W, Feng Z, Levine AJ. The regulation of human reproduction by p53 and its pathway. Cell Cycle 2009; 8:3621-2; PMID:19884797; http://dx.doi.org/10.4161/cc.8.22.9938
- Celardo I, Antonov A, Amelio I, Annicchiarico-Petruzzelli M, Melino G. p63 transcriptionally regulates the expression of matrix metallopeptidase 13. Oncotarget 2014; 5:1279-89; PMID:24658133
- Amelio I, Grespi F, Annicchiarico-Petruzzelli M, Melino G. p63 the guardian of human reproduction. Cell Cycle 2012; 11:4545-51; PMID:23165243; http://dx.doi.org/10.4161/cc.22819
- Shetzer Y, Kagan S, Koifman G, Sarig R, Kogan-Sakin I, Charni M, Kaufman T, Zapatka M, Molchadsky A, Rivlin N, et al. The onset of p53 loss of heterozygosity is differentially induced in various stem cell types and may involve the loss of either allele. Cell Death Differ 2014; 21:1419-31; PMID:24832469; http://dx.doi.org/10.1038/cdd.2014.57
- Buckley NE, D'Costa Z, Kaminska M, Mullan PB. S100A2 is a BRCA1/p63 coregulated tumour suppressor gene with roles in the regulation of mutant p53 stability. Cell Death Dis 2014; 5:e1070; PMID:24556685; http://dx.doi.org/10.1038/cddis.2014.31
- Mello SS, Attardi LD. Not all p53 gain-of-function mutants are created equal. Cell Death Differ 2013; 20:855-7; PMID:23749181; http://dx.doi.org/10.1038/cdd.2013.53
- Rufini A, Tucci P, Celardo I, Melino G. Senescence and aging: the critical roles of p53. Oncogene 2013; 32:5129-43; PMID:23416979; http://dx.doi.org/10.1038/onc.2012.640
- Li T, Kon N, Jiang L, Tan M, Ludwig T, Zhao Y, Baer R, Gu W. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell 2012; 149:1269-83; PMID:22682249; http://dx.doi.org/10.1016/j.cell.2012.04.026
- Wang J, Qian J, Hu Y, Kong X, Chen H, Shi Q, Jiang L, Wu C, Zou W, Chen Y, et al. ArhGAP30 promotes p53 acetylation and function in colorectal cancer. Nat Commun 2014; 5:4735; PMID:25156493; http://dx.doi.org/10.1038/ncomms5735
- Li D, Yallowitz A, Ozog L, Marchenko N. A gain-of-function mutant p53-HSF1 feed forward circuit governs adaptation of cancer cells to proteotoxic stress. Cell Death Dis 2014; 5:e1194; PMID:24763051; http://dx.doi.org/10.1038/cddis.2014.158
- Weilbacher A, Gutekunst M, Oren M, Aulitzky WE, van der Kuip H. RITA can induce cell death in p53-defective cells independently of p53 function via activation of JNK/SAPK and p38. Cell Death Dis 2014; 5:e1318; PMID:25010984; http://dx.doi.org/10.1038/cddis.2014.284
- Blasius M, Bartek J. ATM targets hnRNPK to control p53. Cell Cycle 2013; 12:1162-3; PMID:23549171; http://dx.doi.org/10.4161/cc.24485
- Xu J, Wang J, Hu Y, Qian J, Xu B, Chen H, Zou W, Fang JY. Unequal prognostic potentials of p53 gain-of-function mutations in human cancers associate with drug-metabolizing activity. Cell Death Dis 2014; 5:e1108; PMID:24603336; http://dx.doi.org/10.1038/cddis.2014.75
- Celardo I, Grespi F, Antonov A, Bernassola F, Garabadgiu AV, Melino G, Amelio I. Caspase-1 is a novel target of p63 in tumor suppression. Cell Death Dis 2013; 4:e645; PMID:23703390; http://dx.doi.org/10.1038/cddis.2013.175
- Guerrieri F, Piconese S, Lacoste C, Schinzari V, Testoni B, Valogne Y, Gerbal-Chaloin S, Samuel D, Brechot C, Faivre J, et al. The sodium/iodide symporter NIS is a transcriptional target of the p53-family members in liver cancer cells. Cell Death Dis 2013; 4:e807; PMID:24052075; http://dx.doi.org/10.1038/cddis.2013.302
