Abstract
DNA double-strand break repair by the error-free pathway of homologous recombination (HR) requires the concerted action of several factors. Among these, EXO1 and DNA2/BLM are responsible for the extensive resection of DNA ends to produce 3′-overhangs, which are essential intermediates for downstream steps of HR. Here we show that EXO1 is a SUMO target and that sumoylation affects EXO1 ubiquitylation and protein stability. We identify an UBC9-PIAS1/PIAS4-dependent mechanism controlling human EXO1 sumoylation in vivo and demonstrate conservation of this mechanism in yeast by the Ubc9-Siz1/Siz2 using an in vitro reconstituted system. Furthermore, we show physical interaction between EXO1 and the de-sumoylating enzyme SENP6 both in vitro and in vivo, promoting EXO1 stability. Finally, we identify the major sites of sumoylation in EXO1 and show that ectopic expression of a sumoylation-deficient form of EXO1 rescues the DNA damage-induced chromosomal aberrations observed upon wt-EXO1 expression. Thus, our study identifies a novel layer of regulation of EXO1, making the pathways that regulate its function an ideal target for therapeutic intervention.
Introduction
Faithful repair of DNA lesions is essential to the maintenance of genome stability.Citation1 Double-strand breaks (DSBs) are the most toxic DNA lesions generated by ionizing radiation (IR), certain chemotherapeutic drugs, collapse of stalled DNA replication forks, endogenous metabolic processes or during meiotic recombination.Citation2–4 Inappropriate repair of DSBs may cause gross chromosomal aberrationsCitation5 resulting in carcinogenesis through activation of oncogenes or inactivation of tumor suppressor genes.Citation1 Cells utilize 2 main mechanisms to repair DSBs: non-homologous end-joining (NHEJ) and homologous recombination (HR).Citation1 Rejoining of DSBs by NHEJ takes place throughout the cell cycle, whereas HR is restricted to the S and G2 phases, where sister chromatids are available as templates. HR is initiated by 5′-3′ resection of DSBs to produce single-stranded DNA (ssDNA) tails that function not only as signal for the ATR-mediated DNA damage checkpoint but also to allow formation of RAD51 filaments and recruitment of recombination proteins.Citation6 Studies conducted in yeast and mammalian cells led to the proposal of a mechanism according to which MRN and CtIP (MRX and Sae2 in yeast) orchestrate the initial trimming of DNA-ends, which is followed by processive resection carried out by 2 alternative pathways that involve either EXO1 or BLM/DNA2.Citation7,8
EXO1 was originally identified in S. pombeCitation9 and subsequently in humans.Citation10 It belongs to the Rad2 family of DNA repair nucleases and is able to remove mononucleotides from the 5′ end of the DNA duplex.Citation11 EXO1 is implicated in several DNA repair pathways including mismatch repair, post-replication repair, meiotic and mitotic recombination and double-strand break repair.Citation12-16 S. cerevisiae Exo1 acts redundantly with Rad27 in processing Okazaki fragments during DNA replication.Citation17 More recently, Exo1 was shown to be recruited to stalled replication forks where it counteracts fork reversal,Citation18 making it a multifaceted protein in pathways controlling genome stability.
