Abstract
Tumor cellular senescence induced by genotoxic treatments has recently been found to be paradoxically linked to the induction of “stemness.” This observation is critical as it directly impinges upon the response of tumors to current chemo-radio-therapy treatment regimens. Previously, we showed that following etoposide (ETO) treatment embryonal carcinoma PA-1 cells undergo a p53-dependent upregulation of OCT4A and p21Cip1 (governing self-renewal and regulating cell cycle inhibition and senescence, respectively). Here we report further detail on the relationship between these and other critical cell-fate regulators. PA-1 cells treated with ETO display highly heterogeneous increases in OCT4A and p21Cip1 indicative of dis-adaptation catastrophe. Silencing OCT4A suppresses p21Cip1, changes cell cycle regulation and subsequently suppresses terminal senescence; p21Cip1-silencing did not affect OCT4A expression or cellular phenotype. SOX2 and NANOG expression did not change following ETO treatment suggesting a dissociation of OCT4A from its pluripotency function. Instead, ETO-induced OCT4A was concomitant with activation of AMPK, a key component of metabolic stress and autophagy regulation. p16ink4a, the inducer of terminal senescence, underwent autophagic sequestration in the cytoplasm of ETO-treated cells, allowing alternative cell fates. Accordingly, failure of autophagy was accompanied by an accumulation of p16ink4a, nuclear disintegration, and loss of cell recovery. Together, these findings imply that OCT4A induction following DNA damage in PA-1 cells, performs a cell stress, rather than self-renewal, function by moderating the expression of p21Cip1, which alongside AMPK helps to then regulate autophagy. Moreover, this data indicates that exhaustion of autophagy, through persistent DNA damage, is the cause of terminal cellular senescence.
Abbreviations
| AMPK | = | AMP-activated protein kinase |
| Baf | = | bafilomycin |
| EC | = | embryonal carcinoma |
| ES | = | embryonic stem |
| ETO | = | Etoposide |
| IF | = | immunofluorescent |
| LC3 | = | microtubule associated protein 1 light chain 3 |
| NT | = | non-treated |
| NT2 | = | NTera 2 |
| ntg | = | non-target |
| pCHK2 | = | phosphorylated CHK2 |
| PI | = | propidium iodide |
| siRNA | = | small interfering RNA |
| shRNA | = | small hairpin RNA |
| Sa-b-gal | = | senescence associated β-galactosidase. |
Introduction
The relationship between cancer cells, normal stem cells, and cancer stem cells represents a question of substantial current interest.Citation1 It has been proposed that transcription networks that confer stem cell properties such as self-renewal, plasticity, or an increased resistance to genotoxic stimuli in normal stem cells may perform a similar function in cancer cells.Citation2 This hypothesis is supported by the growing clinical evidence that expression of key embryonal stem cell (ESC) transcription factors POU1F5 (OCT4A), NANOG and SOX2, are associated with poorer prognosis through tumor resistance, recurrence and progression in a wide variety of cancers.Citation3-9 Furthermore, it has been demonstrated by several groups that ESC transcription factors can be upregulated in response to DNA damage where they likely play a role in regulating survival.Citation10-12
Conversely, accelerated cellular senescence is a phenomenon that has also been shown to be induced by genotoxic treatments of cancer cells.Citation13 Cellular senescence has traditionally been considered a terminal cell fate.Citation13,14 However, more recently it has been shown to be reversible at early stages, at least in tumor cells.Citation15-18 Furthermore, a direct link between senescence and “stemness,” essential cytological characteristics of a stem cell that distinguishes it from ordinary somatic cells, emerged in experiments where pluripotency is induced in normal cells.Citation19,20
The molecular regulators of these processes in normal embryonal development, such as p21Cip1, are slowly becoming discerned.Citation21 One intriguing observation is that embryonal cellular senescence is associated with upregulation of the same pathways which govern the epithelial-mesenchymal transition (EMT).22 This, apparently paradoxical, link between opposites in cell fate provides a challenge for scientific reasoning.
We have previously observed in IMR90 fibroblasts that a pre-senescent phenotype is associated with the appearance of self-renewal and senescence markers coupled to DNA damage.Citation23 We also demonstrated co-incident p53-dependent upregulation of 2 opposing cell fate regulators, p21Cip1 and OCT4A in embryonal carcinoma PA-1 cells treated with Etoposide (ETO).Citation24 We hypothesized that this bi-potential state favors DNA damage repair (DDR) while preventing full commitment to either senescence or self-renewal. In this system, p53 silencing promoted terminal senescence and premature mitosis. Together these data support the presence of a pre-senescent cell state which can arise in response to both senescence and stemness programmes being coactivated in response to genotoxic damage.
In the present study, we asked how key regulators of stemness (OCT4A, SOX2 and NANOG) and senescence (p16inka4a) behave in individual PA-1 cells during the response of ETO-induced DNA damage. Using siRNA silencing approaches we addressed the effect of OCT4A and p21Cip1 expression on each other and subsequent cell fates, determining the role of autophagy and how OCT4A activation impacts on the energy and genomic stress sensor and master metabolic regulator and activator of autophagy AMP-activated protein kinase (AMPK).
Results
Etoposide-treatment elicits a senescence-like phenotype in PA-1 cells
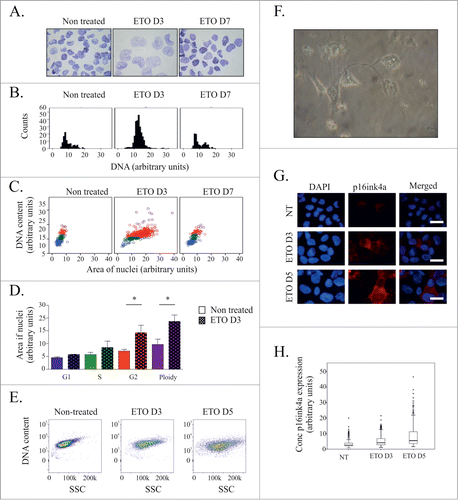
Following ETO treatment many PA-1 cells undergo gradual cell apoptosis and anoikis, while the remainder arrest in G2M and upregulate p53 and p21Cip.Citation24 G2M arrest and nuclear swelling were evident on day 3 after ETO treatment, with restoration of normal cell cycle and nuclear size 4 d later (). Concurrent nuclear area assessments and DNA content measurements demonstrate that the nuclei of ETO treated cells increased in size irrespective of the stage of the cell cycle, but was most evident in G2M and polyploid cells (). Increased nuclear area and DNA content were also accompanied by an increase in cellular granularity, as determined by an increase in side scatter detected by flow cytometry analysis (), and autophagy (see below). The majority of cells displayed flattened morphology (), but only a proportion displayed p16Ink4a nuclear positivity (). Furthermore, p16Ink4a expression was largely confined to the cytoplasm (). All of these features are indicative of senescence.
Figure 1. Senescent-like phenotype of ETO-treated PA-1 cells. PA-1 cells were treated with 8 µM ETO for 20 h before being replaced with fresh media. (A-D) DNA content and nuclear area were assessed using toluidine blue staining at indicated time points. (A) Images of toluidine blue stained nuclei at indicated time points. (B) Histograms of DNA content were produced and used to determine the cell cycle stage of individual cells. (C) Dot-plots of DNA content versus nuclei area with each cell colored according to cell cycle stage as determined by DNA content histograms: G1 (blue); S phase (green); G2 (red); purple (polyploid). A statistically significant difference in nuclear size between treated and non-treated cells was observed in G2 and polyploid fractions (*p < 0.05, n = 3 ). (E) Granularity of cells (SSC) was measured at indicated time points by flow cytometry. (F) Phase contrast microscopy of PA-1 cells displaying a flattened morphology. (G) Immunofluorescence staining for p16ink4a (red) and counterstained with DAPI at indicated time points demonstrating an increase in p16ink4a following ETO treatment. (H) Quantification of p16ink4a expression determined in individual cells by image cytometry displayed as a box plot showing its increase after ETO treatment. Images are representative for at least 3 individual experiments.