- Del Nagro CJ, Choi J, Xiao Y, Rangell L, Mohan S, Pandita A, Zha J, Jackson PK, O'Brien T. Chk1 inhibition in p53-deficient cell lines drives rapid chromosome fragmentation followed by caspase-independent cell death. Cell Cycle 2014; 13:303-14; PMID:24247149; http://dx.doi.org/10.4161/cc.27055
- Rufini S, Lena AM, Cadot B, Mele S, Amelio I, Terrinoni A, Desideri A, Melino G, Candi E. The sterile α-motif (SAM) domain of p63 binds in vitro monoasialoganglioside (GM1) micelles. Biochem Pharmacol 2011; 82:1262-8; PMID:21820419; http://dx.doi.org/10.1016/j.bcp.2011.07.087
- Fan YH, Cheng J, Vasudevan SA, Dou J, Zhang H, Patel RH, Ma IT, Rojas Y, Zhao Y, Yu Y, et al. USP7 inhibitor P22077 inhibits neuroblastoma growth via inducing p53-mediated apoptosis. Cell Death Dis 2013; 4:e867; PMID:24136231; http://dx.doi.org/10.1038/cddis.2013.400
- Melino G, De Laurenzi V, Vousden KH. p73: Friend or foe in tumorigenesis. Nat Rev Cancer 2002; 2:605-15; PMID:12154353; http://dx.doi.org/10.1038/nrc861
- Tomasini R, Mak TW, Melino G. The impact of p53 and p73 on aneuploidy and cancer. Trends Cell Biol 2008; 18:244-52; PMID:18406616; http://dx.doi.org/10.1016/j.tcb.2008.03.003
- Fatt MP, Cancino GI, Miller FD, Kaplan DR. p63 and p73 coordinate p53 function to determine the balance between survival, cell death, and senescence in adult neural precursor cells. Cell Death Differ 2014; 21:1546-59; PMID:24809925; http://dx.doi.org/10.1038/cdd.2014.61
- Rosenbluth JM, Pietenpol JA. The jury is in: p73 is a tumor suppressor after all. Genes Dev 2008; 22:2591-5; PMID:18832062; http://dx.doi.org/10.1101/gad.1727408
- Amelio I, Inoue S, Markert EK, Levine AJ, Knight RA, Mak TW, Melino G. TAp73 opposes tumor angiogenesis by promoting hypoxia-inducible factor 1alpha degradation. Proc Natl Acad Sci U S A 2015; 112:226-31; PMID:25535359; http://dx.doi.org/10.1073/pnas.1410609111
- Stantic M, Sakil HA, Zirath H, Fang T, Sanz G, Fernandez-Woodbridge A, Marin A, Susanto E, Mak TW, Arsenian Henriksson M, et al. TAp73 suppresses tumor angiogenesis through repression of proangiogenic cytokines and HIF-1alpha activity. Proc Natl Acad Sci U S A 2015; 112:220-5; PMID:25535357; http://dx.doi.org/10.1073/pnas.1421697112
- Wilhelm MT, Rufini A, Wetzel MK, Tsuchihara K, Inoue S, Tomasini R, Itie-Youten A, Wakeham A, Arsenian-Henriksson M, Melino G, et al. Isoform-specific p73 knockout mice reveal a novel role for delta Np73 in the DNA damage response pathway. Genes Dev 2010; 24:549-60; PMID:20194434; http://dx.doi.org/10.1101/gad.1873910
- Sayan BS, Yang AL, Conforti F, Tucci P, Piro MC, Browne GJ, Agostini M, Bernardini S, Knight RA, Mak TW, et al. Differential control of TAp73 and DeltaNp73 protein stability by the ring finger ubiquitin ligase PIR2. Proc Natl Acad Sci U S A 2010; 107:12877-82; PMID:20615966; http://dx.doi.org/10.1073/pnas.0911828107
- Venkatanarayan A, Raulji P, Norton W, Chakravarti D, Coarfa C, Su X, Sandur SK, Ramirez MS, Lee J, Kingsley CV, et al. IAPP-driven metabolic reprogramming induces regression of p53-deficient tumours in vivo. Nature 2015; 517:626-30; PMID:25409149; http://dx.doi.org/10.1038/nature13910