The recruitment of proteins marking sites of DNA damage and the choice of pathways addressing their repair heavily relies on control by post-translational modifications (PTMs) that occur in a defined hierarchy.Citation19,20 An archetypal example is the phosphorylation-dependent binding of MDC1 to γH2AX, followed by recruitment of the E3-ubiquitin ligases RNF8 and RNF168 whose activity is enhanced by PIAS1- and PIAS4-mediated sumoylation, resulting in efficient H2A and H2B mono-ubiquitylation.Citation19,21 An interesting feature of PTMs is not only the hierarchical order at which they occur but also their reciprocal influence.Citation19
We have previously shown that human and yeast EXO1 are tightly regulated by interaction with CtIP/RBBP8 and 14–3–3 proteins at DSBs and stalled forks, respectively.Citation5,22 Additionally, human EXO1 is controlled by PTMs, with ATR-dependent phosphorylation targeting it to ubiquitin-mediated degradation upon replication fork stalling,Citation23,24 and ATM-dependent phosphorylation restraining its activity during homologous recombination.Citation25 Analogously, Mec1-dependent phosphorylation was shown to inhibit yeast Exo1 activity at uncapped telomeres.Citation26 Recently, evidence was provided for a role of CDK-dependent phosphorylation of human EXO1 in the pathway choice of DSB repair.Citation27
In this study we have focused on elucidating the molecular mechanism of EXO1 regulation upon stalled DNA replication. We have performed an RNAi-based screen of human E2-conjugating enzymes and identified UBE2I, the human homolog of yeast Ubc9, as major effector of EXO1 stability. We show that EXO1 is sumoylated in vivo in a PIAS1- and PIAS4-dependent manner and in vitro using a reconstituted system. We also provide evidence that SENP6 physically interacts with EXO1 and that depletion of SENP6 promotes EXO1 degradation. We conclude with the identification of the major SUMO-conjugation sites in EXO1 and report that the high rate of chromosome breaks caused by camptothecin in cells ectopically expressing wild-type EXO1 did not occur upon expression of a SUMO-deficient EXO1 mutant.
Results
EXO1 is degraded in response to camptothecin
In response to agents that cause stalling of DNA replication forks, such as hydroxyurea (HU), an inhibitor of the enzyme ribonucleotide reductase (RNR), () or aphidicolin (APH), a catalytic inhibitor of B-family DNA polymerases (), endogenous or ectopically expressed human EXO1 undergoes ubiquitin-dependent proteasomal degradation.Citation23,24 To substantiate and extend this observation, we also tested the effect of the topoisomerase-1 inhibitor camptothecin (CPT). Treatment with CPT led to a decrease of the endogenous EXO1 protein level and this effect was rescued by the presence of the proteasome inhibitor MG-132 (). Given the low level of EXO1 expression in mammalian cells, which requires combined immunoprecipitation and Western blot to visualize the protein (, and ref.Citation23), we performed all subsequent studies on HEK-293T cells transiently expressing GFP-EXO1 or on U2OS and HEK-293 stably expressing GFP-EXO1. In analogy with the endogenous protein, exogenous GFP-EXO1 was sensitive to CPT-treatment (, left panel) already at the lowest concentration tested (Fig. S1). The ability of CPT to cause a decrease in EXO1 protein level was also confirmed in U2OS cells stably expressing GFP-EXO1 (U2OS-GFP-EXO1)Citation5 by either Western blot analysis (, right panel) or immunofluorescence (). Addition of the proteasome inhibitor MG-132 rescued CPT-induced EXO1 degradation to a significant extent ().
Figure 1. Camptothecin targets EXO1 for proteasome-mediated degradation. (A) HEK-293T cells were treated with hydroxyurea (HU, 2 mM) for 16h. Endogenous EXO1 was immunoprecipitated with a rabbit polyclonal antibody and visualized with a specific monoclonal antibody. Whole cell extracts (WCEs, inputs) were analyzed using the indicated antibodies. (B) HEK-293T cells ectopically expressing GFP-EXO1 were treated with HU (2 mM) and MG-132 (10 μM) for 16h. WCEs were analyzed using the indicated antibodies. Mock transfected HEK-293T were used as control. (C HEK-293T cells were treated with camptothecin (CPT, 1 μM), aphidicolin (APH, 15 μM) and MG-132 (10 μM) for 4h, as indicated. Endogenous EXO1 was visualized as described in A. IgG(H) were used as control for the quality of the immunoprecipitation (IP). WCEs (inputs) were analyzed using the indicated antibodies. The EXO1 signal was quantified upon normalization on IgG(H): CTRL, lane 1 = 1; CPT, lane 2 = 0.19; APH, lane 3 = 0.77. (D) HEK-293T cells ectopically expressing GFP-EXO1 (left) or stable U2OS-GFP-EXO1 cells (right) were treated with CPT (1 μM) and MG-132 (10 μM) for 4h. WCEs were analyzed using the indicated antibodies. Mock transfected HEK-293T and wild-type U2OS were used as controls. (E) Indirect immunofluorescence analysis of wild-type and stable U2OS-GFP-EXO1 cells shown in (D).