Heterogeneous expression of OCT4A and p21Cip1 following Etoposide treatment
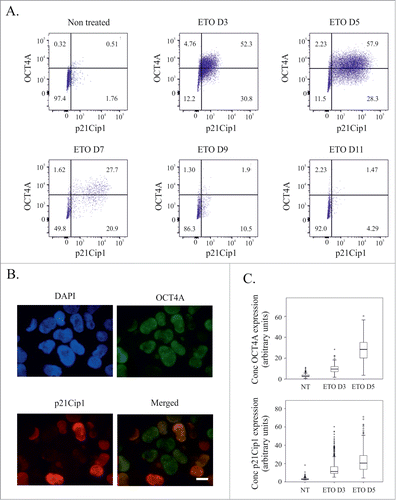
We previously showed that the senescence marker p21Cip1 and the stemness marker OCT4A simultaneously increase and co-exist in the same cells following ETO treatment.Citation24 After confirming the pre-senescent phenotype of these cells, we next wished to explore how these critical factors for cell fate were dynamically regulated, integrated and diversified. As judged by flow cytometry ˜60% of the cells become positive for both markers at the peak of the response (day 5), thereafter the expression of both markers and the proportion of double positive cells gradually decreases () as clonogenic, mitotic division ensues. To exclude the possible impact of the senescence-associated cytoplasmic and nuclear area changes on the flow cytometry measurements, semi-automatic image cytometry was undertaken, measuring the expression of both factors in the cell nucleus (apoptotic cells possessing very high p21Cip1 positivity lacking OCT4A were deliberately excluded from this analysis as distorting stoichiometry of measurements). Both the expression and variation of expression of OCT4A and p21Cip1 increased on day 3 after ETO treatment; and increased further on day 5 (, Fig. S1) and a high number of double-positive cells confirmed ().
Figure 2. Expression of OCT4A and p21Cip1 following ETO treatment of PA-1 cells. (A) Dual staining for OCT4A and p21cip1 by immunofluorescence 3 d after ETO treatment. (B) Quantitation of OCT4A and p21Cip1 expression on day 3 and day 5 after ETO treatment as measured by image cytometry. (C) Detection of OCT4A and p21Cip1 by flow cytometry after ETO treatment. Data collected over 11 d indicate an increasing expression and heterogeneity, with numerous double positive cells at the time of maximal increase (day 5). Starting from day 7 their expression, double-positive population and degree of heterogeneity gradually decreases during the recovery of clonogenic growth.
OCT4A upregulation is not accompanied by changes in SOX2 or NANOG
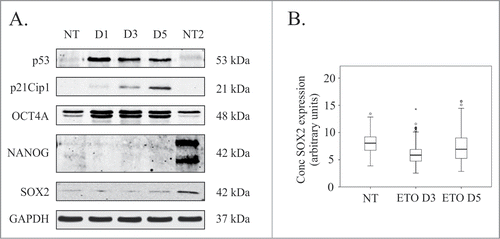
In its function as a master regulator of pluripotency and self-renewal, OCT4A cooperates with SOX2 and NANOG whose expression are therefore required for maintaining the embryonic stem cell phenotype.Citation25 We thus explored the expression of these transcription factors in the DNA damage response of PA-1 cells. Immunoblotting confirmed that p53 increased in response to ETO from day 1, resulting in the induction of p21Cip1, which reached its maximum on day 5 (). As documented previously, OCT4A expression was present in control cells and increased following ETO treatment, coincident with p53 upregulation.Citation24 Weak expression of SOX2 was found in untreated cells but did not alter following ETO-treatment (); a result confirmed by image cytometry measurements in individual cells (, Fig. S1). NANOG was not detected in PA-1 cells by immunoblotting () although rare NANOG positive cells (less than 5%) were found in control and ETO-treated samples by 3 different antibodies (Supplementary Fig. 1). Intriguingly, SOX2 failed to show unambiguous antagonism with p21Cip1, which has previously been reported as directly inhibiting SOX2 in neurodifferentiation.Citation26 Furthermore, SOX2 expression did not show significant correlation with its self-renewal partner OCT4A (data not shown). Together, these data suggest a dissociation of OCT4A from its self-renewal capacity when it is induced by p53 (in the presence of p21Cip1) in response to DNA damage.
Figure 3. Analysis of pluripotency transcription factors in response to ETO treatment in PA-1 cells. (A) Protein expression of p53, p21Cip1, OCT4A, SOX2 and NANOG were analyzed by immunoblotting at indicated time points after ETO-treatment. GAPDH was used as a loading control NT2 cells were used as a positive control for pluripotency genes OCT4A, SOX2 and NANOG. p53 and p2Cip1 protein expression was upregulated in the DNA damage response. An upregulation of the OCT4A protein was detected from day 1 post ETO treatment, while SOX2 remained low and unchanged post ETO expression. NANOG was not detected in NT or ETO treated PA-1 cells. Results are representative of 3 independent experiments. (B) Semi-automatic image cytometry of SOX2 expression was performed on the nuclei of individual cells. Box-plots indicate the heterogeneity and expression levels measured. The data show that SOX2 remains unresponsive to ETO-treatment.
OCT4A suppresses p21Cip1 and promotes an enhanced senescence phenotype
Given its potential lack of self-renewal capacity, we next investigated the role of OCT4A in PA-1 cells following ETO treatment using siRNA. Prior to this analysis, we first established which OCT4 isoforms were expressed in PA-1 cells. Using a panel of specific mAb, immunoblotting revealed that OCT4B was undetectable in these cells both before and after ETO treatment (Fig. S2), consistent with published previously RT-PCR data.Citation24
Having established that OCT4B was not expressed in PA-1 cells, siRNA silencing was used to assess the role of OCT4A in their response to ETO treatment. Efficient (80–95%) knock-down of OCT4A was confirmed with 3 different siRNA, in addition to a 3 siRNA pool (Fig. S3) and the molecular response to ETO was then examined by immunoblotting () for key regulators of DNA damage, (pCHK2, p53, p21Cip1, and RAD51). These experiments revealed p53, RAD51 and pCHK2 were all upregulated from day 1 independently of OCT4A expression. p21Cip1 expression rose gradually from day 1 throughout the course of experiment as seen in previous experiments. Silencing of OCT4A resulted in a marked increase in p21Cip1 expression from day 1. These results imply that the presence of OCT4A induced by DNA damage suppresses p21Cip1 expression despite comparable levels of p53 expression and equivalent DNA damage as judged by RAD51 and pCHK2 expression.
Figure 4. OCT4A suppresses p21, G2 arrest and induces senescence. PA-1 cells were treated with ntg-siRNA or OCT4-siRNA for 24 h before treatment with 8 µM ETO for 20 h and replacement with fresh media. (A) Protein expression was assessed at the indicated time points by immunoblotting. Cell lysates were made and assessed by immunoblotting for pCHK2, RAD51, p53, p21Cip1, OCT4A and GAPDH as a loading control. (B) Cells were fixed, stained with PtdIns and analyzed by flow cytometry. Both ntg-siRNA treated and OCT4-siRNA treated cells underwent a G2M arrest followibg ETO treatment, which is more profound at day 3 and 5 in OCT4A-siRNA-treated cells. There is subsequently a much lower extent of cell death (sub G1) in OCT4A-siRNA treated cells. (C) The G2M arrest was calculated from the DNA histograms shown in (B) and demonstrate significantly higher levels in OCT4A-silenced cells (*p < 0.05, n = 3). (D) PA-1 cells were treated with ETO as above and assessed for Sa-β-gal activity after 5 d Senescence, as monitored by Sa-β-gal activity, was increased by OCT4A-silencing. Both ntg-siRNA and OCT4A-siRNA treated cells became large following ETO treatment but non-silenced cells lacked Sa-β-gal activity.
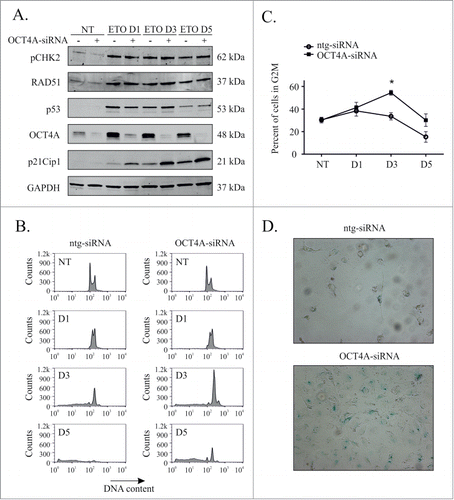
When investigating the cell cycle response, loss of OCT4A caused an almost identical phenotype to that previously reported with p53 silencingCitation24 i.e. OCT4A-silenced cells underwent a profound G2M arrest with apoptosis being greatly reduced (). The cells displayed the large, flattened morphology and Sa-β-gal staining associated with senescence on day 5 (), identical to that previously reported when silencing p53. Thus, the increase in G2M arrest and increased accelerated senescence previously observed with p53 silencing appears to be entirely due to the loss of the induction of OCT4A. Prolonged loss of OCT4A lead to the polyploidisation of these senescent cells and a reduction in clonal recovery (). The absence of off target effects was confirmed by repeating silencing and cell cycle analysis experiments with 3 separate siRNA molecules targeting different sequences of OCT4A mRNA (Supplementary Fig. 4).