- Di C, Yang L, Zhang H, Ma X, Zhang X, Sun C, Li H, Xu S, An L, Li X, et al. Mechanisms, function and clinical applications of DNp73. Cell Cycle 2013; 12:1861-7; PMID:23708520; http://dx.doi.org/10.4161/cc.24967
- Fillippovich I, Sorokina N, Gatei M, Haupt Y, Hobson K, Moallem E, Spring K, Mould M, McGuckin MA, Lavin MF, et al. Transactivation-deficient p73alpha (p73Deltaexon2) inhibits apoptosis and competes with p53. Oncogene 2001; 20:514-22; PMID:11313982; http://dx.doi.org/10.1038/sj.onc.1204118
- Stiewe T, Theseling CC, Putzer BM. Transactivation-deficient Delta TA-p73 inhibits p53 by direct competition for DNA binding: implications for tumorigenesis. J Biol Chem 2002; 277:14177-85; PMID:11844800; http://dx.doi.org/10.1074/jbc.M200480200
- Stiewe T, Zimmermann S, Frilling A, Esche H, Putzer BM. Transactivation-deficient DeltaTA-p73 acts as an oncogene. Cancer Res 2002; 62:3598-602; PMID:12097259
- Zaika AI, Slade N, Erster SH, Sansome C, Joseph TW, Pearl M, Chalas E, Moll UM. DeltaNp73, a dominant-negative inhibitor of wild-type p53 and TAp73, is up-regulated in human tumors. J Exp Med 2002; 196:765-80; PMID:12235210; http://dx.doi.org/10.1084/jem.20020179
- Killick R, Niklison-Chirou M, Tomasini R, Bano D, Rufini A, Grespi F, Velletri T, Tucci P, Sayan BS, Conforti F, et al. p73: a multifunctional protein in neurobiology. Mol Neurobiol 2011; 43:139-46; PMID:21380933; http://dx.doi.org/10.1007/s12035-011-8172-6
- Agostini M, Tucci P, Killick R, Candi E, Sayan BS, Rivetti di Val Cervo P, Nicotera P, McKeon F, Knight RA, Mak TW, et al. Neuronal differentiation by TAp73 is mediated by microRNA-34a regulation of synaptic protein targets. Proc Natl Acad Sci U S A 2011; 108:21093-8; PMID:22160687; http://dx.doi.org/10.1073/pnas.1112061109
- Alexandrova EM, Talos F, Moll UM. p73 is dispensable for commitment to neural stem cell fate, but is essential for neural stem cell maintenance and for blocking premature differentiation. Cell Death Differ 2013; 20:368; PMID:23099852; http://dx.doi.org/10.1038/cdd.2012.134
- Niklison-Chirou MV, Steinert JR, Agostini M, Knight RA, Dinsdale D, Cattaneo A, Mak TW, Melino G. TAp73 knockout mice show morphological and functional nervous system defects associated with loss of p75 neurotrophin receptor. Proc Natl Acad Sci U S A 2013; 110:18952-7; PMID:24190996; http://dx.doi.org/10.1073/pnas.1221172110
- Jeong K, Kim H, Kim K, Kim SJ, Hahn BS, Jahng GH, Yoon KS, Kim SS, Ha J, Kang I, et al. Cyclophilin B is involved in p300-mediated degradation of CHOP in tumor cell adaptation to hypoxia. Cell Death Differ 2014; 21:438-50; PMID:24270407; http://dx.doi.org/10.1038/cdd.2013.164
- Carmeliet P, Dor Y, Herbert JM, Fukumura D, Brusselmans K, Dewerchin M, Neeman M, Bono F, Abramovitch R, Maxwell P, et al. Role of HIF-1alpha in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature 1998; 394:485-90; PMID:9697772; http://dx.doi.org/10.1038/28867
- Leek RD, Talks KL, Pezzella F, Turley H, Campo L, Brown NS, Bicknell R, Taylor M, Gatter KC, Harris AL. Relation of hypoxia-inducible factor-2 α (HIF-2 α) expression in tumor-infiltrative macrophages to tumor angiogenesis and the oxidative thymidine phosphorylase pathway in Human breast cancer. Cancer Res 2002; 62:1326-9; PMID:11888900