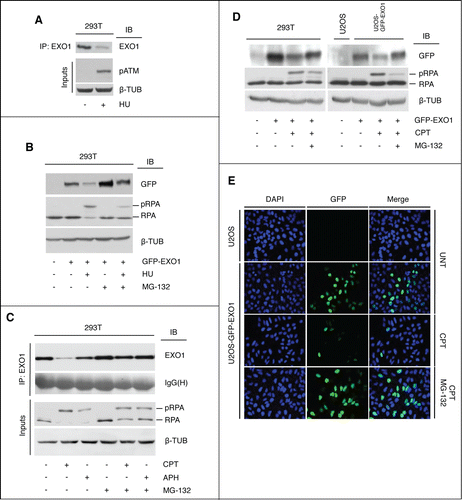
Taken together, these data show that both endogenous and exogenous EXO1 are targets for ubiquitin-dependent proteasomal degradation in response to topoisomerase-I inhibition.
EXO1 degradation depends on SUMO pathways
In order to shed light on the mechanism controlling EXO1 protein stability, we set out to identify the pathway responsible for EXO1 degradation. To this end, we performed an immunofluorescence-based high-throughput screen in U2OS-GFP-EXO1 cells using a siRNA library encompassing all 37 human E2-conjugating enzymes, followed by image and computational analysis of the acquired data. The most significant increase in green fluorescence, taken as read-out for GFP-EXO1 protein stabilization, was obtained upon depletion of UBE2I (the human homolog of the yeast Ubc9 enzyme, referred to as UBC9), both in basal and damaging conditions (). We could exclude any indirect effect of UBC9 depletion on cell size and cell cycle phases (Fig. S2). To corroborate the role of UBC9, we examined EXO1 stability in U2OS cells depleted for UBC9 with a single siRNA. Under these conditions, indirect immunofluorescence confirmed an increase of GFP-EXO1 protein level in both untreated and CPT-treated cells (). UBC9 depletion led to ∼2.5- and ∼3.5-fold increase in the number of GFP-positive cells in untreated and CPT-treated conditions, respectively (Fig. S3). HEK-293T cells ectopically expressing GFP-EXO1 confirmed the pattern of EXO1 protein stabilization upon depletion of UBC9 ().
Figure 2 (See previous page). UBC9 controls EXO1 protein level. (A) UBC9 depletion causes stabilization of EXO1. An E2 conjugating enzymes siRNA library was screened on untreated or HU-treated U2OS cells stably expressing GFP-EXO1. Mean intensity of the green signal averaged over all cells per gene was plotted (left of the panel). Single cell distribution of the green signal for Ctrl- and UBC9- (UBE2I in the figure) depleted cells was plotted (right of the panel). Entire well images and magnified indicative fields (greyscale) of Ctrl and UBC9 siRNA-depleted cells are shown. Scale bar entire field = 350 μm. Scale bar enlargement = 50 μm. (B) Indirect immunofluorescence of stable U2OS-GFP-EXO1 cells that were transfected with CTRL or UBC9 siRNA oligonucleotides and left untreated or treated with CPT. (C) Western blot analysis of HEK-293T cells ectopically expressing GFP-EXO1 that were transfected with CTRL or UBC9 siRNA oligonucleotides and treated with CPT. WCEs were analyzed using the indicated antibodies. (D) UBC9 depletion impairs EXO1 ubiquitylation. HEK-293T cells ectopically expressing GFP-EXO1 were transfected with CTRL or UBC9 siRNA oligonucleotides and treated with CPT in the presence of MG-132 (10 μM). WCEs were immunoprecipitated and analyzed with the indicated antibodies.