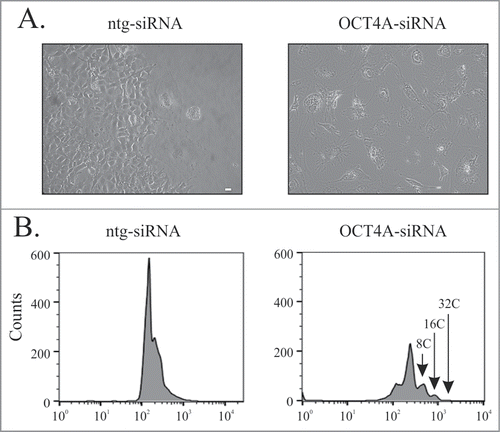
Figure 5. OCT4A silencing inhibits clonal recovery of ETO-treated PA-1 cells. PA-1 cells were treated with ntg-siRNA or OCT4A-siRNA for 24 h before treatment with 8 µM ETO for 20 h and replacement with fresh media. Cells were maintained until day 11. Clonal recovery was observed in non-silenced cells but not in OCT4A-silenced cells, which maintained a flat senescent like morphology. (B) Cells were fixed, stained with PI and analyzed by flow cytometry. A return of the G1 peak was observed in non-silenced ntg cells whereas OCT4A-silenced cells maintained a profound G2 arrest. OCT4A-silenced cells entered rounds of endoreduplication with genomes as large as 32C observed.
p21Cip1 silencing does not affect OCT4A expression or cell cycle arrest
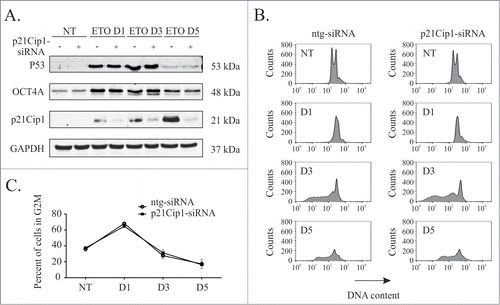
Having established that OCT4A suppresses p21Cip1 and a senescent phenotype we sought to investigate the possibility of a reciprocal suppression of OCT4A by p21Cip1. To do this p21Cip1 was silenced using siRNA. Silencing of p21Cip1 was efficient as judged by immunoblotting but was not found to affect the expression of OCT4A in the presence or absence of ETO treatment (). Furthermore, we detected no difference in the cell cycle profiles of p21Cip1-silenced cells compared to control siRNA treated cells following ETO treatment.
Figure 6. p21Cip1 expression does not affect G2M arrest in response to ETO treatment of PA-1 cells. PA-1 cells were treated with ntg-siRNA or p21Cip1-siRNA for 24 h before treatment with 8 µM ETO for 20 h and replacement with fresh media. (A) Expression of OCT4A, p21Cip1 and p53 were assessed at the indicated time points by immunoblotting. GAPDH was used as a loading control. (B) Cells were fixed, stained with PtdIns and analyzed by flow cytomtery. Both ntg-siRNA treated and p21Cip1-siRNA treated cells underwent a comparable G2M arrest after ETO treatment. (C) There was no significant difference in G2M arrest between p21Cip1 silenced and non-silenced cells.
In summary, we found that OCT4A tempers p21Cip1 upregulation, restricting entry into senescence, indicating that co-expression defines the pre-senescent state. While p21Cip1 did not regulate OCT4A expression or change the cell cycle, depletion of OCT4A profoundly impacted it. We also found that p53-activated OCT4A does not support the pluripotency network in pre-senescent cells (owing to the lack of SOX2/NANOG co-ordination), leaving its biological role unclear. We, therefore, interrogated its possible role as a stress responder where one of these stresses may be metabolic. AMPK is a key sensor of metabolism and energy status.Citation27 Therefore, we next explored the activation of AMPK by virtue of its phosphorylation at Threonine 172 (pAMPKThr172), in relation to OCT4A expression.
AMPK and OCT4A are coordinately activated in response to ETO-treatment
The results of this comparative study are shown in . Microscopy of control, untreated, cells revealed heterogeneous pAMPKThr172 activation with expression in some cell nuclei and also in centrosomes, central spindle and mid-bodies of mitotic cells as described previously,Citation28 and rarely ( <1% cells), in the cytoplasm (). In ETO-treated cells the staining for AMPK increased and strikingly coincided with the localization and intensity of staining for OCT4A in most cells () as also confirmed by IF measurements, where clear correlation was shown (). However, while OCT4A is seen homogenously throughout the nucleus pAMPKThr172 staining revealed a fine (euchromatic) nuclear pattern, which indicates binding to chromatin, unlike OCT4A.
Figure 7. Activation of AMPK and OCT4A in response to ETO-treatment. PA-1 cells were treated with 8 µM ETO for 20 h and replacement with fresh media. Cells were harvested after 4 d before immunofluorescent staining for OCT4A, pAMPKThr172 and DAPI. (A) Non-treated (upper panel) and ETO-treated (lower panel) samples were compared. AMPK and OCT4 are both activated by ETO (day 4) in interphase cells, however pAMPK is located in multiple centrosomes of arrested metaphases, while OCT4A does not; (B) scatterplot of image cytometry of OCT4A and pAMPK in individual cells assessed on day 4. There is a clear correlation between enhanced expression of OCT4A and pAMPKThr172 of ETO-treated cells; (C) OCT4A and pAMPKThr172 immunofluorescence in PA-1 cells 4 d after ETO-treatment. OCT4A and pAMPKThr172 are mostly expressed in the same interphase nuclei, but an extremely high signal of pAMPK but not OCT4A is found within apoptotic cells. Staining of pAMPKThr172 is associated with euchromatin, while the pattern of OCT4A staining is karyoplasmic (not bound to the chromatin). BRG – a 3-band optical filter. Bars =20 µm.
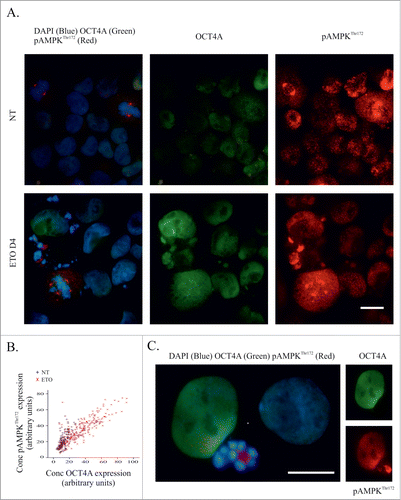
Intense staining of pAMPKThr172 was also found in apoptotic cell nuclei and bodies, while OCT4A was not (). In arrested metaphases appearing after ETO on day 4, pAMPKThr172 was localized to multiple centrosomes, while OCT4A was not (). This difference in staining was confirmed by co-staining for γ-tubulin and AURORA B kinase (not shown). Together, these data indicate that OCT4A is co-ordinately regulated alongside pAMPKThr172 in interphase cells but that in apoptotic cells their functions may differ. Moreover, the concordant expression of OCT4A and pAMPKThr172 in interphase cells suggests that the p53-dependent OCT4A induction is a result of a stress response following ETO treatment.
As a part of its role as a central metabolic regulator, AMPK is directly involved in the regulation of autophagy.Citation29 In this regard, previous studies have shown that autophagy is implicated in the chemo-resistance to ETO and this effect is reversed by inhibition of AMPK, which led to cell death by apoptosis.Citation30
Conversely, cellular senescence is also associated with autophagyCitation30 representing another potential cell fate phenotype following ETO-treatment.Citation24 p16ink4a is believed to drive irreversible senescence. Citation31 Therefore, we next decided to explore the relationship of p16ink4a and macroautophagy.
p16ink4a is sequestered by autophagy in ETO-treated cells
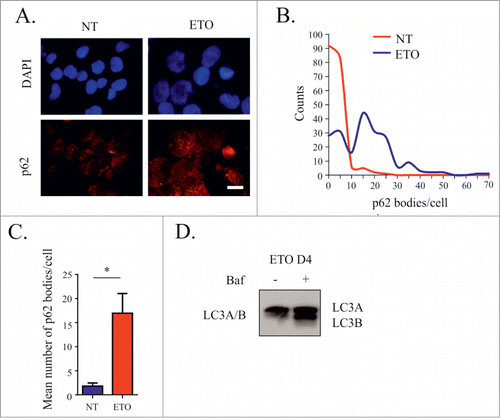
Autophagy was verified using immunofluorescence for p62 as a marker of autophagy. Post ETO treatment, an increase in the number of p62 foci/cell was observed by immunofluorescence () and mean number of p62 foci/cell in 3 independent experiments was shown to increase > 5-fold (). Bafilomycin A (Baf) halted autophagic flux in ETO-treated cells (shown as an accumulation of LC3B in relation to LC3A), confirming functional autophagy and its suppression (). Baf treatment also enhanced cell death preventing clonogenic recovery only in ETO-treated cells (Fig. S5), which indicates the importance of autophagy for cell survival after DNA damage.