- Vincent KA, Feron O, Kelly RA. Harnessing the response to tissue hypoxia: HIF-1 α and therapeutic angiogenesis. Trends Cardiovasc Med 2002; 12:362-7; PMID:12536123; http://dx.doi.org/10.1016/S1050-1738(02)00186-X
- Krick S, Hanze J, Eul B, Savai R, Seay U, Grimminger F, Lohmeyer J, Klepetko W, Seeger W, Rose F. Hypoxia-driven proliferation of human pulmonary artery fibroblasts: cross-talk between HIF-1alpha and an autocrine angiotensin system. FASEB J 2005; 19:857-9; PMID:15718424
- Sun CL, Kim E, Crowder CM. Delayed innocent bystander cell death following hypoxia in Caenorhabditis elegans. Cell Death Differ 2014; 21:557-67; PMID:24317200; http://dx.doi.org/10.1038/cdd.2013.176
- Feng Y, Zhu M, Dangelmajer S, Lee YM, Wijesekera O, Castellanos CX, Denduluri A, Chaichana KL, Li Q, Zhang H, et al. Hypoxia-cultured human adipose-derived mesenchymal stem cells are non-oncogenic and have enhanced viability, motility, and tropism to brain cancer. Cell Death Dis 2014; 5:e1567; PMID:25501828; http://dx.doi.org/10.1038/cddis.2014.521
- Gordan JD, Bertout JA, Hu CJ, Diehl JA, Simon MC. HIF-2alpha promotes hypoxic cell proliferation by enhancing c-myc transcriptional activity. Cancer Cell 2007; 11:335-47; PMID:17418410; http://dx.doi.org/10.1016/j.ccr.2007.02.006
- Chen Q, Xu J, Li L, Li H, Mao S, Zhang F, Zen K, Zhang CY, Zhang Q. MicroRNA-23a/b and microRNA-27a/b suppress Apaf-1 protein and alleviate hypoxia-induced neuronal apoptosis. Cell Death Dis 2014; 5:e1132; PMID:24651435; http://dx.doi.org/10.1038/cddis.2014.92
- Gordan JD, Thompson CB, Simon MC. HIF and c-Myc: sibling rivals for control of cancer cell metabolism and proliferation. Cancer Cell 2007; 12:108-13; PMID:17692803; http://dx.doi.org/10.1016/j.ccr.2007.07.006
- Luther J, Ubieta K, Hannemann N, Jimenez M, Garcia M, Zech C, Schett G, Wagner EF, Bozec A. Fra-2/AP-1 controls adipocyte differentiation and survival by regulating PPARgamma and hypoxia. Cell Death Differ 2014; 21:655-64; PMID:24464219; http://dx.doi.org/10.1038/cdd.2013.198
- Chen L, Qiu JH, Zhang LL, Luo XD. Adrenomedullin promotes human endothelial cell proliferation via HIF-1alpha. Mol Cell Biochem 2012; 365:263-73; PMID:22406979; http://dx.doi.org/10.1007/s11010-012-1267-1
- Volm M, Koomagi R. Hypoxia-inducible factor (HIF-1) and its relationship to apoptosis and proliferation in lung cancer. Anticancer Res 2000; 20:1527-33; PMID:10928066
- Raz S, Sheban D, Gonen N, Stark M, Berman B, Assaraf YG. Severe hypoxia induces complete antifolate resistance in carcinoma cells due to cell cycle arrest. Cell Death Dis 2014; 5:e1067; PMID:24556682; http://dx.doi.org/10.1038/cddis.2014.39
- Huang CY, Kuo WT, Huang YC, Lee TC, Yu LC. Resistance to hypoxia-induced necroptosis is conferred by glycolytic pyruvate scavenging of mitochondrial superoxide in colorectal cancer cells. Cell Death Dis 2013; 4:e622; PMID:23640464; http://dx.doi.org/10.1038/cddis.2013.149
- Son TW, Yun SP, Yong MS, Seo BN, Ryu JM, Youn HY, Oh YM, Han HJ. Netrin-1 protects hypoxia-induced mitochondrial apoptosis through HSP27 expression via DCC- and integrin alpha6beta4-dependent Akt, GSK-3beta, and HSF-1 in mesenchymal stem cells. Cell Death Dis 2013; 4:e563; PMID:23538444; http://dx.doi.org/10.1038/cddis.2013.94