Figure 8. Assessment of macroautophagy following ETO treatment (day 4). PA-1 cells were treated with 8 µM ETO for 20 h and replacement with fresh media in the presence or absence of Baf. Cells were then stained for p62 and the number of p62 positive bodies per cell counted. (A) Immunoflurorescent examples of staining for p62 showing foci (p62 bodies) in control and ETO-treated cells. (B) Representative histograms of p62 bodies/cell. (C) The average number of p62 bodies per cell was compared for NT and ETO treated cells from 3 independent experiments (* p< 0.05, n = 3 ) indicating activation by autophagy. (D) Immunoblotting analysis of the ratio of LC3A and LC3B with and without Baf treatment post ETO treatment. Enhanced levels of LC3B in relation to LC3A as a result of Baf treatment indicates interruption of autophagic flux in ETO treated cells. Bars = 20µm.
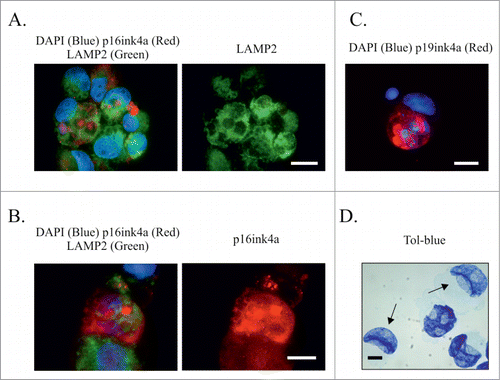
We then assessed immunostaining for p16ink4a and LAMP2 in the presence and absence of Baf post ETO treatment. LAMP2 is an important regulator of lysosomal biogenesis, required for the maturation of functional autophagosomes. In control, untreated cells, p16ink4a was observed in ˜5% of cells as small aggresomes, surrounded by LAMP2-positive foci (data not shown). In ETO-treated cells (day 4), the cytoplasm was highly enriched with LAMP2, while p16ink4a-positive aggresomes (larger and more numerous than in control) were sequestered in autophago-lysosomes (). Approximately 30% of cells were seen to accumulate large amounts of p16ink4a in multiple aggresomes, as measure by >3 aggresomes and/or diffusely detected within cytoplasm, or in large vacuoles (), and often showed DNA in cytoplasm (). The proportion of such cells, with high levels of p16ink4a, increased fold2- (29 ± 6 .5 and 58 ± 3 .5, +/− Baf, respectively) as measured on day 4. These observations indicate the important role of autophagy in sequestering and degrading p16ink4a in ETO-treated PA-1 cells, thus preventing senescence. They also show that a considerable proportion of ETO-treated cells suffer from exhaustion of autophagy.
Figure 9. The relationship between autophagy functionality and senescence marker p16ink4ain ETO-treated PA-1 cells. PA-1 cells were treated with 8 µM ETO for 20 h without or with Baf, prior to media being removed and replaced with fresh media; cells were harvested 48h later (day 4). (A, B) imunofluorescent staining for p16ink4a (red), LAMP2 (green) or DAPI (blue). (A) As shown via the BRG optical filter, (B) only the green filter for LAMP2 demonstrating high level of functional autophagy sequestrating p16ink4a- containing aggresomes. (B, C) show examples of autophagic failure, where on (B) sequestration of p16ink4a is partly lost, diffusing in to the cytoplasm, while on (C) the dysfunction of autophagy is evidenced by a large vacuole overloaded with undigested p16ink4a. In all 3 cases, the nuclei are impaired: (B) ‘laced’; (C) displaced and twisted by a large autophagic vacuole with some DNA lost from cell nucleus observed. These processes were observed both in ETO-treated (B) and more commonly in ETO+BAF(c) treated cells. (D) An image showing cell nuclei of normal and sickle-type shape as stained specifically for DNA with Toluidine bluein the ETO-teated specimen (sickle-like shaped nuclei are shown with arrows).
Failure of autophagy to sequester p16ink4a in the cytoplasm is accompanied by drastic nuclear changes and loss of DNA
In ETO-treated samples counterstained with DAPI, we noted that the failure of macroautophagy to control and sequester p16ink4a in the cytoplasm coincided with prominent changes in cell nuclei, particularly following Baf treatment. Nuclei became thinner, less intensely DAPI-stained (e.g. ), or, more frequently, displaced to one of the cell poles by large autophagic vacuoles (). Twisting, thinning, and disintegrating cell nuclei were the main phenotypes of ETO+Baf-treated cells. As detailed above, in their extreme distortions, the cell nuclei were displaced and squeezed by large autophagic vacuoles and attained a sickle-like shape (). It remains possible that the loss of sequestration of p16ink4a, distortion of cell nuclei and loss of DNA are all linked, belonging to a related series of processes associated with exhausted autophagy, which is further exaggerated by Baf where full disintegration of the cell nucleus and alveolar LC3-positive cytoplasm are evident ().
Figure 10. ETO and ETO+Baf impair cell nuclear integrity causing a delay in late S-phase in a proportion of cells with persistent DNA damage. (A) Disintegration of the nucleus is observed in ETO treated cells following prolonged Baf exposure (72h), where impaired autophagic flux is confirmed by alveolar structure of LC3B-positive cytoplasm in the absence of LC3 foci; Bars = 20 µm. (B) Persistent DNA damage leads to destruction of the epichromatin ternary complex (a specific conformation of nucleosomes attached to lamin B1) seen by a decrease in staining by the Pl2–6 antibody where Chk2-positive foci is detected in ETO-treated PA1 cells at day 4. (C) Representative DNA histograms obtained by image cytometry of ETO-treated and ETO+BAF-treated cells samples (day 4 after treatment); one of 3 independent experiments. BafA alone causes partial arrest in G1 in control (NT)cells and a subsequent decrease of the G2 fraction; ETO-treatment induces G2-arrest, however, a proportion of cells remain delayed in late S-phase (<4C). Cells with the DNA content <4C and 4C(G2) still reach peaks of corresponding doubling, <8C and 8C. Suppression of autophagy by Baf in ETO-treated cells favors an increase of the proportion of cells in the <4C fraction and reduces the number of cells.
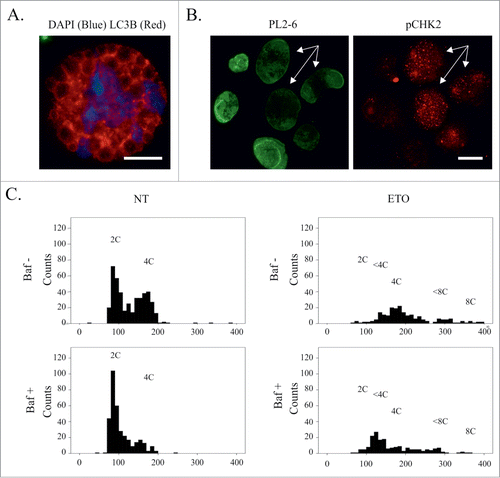
Trying to understand how the penetrability of the cell nuclei was related to DNA damage we stained with an antibody (PL2–6) which detects a unique epitope involving a ternary complex of histones H2A and H2B and DNA at the site of the chromatin attachment to lamin B1.Citation32 We applied it to ETO-treated cell nuclei (day 4) combining it with a label for DNA damage (pCHK2). The result shows () bright nuclear staining with PL2–6 in cell nuclei without DNA damage and its reduction in cell nuclei containing DNA damage, possibly suggesting that binding to lamin B receptors in compromised.
In view of this data, we decided to use detailed stoichiometric DNA image cytometry following ETO treatment in the presence or absence of Baf. Typical histograms from one representative experiment are shown in , while in Fig. S6 comparative DNA histograms of selected normal- and sickle-shaped cell nuclei from ETO+Baf treatment are presented. Measurements from ETO-treated cells revealed a peak of delay in late S-phase, which was augmented after Baf treatment showing apparent redistribution from G2 (4C) into <4C, which was exaggerated in selected sickle-type nuclei of ETO+Baf-treated cells (Fig. S6). These data support our microscopic observations that DNA is preferentially lost from the sickle-like shape nuclei in the cells as a result of failed autophagic processing with the proportion of sickle-like nuclei cells with sickle-like nuclei in ETO-treated samples increased more than 2-fold (2.38 ± 0.2) following Baf treatment (n = 3).