- Koivunen P, Hirsila M, Remes AM, Hassinen IE, Kivirikko KI, Myllyharju J. Inhibition of hypoxia-inducible factor (HIF) hydroxylases by citric acid cycle intermediates: possible links between cell metabolism and stabilization of HIF. J Biol Chem 2007; 282:4524-32; PMID:17182618; http://dx.doi.org/10.1074/jbc.M610415200
- Esteban MA, Maxwell PH. HIF, a missing link between metabolism and cancer. Nat Med 2005; 11:1047-8; PMID:16211034; http://dx.doi.org/10.1038/nm1005-1047
- Lall R, Ganapathy S, Yang M, Xiao S, Xu T, Su H, Shadfan M, Asara JM, Ha CS, Ben-Sahra I, et al. Low-dose radiation exposure induces a HIF-1-mediated adaptive and protective metabolic response. Cell Death Differ 2014; 21:836-44; PMID:24583639; http://dx.doi.org/10.1038/cdd.2014.24
- Guo K, Searfoss G, Krolikowski D, Pagnoni M, Franks C, Clark K, Yu KT, Jaye M, Ivashchenko Y. Hypoxia induces the expression of the pro-apoptotic gene BNIP3. Cell Death Differ 2001; 8:367-76; PMID:11550088; http://dx.doi.org/10.1038/sj.cdd.4400810
- Kothari S, Cizeau J, McMillan-Ward E, Israels SJ, Bailes M, Ens K, Kirshenbaum LA, Gibson SB. BNIP3 plays a role in hypoxic cell death in human epithelial cells that is inhibited by growth factors EGF and IGF. Oncogene 2003; 22:4734-44; PMID:12879018; http://dx.doi.org/10.1038/sj.onc.1206666
- Maes H, Van Eygen S, Krysko DV, Vandenabeele P, Nys K, Rillaerts K, Garg AD, Verfaillie T, Agostinis P. BNIP3 supports melanoma cell migration and vasculogenic mimicry by orchestrating the actin cytoskeleton. Cell Death Dis 2014; 5:e1127; PMID:24625986; http://dx.doi.org/10.1038/cddis.2014.94
- Fan Q, Gao F, Zhang L, Christopher TA, Lopez BL, Ma XL. Nitrate tolerance aggravates postischemic myocardial apoptosis and impairs cardiac functional recovery after ischemia. Apoptosis 2005; 10:1235-42; PMID:16215686; http://dx.doi.org/10.1007/s10495-005-1455-5
- Roberts DJ, Miyamoto S. Hexokinase II integrates energy metabolism and cellular protection: Akting on mitochondria and TORCing to autophagy. Cell Death Differ 2015; 22:248-57; PMID:25323588; http://dx.doi.org/10.1038/cdd.2014.173
- Diwan A, Krenz M, Syed FM, Wansapura J, Ren X, Koesters AG, Li H, Kirshenbaum LA, Hahn HS, Robbins J, et al. Inhibition of ischemic cardiomyocyte apoptosis through targeted ablation of Bnip3 restrains postinfarction remodeling in mice. J Clin Invest 2007; 117:2825-33; PMID:17909626; http://dx.doi.org/10.1172/JCI32490
- Seo Y, Ji YW, Lee SM, Shim J, Noh H, Yeo A, Park C, Park MS, Chang EJ, Lee HK. Activation of HIF-1alpha (hypoxia inducible factor-1alpha) prevents dry eye-induced acinar cell death in the lacrimal gland. Cell Death Dis 2014; 5:e1309; PMID:24967971; http://dx.doi.org/10.1038/cddis.2014.260
- Bianchi P, Kunduzova O, Masini E, Cambon C, Bani D, Raimondi L, Seguelas MH, Nistri S, Colucci W, Leducq N, et al. Oxidative stress by monoamine oxidase mediates receptor-independent cardiomyocyte apoptosis by serotonin and postischemic myocardial injury. Circulation 2005; 112:3297-305; PMID:16286591; http://dx.doi.org/10.1161/CIRCULATIONAHA.104.528133
- Qi Y, Liu J, Saadat S, Tian X, Han Y, Fong GH, Pandolfi PP, Lee LY, Li S. PTEN induces apoptosis and cavitation via HIF-2-dependent Bnip3 upregulation during epithelial lumen formation. Cell Death Differ 2015; 22(5):875-84.
- Boyd JM, Malstrom S, Subramanian T, Venkatesh LK, Schaeper U, Elangovan B, D'Sa-Eipper C, Chinnadurai G. Adenovirus E1B 19 kDa and Bcl-2 proteins interact with a common set of cellular proteins. Cell 1994; 79:341-51; PMID:7954800; http://dx.doi.org/10.1016/0092-8674(94)90202-X
- Schmidt-Kastner R, Aguirre-Chen C, Kietzmann T, Saul I, Busto R, Ginsberg MD. Nuclear localization of the hypoxia-regulated pro-apoptotic protein BNIP3 after global brain ischemia in the rat hippocampus. Brain Res 2004; 1001:133-42; PMID:14972662; http://dx.doi.org/10.1016/j.brainres.2003.11.065
- Burton TR, Eisenstat DD, Gibson SB. BNIP3 (Bcl-2 19 kDa interacting protein) acts as transcriptional repressor of apoptosis-inducing factor expression preventing cell death in human malignant gliomas. J Neurosci 2009; 29:4189-99; PMID:19339613; http://dx.doi.org/10.1523/JNEUROSCI.5747-08.2009
- Hanna RA, Quinsay MN, Orogo AM, Giang K, Rikka S, Gustafsson AB. Microtubule-associated protein 1 light chain 3 (LC3) interacts with Bnip3 protein to selectively remove endoplasmic reticulum and mitochondria via autophagy. J Biol Chem 2012; 287:19094-104; PMID:22505714; http://dx.doi.org/10.1074/jbc.M111.322933
- Quinsay MN, Thomas RL, Lee Y, Gustafsson AB. Bnip3-mediated mitochondrial autophagy is independent of the mitochondrial permeability transition pore. Autophagy 2010; 6:855-62; PMID:20668412; http://dx.doi.org/10.4161/auto.6.7.13005
- Kubli DA, Ycaza JE, Gustafsson AB. Bnip3 mediates mitochondrial dysfunction and cell death through Bax and Bak. Biochem J 2007; 405:407-15; PMID:17447897; http://dx.doi.org/10.1042/BJ20070319
- Burton TR, Henson ES, Azad MB, Brown M, Eisenstat DD, Gibson SB. BNIP3 acts as transcriptional repressor of death receptor-5 expression and prevents TRAIL-induced cell death in gliomas. Cell Death Dis 2013; 4:e587; PMID:23579274; http://dx.doi.org/10.1038/cddis.2013.100
- Feng X, Liu X, Zhang W, Xiao W. p53 directly suppresses BNIP3 expression to protect against hypoxia-induced cell death. EMBO J 2011; 30:3397-415; PMID:21792176; http://dx.doi.org/10.1038/emboj.2011.248
- Tomasini R, Tsuchihara K, Wilhelm M, Fujitani M, Rufini A, Cheung CC, Khan F, Itie-Youten A, Wakeham A, Tsao MS, et al. TAp73 knockout shows genomic instability with infertility and tumor suppressor functions. Genes Dev 2008; 22:2677-91; PMID:18805989; http://dx.doi.org/10.1101/gad.1695308
- Flores ER, Sengupta S, Miller JB, Newman JJ, Bronson R, Crowley D, Yang A, McKeon F, Jacks T. Tumor predisposition in mice mutant for p63 and p73: evidence for broader tumor suppressor functions for the p53 family. Cancer Cell 2005; 7:363-73; PMID:15837625; http://dx.doi.org/10.1016/j.ccr.2005.02.019
- Vakkila J, Lotze MT. Inflammation and necrosis promote tumour growth. Nat Rev Immunol 2004; 4:641-8; PMID:15286730; http://dx.doi.org/10.1038/nri1415
- Rosenfeldt MT, Ryan KM. The role of autophagy in tumour development and cancer therapy. Expert Rev Mol Med 2009; 11:e36; PMID:19951459; http://dx.doi.org/10.1017/S1462399409001306