Discussion
In this study, we expanded our investigation of the response of PA-1 embryonal carcinoma cells to DNA damaging chemotherapy. We previously observed,Citation24 a concurrent p53-dependent increase in OCT4A and p21Cip1 protein expression after ETO-treatment, with the resulting cells transiting through an apparently bi-potential state with hallmarks of both premature multi-centriolar mitoses and senescence. Herein, we extended our analysis and highlight a hitherto unobserved DNA damage response (DDR) function of OCT4A following ETO treatment, which is different from its established pluripotency and self-renewal functions. This p53-dependant DDR-induced OCT4A is not coordinated with SOX2 or NANOG activity, but does serve to suppress p21Cip1. This finding is in accord with the previously shown observation that OCT4A can directly repress p21Cip1 expression by binding its promoter in the absence of SOX2 and NANOG. Citation33 Although not yet confirmed in other cell types or following different stimuli, this phenomenon likely relates to properties evidenced during embryonal ‘stemness’, as similar nuclear accumulation of OCT4A was similarly recently reported following laser beam damage of live ESC cells.Citation34
Our measurements of OCT4A and p21Cip1 expression in individual cells revealed that cell fate, in response to extensive DNA damage, occurs with high levels of heterogeneity. Both factors increase coordinately with huge variations in expression, covering (at day 5) a wide spectrum of p21Cip1 and OCT4A expression.Citation35 These observations demonstrate a typical characteristic of self-organizing, adapting systems, using heterogeneity over time to provide bifurcations of cell fate choice.Citation36,37 The increased variance in both factors – from day 3 until day 7 – corresponds to the period with most prominent cell death (99% cells will die and only <1% has the chance to survive, likely representing the degree of challenge that they must overcome).Citation24 When OCT4A is silenced the level of p21Cip1 increases, more cells enter senescence and mitotic recovery is reduced, abrogating fragile balance in bifurcation point and removing the possibility for rare cells to choose survival as their fate. It is worth noting that this response was phenotypically identical to that seen when silencing p53.Citation24 We hypothesize that the loss of p53 or OCT4A in silencing experiments causes the loss of the embryonal G2M checkpoint leading to increased mitotic slippage, ultimately ending in terminal senescence rather than executing the G2M checkpoint function via apoptosis. Continued silencing of OCT4A inhibited clonogenic recovery post ETO treatment highlighting the importance of the pre-senescent G2M arrest for clonogenic recovery of the small percentage of cells that are able to repair their DNA damage. Silencing of OCT4A also leads to endoreplication in senescent cells further eluding to the presence of mitotic slippage.Citation38 p21Cip1 silencing experiments demonstrated that the observed G2M arrest was independent of p21Cip1 expression.
Mitotic survival of the ETO-treated cells ultimately originates from metastable pre-senescence. Theoretically, this means that the self-renewal function of OCT4A is finally restored. The attractor of self-renewal fate in individual cells is high NANOG,Citation39 released from direct suppression by p53Citation40 and activated by OCT4A-SOX2 heterodimer. Although NANOG is only found in a very small number of PA-1 cells, its role requires further exploration.
This moderator function of DDR-induced OCT4A occurs alongside activation of AMPK. AMPK is an evolutionary conserved low-energy checkpoint that operates as a master regulator of cellular metabolism and activator of autophagy. AMPK also acts as a genomic stress sensor that is required for the adaptation of metabolism switches and the DDR for efficient repair of DNA.Citation41 We previously observed a bidirectional relationship between AMPK and ATM/CHK2 pathways that integrates both the metabolic and DNA damage stress signaling.Citation42 AMPK activation was previously shown as necessary for maximal stress-induced transcription of p53-dependent genes that promote cell survival.Citation43 Interestingly, AMPK has also been shown to activate and stabilize p53.Citation44,45 Thus, the energetic stress-response of AMPK is tightly linked to the DDR of p53. The role of AMPK in the autophagy and cell death response after ETO has previously been reported.Citation30
We previously revealed the p53-dependant DDR-induced elevation in OCT4A in PA-1 cells following ETO treatment. Therefore, it was not unexpected that DDR-induced OCT4A and AMPK activation should be coordinately regulated by ETO as we observed here. We found that while autophagy apparently remains functional, cells retaining both AMPK and DDR-induced OCT4A in cell nuclei are capable of preventing terminal senescence. This data are in accordance with the report by Chitikova and colleagues, Citation46 who demonstrated that silencing mTOR (the antagonist of AMPK suppressing autophagy) in irradiated, apoptosis-resistant cells caused a reversal of senescence. Thus OCT4A moderating p21Cip1 in collaboration with pAMPKThr172, supporting cells with metabolites and energy through autophagy and other means, channel cells through disadaptation chaos caused by high DNA damage toward a survival phenotype. ETO-treated PA-1 cells struggle against terminal senescence in particular by autophagic sequestration and removal of its regulator p16ink4a.
There is much controversy regarding markers of terminal senescence and even what the process really is. It has become recently apparent that terminal senescence is not only growth arrest,Citation47 moreover, it is intrinsically associated with the origin of EMTCitation22 and predicts poor clinical outcome after cancer treatment.Citation48
In our previous and current article we have partly unravelled a mechanism showing that in response to DNA damage p53 induces 2 regulators of opposite cell fates. The setting is interesting: although they both are positively regulated by the same up-stream inducer, OCT4A suppresses p21Cip1, while the latter does not directly suppress OCT4A. Nevertheless, the scattering of the cells in expression of both through the phase space ending up in one of 2 cell fates presumes their reciprocity. The observation of cell-to-cell variability can also be a result from biological oscillations in time.Citation49 The best documented example of this is the p53-mdm2 oscillating system in which p53 oscillations are, indeed, crucial for cell recovery after DNA damage.Citation50 It is, therefore, possible that heterogeneity in p53 due to oscillations results in downstream heterogeneity of both OCT4A and p21Cip1 in PA-1 cells.
Senescence is intrinsically associated with macroautophagy induction (regulated by AMPK and persistent DNA damage). Citation51 Here, we found that persistent DNA damage was associated with destruction of the ternary complex of histones (H2A and H2B) and DNA in their attachment to nuclear lamin B1 using a unique antibody reagent.Citation32 This observation is in keeping with data of the thinning and disassembly of lamin B1 and leakage of DNA from cells as a feature of terminal senescence.Citation52
Our DNA cytometric observations revealed a delay in late S-phase after ETO, augmented by BafA1. The consequence of this delay would be under-replication of heterochromatin,Citation53 appearance of single strand DNA breaks, demethylation and activation of the transposable elements (TE) ALU/LINE 1, whose mobility after ETO has already been described.Citation54 As sequestering ALU mRNA and LINE 1 by autophagy in the cytoplasm has been reported as a mechanism for protecting against transposition,Citation55 it therefore remains possible, (although purely speculative) that functional autophagy prevents terminal senescence in this manner also. The model with the scenario of terminal senescence occurring due to activation of TE as discussed in several other studies.Citation56-58 It follows then that terminal senescence may in fact be mainly triggered by several mechanisms, which potentiate each other and result from failure of autophagy. Thus, autophagy should be a key target for preventing resistance of cancer stem cells to genotoxic therapies, a supposition supported by others.Citation59,60
Materials and Methods
Cells
PA-1 is an ovarian teratocarcinoma (a germline tumor) cell-line, obtained from ATCC, with a stable near-diploid karyotype.Citation61 It possesses functional p53, in spite of possible acquisition of one mutated allele reported at passages over 407.Citation62 Importantly it retains the ability to differentiate into all 3 tissue types, thus possessing the features of embryonal carcinoma.Citation61 The NTera 2 (NT2) cell line, obtained from ATCC and used as a positive control for pluruipotency genes OCT4A, SOX2 and NANOG.
Antibodies
Primary antibodies and their uses are detailed in . Secondary antibodies and their uses are detailed in .
Table 1. Primary antibodies
Table 2. Secondary antibodies
Cell culture and treatment
PA-1 cells were cultured in Dulbecco's modified Eagle's media (DMEM) supplemented with 10% foetal bovine serum (FBS) if not otherwise stated. NT2 cells were cultured in Dulbecco's modified Eagle's media (DMEM) supplemented with 10% foetal bovine serum (FBS). Cells were grown without antibiotics in 5% CO2 at 37°C. Exponentially growing PA-1 cells were incubated with 4–8 μM ETO for 20 h. Following ETO treatment, cells were maintained by replenishing culture medium every 48h. Bafilomycin A (Baf A, Cayman Chemicals) 50 nM was added to the culture medium for 24h between day 3 to 4 after 20h treatment with 8 µM ETO.
Flow cytometry
Cells were harvested at relevant time points, washed in cold PBS and fixed with 70% ethanol for 20 min at room temperature. After two washes in tris buffered saline (TBS), cells were permeabilised with TBS/4% bovine serum albumin (BSA)/0.1% Triton X-100 for 10 min at room temperature. Samples were then incubated with mouse monoclonal anti-OCT4 antibody solution (Santa Cruz) and rabbit polyclonal p21Cip1 (Pierce) in TBS/4% BSA/0.1% Triton X-100 for 1h at room temperature. Following two washes in TBS, cells were incubated with goat anti-rabbit- IgG-Alexa Fluor 488 (A31627, Invitrogen) and Chicken anti-mouse IgG-Alexa Fluor 647 (A21463, Invitrogen) (1:200) in TBS/4% BSA/0.1% Triton × 100, for 30 min in the dark. DNA was counterstained with 10 μg/ml propidium iodide (PtdIns) in PBS, and assessed by flow cytometry using a FACS Aria (BD Biosciences) using Cell Quest Pro Software). Data was analyses using FlowJo analysis software.
Immunofluorescence
Cells were suspended in warm FBS and cytospun onto glass slides. Cytospins were fixed in methanol for 7 min at −20°C and dipped 10 times in ice cold acetone. Slides were then washed thrice in TBS 0.01% Tween 20 (TBST) for 5 min. Slides were subsequently blocked for 15 min in TBS, 0.05% Tween 20%, 1% BSA at room temperature. Samples were covered with TBS, 0.025% Tween 20%, 1% BSA containing primary antibody and incubated overnight at 4°C in a humidified chamber. Samples were then washed thrice in TBST and covered with TBST containing the appropriate secondary antibodies (Goat anti-mouse IgG Alexa Fluor 488 (A31619, Invitrogen) and Goat anti-rabbit- IgG Alexa Fluor 594 (A31631, Invitrogen)) and incubated for 40 min at room temperature in the dark. Slides were washed thrice for 5 min with TBST and once for 2 min in PBS. Samples were then counterstained with 0.25 μg/ml DAPI for 2 min, and finally embedded in Prolong Gold (Invitrogen). When staining for p-AMPKα1/2 (alone or in combination with the antibody for OCT4), fixation in 4% paraformaldehyde for 15 min was used, followed by washing thrice in PBS 0,1% glycine. Primary antibodies and their source are listed in .
Microscopy and immunofluorescence image cytometry
Slides were evaluated using a Leitz Ergolux L03–10 microscope equipped with Sony DXC 390P color video camera, for microscopic observations; in addition to separate optical filters, a 3-band BRG (blue, red, green) optical filter (Leica) was used. Image cytometry was carried out by semi-automatic measuring fluorescence values for each cell nuclei in all 3 channels and analyzed using Image-Pro Plus 4.1 software (Media Cybernetics). Apoptotic cells, determined by nuclear morphology, were omitted from measurements.
DNA image cytometry
For DNA cytometry measurements, stoichiometric toluidine blue DNA staining was performed as previously.Citation63 In brief, cytospins were fixed in ethanol:acetone (1:1) for >30 min at 4°C and air dried. Slides were then treated with 5N HCl for 20 min at room temperature, washed in distilled water (5 × 1 min) and stained for 10 min with 0.05% toluidine blue in 50% citrate-phosphate McIlvain buffer pH 4. Slides were rinsed with distilled water, blotted dry and dehydrated in butanol for 2×3 min at 37°C. Samples were then incubated twice in xylene for 3 min each at room temperature and embedded in DPX (Sigma). DNA content was measured as the integral optical density with a calibrated Sony DXC 390P video camera in the green channel. Nuclear area was calculated using Image-Pro Plus 4.1 software (Media Cybernetics). Apoptotic cells were omitted from measurements. The stoichiometry of DNA staining was verified using the values obtained for metaphases compared to anaphases and telophases (ratio 2.0); the summary error of the method and device was estimated to be less than 5%.
DNA flow cytometry
Cells (including those in media) were harvested at indicated time points, washed in PBS and re-suspended in hypotonic fluorochrome solution [50 µg/ml propidium iodide (PI), 0.1% (w/v) sodium citrate, 0.1% (v/v) Triton-X-100] and stored for at least 1 h in the dark at 4°C. Flow cytometry was performed using a FACScan (BD Biosciences) or Accuri™ C6 Cytometer (BD Biosciences). Data was analyses using FlowJo analysis software.
Detection of sa-β-galactosidase activity
The senescence β-galactosidase (sa-β-gal) staining kit (Cell Signaling Technology, UK) was used to detect sa-β-gal activity in cells according to the manufacturer's protocol.
Immunoblotting
For whole-cell lysates, cells were harvested, washed once with PBS and lysed using RIPA buffer containing protease inhibitor cocktail (Sigma P8340). Protein concentrations were determined by Bradford assay (Thermo Scientific), with 10–30 μg of total protein separated on 20% SDS polyacrylamide gels followed by electrophoretic transfer onto BA85 nitrocellulose membranes (Schleicher & Schuell GmbH) overnight. Equal protein loading in each lane was confirmed by Ponceau S staining. Blots were probed with appropriate antibodies primary and secondary detected using ECL Western Blotting Substrate (Pierce™, 32106).
For comparison of OCT4 isoforms protein was transferred onto immobilon-FC transfer membrane and probed with primary antibodies. Blots were then probed with fluorescent secondary antibodies and the signal was visualized using a LICOR Odyssey imaging system.
Small interfering RNA (siRNA) silencing
FlexiTube siRNA (SI04950274, SI04950267, SI04153835 and SI00690382; Qiagen) was used to silence OCT4A expression, and FlexiTube siRNA (SI00604905, SI00604898 and SI00299810; Qiagen) was used to silence p21Cip1 expression. ON-TARGET plus non-targeting siRNA #1 (Dharmacon) was used as a negative control. Cells were transfected with siRNA using HiPerfect (Qiagen) according to the manufacturer's protocol.
Methods of statistical analysis
Statistical analysis was performed in GraphPad (GraphPad Software Ltd). Student t-test was used to calculate the statistical significance of difference of means where appropriate. Statistical significance was accepted when p < 0.05. Graphs were plotted in GraphPad Prism 5 and IBM SPSS statistics 22.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
1056948_Supplemental_Material.zip
Download Zip (22.2 MB)Acknowledgments
The authors would like to thank Professor Hao and Dr Zhao, Dept of Histology and Embryology, Shandong University School of Medicine for the kind gift of the OCT4A and OCT4A-Psg-1 vector constructs and Ada and Donald Olins for providing the antibody for epichromatin.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Funding
The project was funded through an MRC PhD studentship and the Gerald Kerkut Charitable Trust, the University of Manchester Project Diamond and the Manchester Cancer Research Center and by the Europe Social Fund Project, project No. 2013/0023/1DP/1.1.1.2.0/13/APIA/VIAA/037. The publishing costs associated with this article are in part provided by the Manchester Project Diamond and by the Europe Social Fund Project, project No. 2013/0023/1DP/1.1.1.2.0/13/APIA/VIAA/037.