- Peng X, Gong F, Chen Y, Jiang Y, Liu J, Yu M, Zhang S, Wang M, Xiao G, Liao H. Autophagy promotes paclitaxel resistance of cervical cancer cells: involvement of Warburg effect activated hypoxia-induced factor 1-α-mediated signaling. Cell Death Dis 2014; 5:e1367; PMID:25118927; http://dx.doi.org/10.1038/cddis.2014.297
- Amelio I, Melino G, Knight RA. Cell death pathology: cross-talk with autophagy and its clinical implications. Biochem Biophys Res Commun 2011; 414:277-81; PMID:21963447; http://dx.doi.org/10.1016/j.bbrc.2011.09.080
- Liu K, Shi Y, Guo X, Wang S, Ouyang Y, Hao M, Liu D, Qiao L, Li N, Zheng J, et al. CHOP mediates ASPP2-induced autophagic apoptosis in hepatoma cells by releasing Beclin-1 from Bcl-2 and inducing nuclear translocation of Bcl-2. Cell Death Dis 2014; 5:e1323; PMID:25032846; http://dx.doi.org/10.1038/cddis.2014.276
- Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer 2007; 7:961-7; PMID:17972889; http://dx.doi.org/10.1038/nrc2254
- Ashrafi G, Schwarz TL. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ 2013; 20:31-42; PMID:22743996; http://dx.doi.org/10.1038/cdd.2012.81
- Liu T, Roh SE, Woo JA, Ryu H, Kang DE. Cooperative role of RanBP9 and P73 in mitochondria-mediated apoptosis. Cell Death Dis 2013; 4:e476; PMID:23348590; http://dx.doi.org/10.1038/cddis.2012.203
- Rufini A, Niklison-Chirou MV, Inoue S, Tomasini R, Harris IS, Marino A, Federici M, Dinsdale D, Knight RA, Melino G, et al. TAp73 depletion accelerates aging through metabolic dysregulation. Genes Dev 2012; 26:2009-14; PMID:22987635; http://dx.doi.org/10.1101/gad.197640.112
- He Z, Liu H, Agostini M, Yousefi S, Perren A, Tschan MP, Mak TW, Melino G, Simon HU. p73 regulates autophagy and hepatocellular lipid metabolism through a transcriptional activation of the ATG5 gene. Cell Death Differ 2013; 20:1415-24; PMID:23912709; http://dx.doi.org/10.1038/cdd.2013.104
- Sayan AE, Sayan BS, Gogvadze V, Dinsdale D, Nyman U, Hansen TM, Zhivotovsky B, Cohen GM, Knight RA, Melino G. P73 and caspase-cleaved p73 fragments localize to mitochondria and augment TRAIL-induced apoptosis. Oncogene 2008; 27:4363-72; PMID:18362891; http://dx.doi.org/10.1038/onc.2008.64
- Adamovich Y, Adler J, Meltser V, Reuven N, Shaul Y. AMPK couples p73 with p53 in cell fate decision. Cell Death Differ 2014; 21:1451-9; PMID:24874608; http://dx.doi.org/10.1038/cdd.2014.60
- Su X, Gi YJ, Chakravarti D, Chan IL, Zhang A, Xia X, Tsai KY, Flores ER. TAp63 is a master transcriptional regulator of lipid and glucose metabolism. Cell Metabol 2012; 16:511-25; PMID:23040072; http://dx.doi.org/10.1016/j.cmet.2012.09.006
- Sahin E, DePinho RA. Axis of ageing: telomeres, p53 and mitochondria. Nat Rev Mol Cell Biol 2012; 13:397-404; PMID:22588366; http://dx.doi.org/10.1038/nrm3352
- Zhao Y, Chaiswing L, Velez JM, Batinic-Haberle I, Colburn NH, Oberley TD, St Clair DK. p53 translocation to mitochondria precedes its nuclear translocation and targets mitochondrial oxidative defense protein-manganese superoxide dismutase. Cancer Res 2005; 65:3745-50; PMID:15867370; http://dx.doi.org/10.1158/0008-5472.CAN-04-3835