References
- Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet 2008; 40(5): 499-507; PMID:18443585; http://dx.doi.org/10.1038/ng.127
- Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature 2001; 414(6859): 105-111; PMID:11689955; http://dx.doi.org/10.1038/35102167
- Chiou SH, Yu CC, Huang CY, Lin SC, Liu CJ, Tsai TH, Chou SH, Chien CS, Ku HH, Lo JF. Positive correlations of Oct-4 and Nanog in oral cancer stem-like cells and high-grade oral squamous cell carcinoma. Clin Cancer Res 2008; 14(13): 4085-4095; PMID:18593985; http://dx.doi.org/10.1158/1078-0432.CCR-07-4404
- Meng HM, Zheng P, Wang XY, Liu C, Sui HM, Wu SJ, Zhou J, Ding YQ, Li JM. Overexpression of nanog predicts tumor progression and poor prognosis in colorectal cancer. Cancer Biol Ther 2010; 9(4): 295-302; PMID:20026903; http://dx.doi.org/10.4161/cbt.9.4.10666
- Du LT, Yang YM, Xiao XY, Wang CX, Zhang XH, Wang LL, Zhang X, Li W, Zheng GX, Wang S. others. Sox2 nuclear expression is closely associated with poor prognosis in patients with histologically node-negative oral tongue squamous cell carcinoma. Oral Oncol 2011; 47(8): 709-713; PMID:21689966; http://dx.doi.org/10.1016/j.oraloncology.2011.05.017
- Lengerke C, Fehm T, Kurth R, Neubauer H, Scheble V, Muller F, Schneider F, Petersen K, Wallwiener D, Kanz L. others. Expression of the embryonic stem cell marker SOX2 in early-stage breast carcinoma. Bmc Cancer 2011; 11:42; PMID:21276239; http://dx.doi.org/10.1186/1471-2407-11-42
- He W, Li K, Wang F, Qin YR, Fan QX. Expression of OCT4 in human esophageal squamous cell carcinoma is significantly associated with poorer prognosis. World J Gastroenterol 2012; 18(7): 712-719; PMID:22363145; http://dx.doi.org/10.3748/wjg.v18.i7.712
- Huang P, Chen J, Wang L, Na YQ, Kaku H, Ueki H, Sasaki K, Yamaguchi K, Zhang K, Saika T. others. Implications of transcriptional factor, OCT-4, in human bladder malignancy and tumor recurrence. Med Oncol 2012; 29(2): 829-834; PMID:21533858; http://dx.doi.org/10.1007/s12032-011-9962-4
- Lin T, Ding YQ, Li JM. Overexpression of Nanog protein is associated with poor prognosis in gastric adenocarcinoma. Med Oncol 2012; 29(2): 878-885; PMID:21336986; http://dx.doi.org/10.1007/s12032-011-9860-9
- Lagadec C, Vlashi E, Della Donna L, Dekmezian C, Pajonk F. Radiation-Induced Reprogramming of Breast Cancer Cells. Stem Cells 2012; 30(5): 833-844; PMID:22489015; http://dx.doi.org/10.1002/stem.1058
- Abubaker K, Latifi A, Luwor R, Nazaretian S, Zhu HJ, Quinn MA, Thompson EW, Findlay JK, Ahmed N. Short-term single treatment of chemotherapy results in the enrichment of ovarian cancer stem cell-like cells leading to an increased tumor burden. Mol Cancer 2013; 12; PMID:23537295; http://dx.doi.org/10.1186/1476-4598-12-24
- Salmina K, Jankevics E, Huna A, Perminov D, Radovica I, Klymenko T, Ivanov A, Jascenko E, Scherthan H, Cragg M. others. Up-regulation of the embryonic self-renewal network through reversible polyploidy in irradiated p53-mutant tumour cells. Exp Cell Res 2010; 316(13): 2099-2112
- Roninson IB. Tumor cell senescence in cancer treatment. Cancer Res 2003; 63(11): 2705-2715; PMID:12782571
- Roninson IB, Broude EV, Chang BD. If not apoptosis, then what? - Treatment-induced senescence and mitotic catastrophe in tumor cells. Drug Resistance Updates 2001; 4(5): 303-313; PMID:11991684; http://dx.doi.org/10.1054/drup.2001.0213
- Sherman MY, Meng L, Stampfer M, Gabai VL, Yaglom JA. Oncogenes induce senescence with incomplete growth arrest and suppress the DNA damage response in immortalized cells. Aging Cell 2011; 10(6): 949-961; PMID:21824272; http://dx.doi.org/10.1111/j.1474-9726.2011.00736.x
- Puig PE, Guilly MN, Bouchot A, Droin N, Cathelin D, Bouyer F, Favier L, Ghiringhelli F, Kroemer G, Solary E. others. Tumor cells can escape DNA-damaging cisplatin through DNA endoreduplication and reversible polyploidy. Cell Biol Int 2008; 32(9): 1031-1043.
- Roberson RS, Kussick SJ, Vallieres E, Chen SYJ, Wu DY. Escape from therapy-induced accelerated cellular senescence in p53-null lung cancer cells and in human lung cancers. Cancer Res 2005; 65(7): 2795-2803; PMID:15805280; http://dx.doi.org/10.1158/0008-5472.CAN-04-1270
- Sabisz M. Skladanowski A. Cancer stem cells and escape from drug-induced premature senescence in human lung tumor cells Implications for drug resistance and in vitro drug screening models. Cell Cycle 2009; 8(19): 3208-3217; PMID:19738435; http://dx.doi.org/10.4161/cc.8.19.9758
- Banito A, Rashid ST, Acosta JC, Li S, Pereira CF, Geti I, Pinho S, Silva JC, Azuara V, Walsh M. others. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev 2009; 23(18): 2134-2139; PMID:19696146; http://dx.doi.org/10.1101/gad.1811609
- Jaenisch R. Nuclear cloning and direct reprogramming: the long and the short path to Stockholm. Cell Stem Cell 2012; 11(6): 744-747; PMID:23217419; http://dx.doi.org/10.1016/j.stem.2012.11.005
- Storer M, Mas A, Robert-Moreno A, Pecoraro M, Ortells MC, Di Giacomo V, Yosef R, Pilpel N, Krizhanovsky V, Sharpe J. others. Senescence is a developmental mechanism that contributes to embryonic growth and patterning. Cell 2013; 155(5): 1119-1130; PMID:24238961; http://dx.doi.org/10.1016/j.cell.2013.10.041
- Kishi S, Bayliss PE, Hanai JI. A prospective epigenetic paradigm between cellular senescence and epithelial-mesenchymal transition in organismal development and aging. Transl Res 2015; 165(1): 241-249; PMID:24924348; http://dx.doi.org/10.1016/j.trsl.2014.05.007
- Huna A, Salmina K, Jascenko E, Duburs G, Inashkina I, Erenpreisa J. Self-Renewal Signalling in Presenescent Tetraploid IMR90 Cells. J Aging Res 2011; 2011: 103253; PMID:21629737; http://dx.doi.org/10.4061/2011/103253
- Jackson TR, Salmina K, Huna A, Inashkina I, Jankevics E, Riekstina U, Kalnina Z, Ivanov A, Townsend PA, Cragg MS. others. DNA damage causes TP53-dependent coupling of self-renewal and senescence pathways in embryonal carcinoma cells. Cell Cycle 2013; 12(3): 430-441; PMID:23287532; http://dx.doi.org/10.4161/cc.23285
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JR, Guenther MG, Kumar RM, Murray HL, Jenner RG. others. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 2005; 122(6): 947-956; PMID:16153702; http://dx.doi.org/10.1016/j.cell.2005.08.020
- Marques-Torrejon MA, Porlan E, Banito A, Gomez-Ibarlucea E, Lopez-Contreras AJ, Fernandez-Capetillo O, Vidal A, Gil J, Torres J, Farinas I. Cyclin-Dependent Kinase Inhibitor p21 Controls Adult Neural Stem Cell Expansion by Regulating Sox2 Gene Expression. Cell Stem Cell 2013; 12(1): 88-100; PMID:23260487; http://dx.doi.org/10.1016/j.stem.2012.12.001
- Sun Y, Connors KE, Yang DQ. AICAR induces phosphorylation of AMPK in an ATM-dependent, LKB1-independent manner. Mol Cell Biochem 2007; 306(1-2): 239-245; PMID:17786544; http://dx.doi.org/10.1007/s11010-007-9575-6
- Vazquez-Martin A, Lopez-Bonet E, Oliveras-Ferraros C, Perez-Martinez MC, Bernado L, Menendez JA. Mitotic kinase dynamics of the active form of AMPK (phospho-AMPK α(Thr172)) in human cancer cells. Cell Cycle 2009; 8(5): 788-791; PMID:19221486; http://dx.doi.org/10.4161/cc.8.5.7787
- Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 2011; 13(2): 132-U71; PMID:21258367; http://dx.doi.org/10.1038/ncb2152
- Xie BS, Zhao HC, Yao SK, Zhuo DX, Jin B, Lv DC, Wu CL, Ma DL, Gao C, Shu XM. others. Autophagy inhibition enhances etoposide-induced cell death in human hepatoma G2 cells. Int J Mol Med 2011; 27(4): 599-606; PMID:21274505