- Bergeaud M, Mathieu L, Guillaume A, Moll UM, Mignotte B, Le Floch N, Vayssiere JL, Rincheval V. Mitochondrial p53 mediates a transcription-independent regulation of cell respiration and interacts with the mitochondrial F(1)F0-ATP synthase. Cell Cycle 2013; 12:2781-93; PMID:23966169; http://dx.doi.org/10.4161/cc.25870
- Amelio I, Antonov AA, Catani MV, Massoud R, Bernassola F, Knight RA, Melino G, Rufini A. TAp73 promotes anabolism. Oncotarget 2014; 5:12820-934; PMID:25514460
- Amelio I, Markert EK, Rufini A, Antonov AV, Sayan BS, Tucci P, Agostini M, Mineo TC, Levine AJ, Melino G. p73 regulates serine biosynthesis in cancer. Oncogene 2014; 33:5039-46; PMID:24186203; http://dx.doi.org/10.1038/onc.2013.456
- D'Alessandro A, Amelio I, Berkers CR, Antonov A, Vousden KH, Melino G, Zolla L. Metabolic effect of TAp63alpha: enhanced glycolysis and pentose phosphate pathway, resulting in increased antioxidant defense. Oncotarget 2014; 5:7722-33; PMID:25229745
- Levine AJ, Puzio-Kuter AM. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science 2010; 330:1340-4; PMID:21127244; http://dx.doi.org/10.1126/science.1193494
- Pan LZ, Ahn DG, Sharif T, Clements D, Gujar SA, Lee PW. The NAD+ synthesizing enzyme nicotinamide mononucleotide adenylyltransferase 2 (NMNAT-2) is a p53 downstream target. Cell Cycle 2014; 13:1041-8; PMID:24552824; http://dx.doi.org/10.4161/cc.28128
- Pfister NT, Yoh KE, Prives C. p53, DNA damage, and NAD+ homeostasis. Cell Cycle 2014; 13:1661-2; PMID:24810733; http://dx.doi.org/10.4161/cc.29151
- Velletri T, Romeo F, Tucci P, Peschiaroli A, Annicchiarico-Petruzzelli M, Niklison-Chirou MV, Amelio I, Knight RA, Mak TW, Melino G, et al. GLS2 is transcriptionally regulated by p73 and contributes to neuronal differentiation. Cell Cycle 2013; 12:3564-73; PMID:24121663; http://dx.doi.org/10.4161/cc.26771
- Giacobbe A, Bongiorno-Borbone L, Bernassola F, Terrinoni A, Markert EK, Levine AJ, Feng Z, Agostini M, Zolla L, Agro AF, et al. p63 regulates glutaminase 2 expression. Cell Cycle 2013; 12:1395-405; PMID:23574722; http://dx.doi.org/10.4161/cc.24478
- Kostecka A, Sznarkowska A, Meller K, Acedo P, Shi Y, Mohammad Sakil HA, Kawiak A, Lion M, Krolicka A, Wilhelm M, et al. JNK-NQO1 axis drives TAp73-mediated tumor suppression upon oxidative and proteasomal stress. Cell Death Dis 2014; 5:e1484; PMID:25341038; http://dx.doi.org/10.1038/cddis.2014.408
- He Z, Simon HU. A novel link between p53 and ROS. Cell Cycle 2013; 12:201-2; PMID:23287470; http://dx.doi.org/10.4161/cc.23418
- Antonov AV. BioProfiling.de: analytical web portal for high-throughput cell biology. Nucleic Acids Res 2011; 39:W323-7; PMID:21609949; http://dx.doi.org/10.1093/nar/gkr372
- Dietmann S, Lee W, Wong P, Rodchenkov I, Antonov AV. CCancer: a bird's eye view on gene lists reported in cancer-related studies. Nucleic Acids Res 2010; 38:W118-23; PMID:20529879; http://dx.doi.org/10.1093/nar/gkq515