- Rayess H, Wang MB, Srivatsan ES. Cellular senescence and tumor suppressor gene p16. Int J Cancer 2012; 130(8): 1715-1725; http://dx.doi.org/10.1002/ijc.27316
- Olins AL, Langhans M, Monestier M, Schlotterer A, Robinson DG, Viotti C, Zentgraf H, Zwerger M, Olins DE. An epichromatin epitope Persistence in the cell cycle and conservation in evolution. Nucleus-Austin 2011; 2(1): 47-60; http://dx.doi.org/10.4161/nucl.13655
- Lee J, Go Y, Kang I, Han YM, Kim J. Oct-4 controls cell-cycle progression of embryonic stem cells. Biochem J 2010; 426: 171-181; PMID:19968627; http://dx.doi.org/10.1042/BJ20091439
- Bartova E, Sustackova G, Stixova L, Kozubek S, Legartova S, Foltankova V. Recruitment of Oct4 Protein to UV-Damaged Chromatin in Embryonic Stem Cells. Plos One 2011; 6(12); PMID:22164208; http://dx.doi.org/10.1371/journal.pone.0027281
- Gorban AN, Smirnova EV, Tyukina TA. Correlations, risk and crisis: From physiology to finance. Physica A-Statistical Mechanics Its Applications 2010; 389(16): 3193-3217: http://dx.doi.org/10.1016/j.physa.2010.03.035
- Stuart A. Kauffman, “The origins of order: Self-organization and selection in evolution.,”in (Oxford university press, 2013)
- Huang S. Non-genetic heterogeneity of cells in development: more than just noise. Development 2009; 136(23): 3853-3862; PMID:19906852; http://dx.doi.org/10.1242/dev.035139
- Edgar BA, Orr-Weaver TL. Endoreplication cell cycles: More for less. Cell 2001; 105(3): 297-306; PMID:11348589; http://dx.doi.org/10.1016/S0092-8674(01)00334-8
- Kalmar T, Lim C, Hayward P, Munoz-Descalzo S, Nichols J, Garcia-Ojalvo J, Arias AM. Regulated Fluctuations in Nanog Expression Mediate Cell Fate Decisions in Embryonic Stem Cells. Plos Biol 2009; 7(7); PMID:19582141; http://dx.doi.org/10.1371/journal.pbio.1000149
- Lin TX, Chao C, Saito S, Mazur SJ, Murphy ME, Appella E, Xu Y. P53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat Cell Biol 2005; 7(2): 165-U80; PMID:15619621; http://dx.doi.org/10.1038/ncb1211
- Sanli T, Steinberg GR, Singh G, Tsakiridis T. AMP-activated protein kinase (AMPK) beyond metabolism A novel genomic stress sensor participating in the DNA damage response pathway. Cancer Biol Ther 2014; 15(2): 156-169; PMID:24100703; http://dx.doi.org/10.4161/cbt.26726
- Vazquez-Martin A, Oliveras-Ferraros C, Cufi S, Martin-Castillo B, Menendez JA. Metformin activates an Ataxia Telangiectasia Mutated (ATM)/Chk2-regulated DNA damage-like response. Cell Cycle 2011; 10(9): 1499-1501; PMID:21566461; http://dx.doi.org/10.4161/cc.10.9.15423
- Bungard D, Fuerth BJ, Zeng PY, Faubert B, Maas NL, Viollet B, Carling D, Thompson CB, Jones RG, Berger SL. Signaling Kinase AMPK Activates Stress-Promoted Transcription via Histone H2B Phosphorylation. Science 2010; 329(5996): 1201-1205; PMID:20647423; http://dx.doi.org/10.1126/science.1191241
- Jones RG, Plas DR, Kubek S, Buzzai M, Mu J, Xu Y, Birnbaum MJ, Thompson CB. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell 2005; 18(3): 283-293; PMID:15866171; http://dx.doi.org/10.1016/j.molcel.2005.03.027
- Lee CW, Wong LLY, Tse EYT, Liu HF, Leong VYL, Lee JMF, Hardie DG, Ng IOL, Ching YP. AMPK Promotes p53 Acetylation via Phosphorylation and Inactivation of SIRT1 in Liver Cancer Cells. Cancer Res 2012; 72(17): 4394-4404; PMID:22728651; http://dx.doi.org/10.1158/0008-5472.CAN-12-0429
- Chitikova ZV, Gordeev SA, Bykova TV, Zubova SG, Pospelov VA, Pospelova TV. Sustained activation of DNA damage response in irradiated apoptosis-resistant cells induces reversible senescence associated with mTOR downregulation and expression of stem cell markers. Cell Cycle 2014; 13(9): 1424-1439; PMID:24626185; http://dx.doi.org/10.4161/cc.28402
- Blagosklonny MV. Cell cycle arrest is not senescence. Aging-Us 2011; 3(2): 94-101.
- Wang Q, Wu PC, Dong DZ, Ivanova I, Chu E, Zeliadt S, Vesselle H, Wu DY. Polyploidy road to therapy-induced cellular senescence and escape. Int J Cancer 2013; 132(7): 1505-1515; http://dx.doi.org/10.1002/ijc.27810
- Brock A, Chang H, Huang S. OPINION Non-genetic heterogeneity - a mutation-independent driving force for the somatic evolution of tumours. Nat Rev Genet 2009; 10(5): 336-342; PMID:19337290; http://dx.doi.org/10.1038/nrg2556
- Purvis JE, Karhohs KW, Mock C, Batchelor E, Loewer A, Lahav G. p53 Dynamics Control Cell Fate. Science 2012; 336(6087): 1440-1444; PMID:22700930; http://dx.doi.org/10.1126/science.1218351
- Capparelli C, Chiavarina B, Whitaker-Menezes D, Pestell TG, Pestell RG, Hulit J, Ando S, Howell A, Martinez-Outschoorn UE, Sotgia F, others. CDK inhibitors (p16/p19/p21) induce senescence and autophagy in cancer-associated fibroblasts, “fueling” tumor growth via paracrine interactions, without an increase in neo-angiogenesis. Cell Cycle 2012; 11(19): 3599-3610; PMID:22935696; http://dx.doi.org/10.4161/cc.21884
- Ivanov A, Pawlikowski J, Manoharan I, van Tuyn J, Nelson DM, Rai TS, Shah PP, Hewitt G, Korolchuk VI, Passos JF, others. Lysosome-mediated processing of chromatin in senescence. J Cell Biol. 2013; 202(1): 129-143; PMID:23816621; http://dx.doi.org/10.1083/jcb.201212110
- Nagl Walter. Endopolyploidy and polyteny in differentiation and evolution. 1978
- Hagan CR, Sheffield RF, Rudin CM. Human Alu element retrotransposition induced by genotoxic stress. Nat Genet 2003; 35(3): 219-220; PMID:14578886; http://dx.doi.org/10.1038/ng1259
- Guo HS, Chitiprolu M, Gagnon D, Meng LR, Perez-Iratxeta C, Lagace D, Gibbings D. Autophagy supports genomic stability by degrading retrotransposon RNA. Nat Commun 2014; 5:5276
- Wang JR, Geesman GJ, Hostikka SL, Atallah M, Blackwell B, Lee E, Cook PJ, Pasaniuc B, Shariat G, Halperin E, others. Inhibition of activated pericentromeric SINE/Alu repeat transcription in senescent human adult stem cells reinstates self-renewal. Cell Cycle 2011; 10(17): 3016-3030; PMID:21862875; http://dx.doi.org/10.4161/cc.10.17.17543
- Baker DJ, Sedivy JM. Probing the depths of cellular senescence. J Cell Biol 2013; 202(1): 11-13; PMID:23816622; http://dx.doi.org/10.1083/jcb.201305155
- Cecco M, Criscione SW, Peckham EJ, Hillenmeyer S, Hamm EA, Manivannan J, Peterson AL, Kreiling JA, Neretti N, Sedivy JM. Genomes of replicatively senescent cells undergo global epigenetic changes leading to gene silencing and activation of transposable elements. Aging Cell 2013; 12(2): 247-256; PMID:23360310; http://dx.doi.org/10.1111/acel.12047
- Sui X, Chen R, Wang Z, Huang Z, Kong N, Zhang M, Han W, Lou F, Yang J, Zhang Q, others. Autophagy and chemotherapy resistance: a promising therapeutic target for cancer treatment. Cell Death Dis 2013; 4:e838
- Dorr JR, Yu Y, Milanovic M, Beuster G, Zasada C, Dabritz JHM, Lisec J, Lenze D, Gerhardt A, Schleicher K, others. Synthetic lethal metabolic targeting of cellular senescence in cancer therapy. Nature 2013; 501(7467):421-5; PMID:23945590; http://dx.doi.org/10.1038/nature12437
- Zeuthen J, Norgaard JOR, Avner P, Fellous M, Wartiovaara J, Vaheri A, Rosen A, Giovanella BC. Characterization of A Human Ovarian Teratocarcinoma-Derived Cell-Line. Int J Cancer 1980; 25(1): 19-32; http://dx.doi.org/10.1002/ijc.2910250104
- Gao C, Miyazaki M, Li JW, Tsuji T, Inoue Y, Namba M. Cytogenetic characteristics and p53 gene status of human teratocarcinoma PA-1 cells in 407–445 passages. Int J Mol Med 1999; 4(6): 597-600; PMID:10567668
- Erenpreisa J, Freivalds T. Anisotropic staining of apurinic acid with toluidine blue. Histochemistry 1979; 60(3): 321-325; PMID:89109; http://dx.doi.org/10.1007/BF00500660