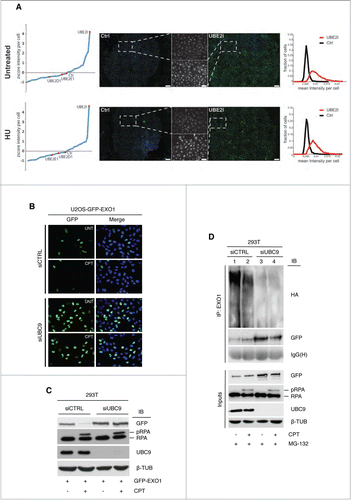
To assess whether sumoylation affects ubiquitylation, we examined the extent of EXO1 ubiquitylation upon down-regulation of UBC9 expression. The pattern of GFP-EXO1 ubiquitylation observed in control siRNA-treated cells (, lanes 1–2) was clearly reduced upon UBC9 depletion (, lanes 3–4). The apparent increase of the protein band corresponding to GFP-EXO1 in UBC9-depleted cells possibly reflects enrichment for the non-ubiquitylated form of the protein migrating in a single band (, lanes 3–4 vs. One–2).
These data indicate that EXO1 protein level is controlled by SUMO pathways both in unperturbed conditions and upon treatment with HU or CPT and establish that sumoylation is a prerequisite for ubiquitin-mediated EXO1 degradation.
EXO1 is a direct target of SUMO pathways
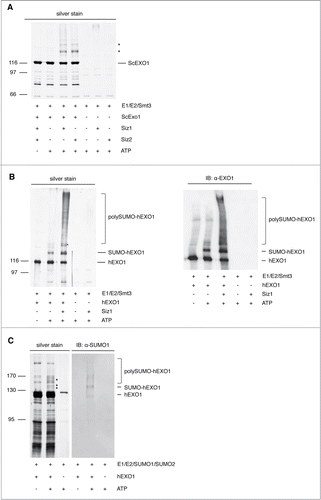
Next, we examined whether EXO1 is a direct target for sumoylation. To this end, we performed in vitro studies with reconstituted sumoylation machinery.28 Using purified recombinant components of the yeast machinery, which is a very robust system for such enzymatic assays,28 we observed that both yeast and human EXO1 are modified by SUMO (). Considering the involvement of EXO1 in DNA repair pathways, we asked whether E3-SUMO ligases with an established role in the DNA damage response would affect EXO1 sumoylation. We observed that the presence of Siz1 or Siz2 was essential for the sumoylation of yeast Exo1 (), significantly increasing the pattern of human EXO1 sumoylation obtained in the absence of a specific E3 enzyme (). Furthermore, in vitro assays performed with purified recombinant human sumoylation components (Fig. S4) confirmed that human EXO1 is target for sumoylation ().
Figure 3. In vitro reconstitution of EXO1 sumoylation. (A) Yeast Exo1 is sumoylated in vitro. Sumoylation of yeast Exo1 was reconstituted in vitro with yeast E1, E2 and Smt3-KR in the presence or the absence of the E3 ligases Siz1 or Siz2, respectively. Samples were resolved by SDS-PAGE and visualized by silver staining. Asterisks indicate the sumoylated forms of Exo1. (B) Human EXO1 is sumoylated in vitro. Left: Sumoylation of human EXO1 was performed as described in (A). Samples were resolved by SDS-PAGE and visualized by silver staining. Right: The samples shown on the left panel were examined by Western blotting using an EXO1-specific monoclonal antibody. Sumoylated forms of EXO1 are indicated. (C) EXO1 sumoylation with a reconstituted human SUMO-machinery. In vitro sumoylation of human EXO1 was performed with human E1, E2, SUMO1 and SUMO2. Samples were resolved by SDS-PAGE and visualized by silver staining or Western blotting using a monoclonal antibody to SUMO1. Asterisks indicate the sumoylated forms of EXO1.
To extend these observations to a cellular system, we ectopically expressed Myc-tagged SUMO1 or SUMO2 along with GFP-EXO1 in HEK-293T cells. Immunoprecipitation of GFP-EXO1 revealed constitutive sumoylation and a slightly reduced preference for SUMO2 over SUMO1 upon CPT treatment (). To follow up the observation on the importance of Siz1 and Siz2 in EXO1 sumoylation in vitro (), we depleted PIAS1 or PIAS4, the human E3-SUMO ligases homolog of Siz enzymes, in HEK-293T cells using established siRNA oligonucleotides.29 Downregulation of PIAS1 effectively increased the level of EXO1 in untreated and CPT-treated cells (). An increase of EXO1 protein level was also observed upon PIAS4 depletion, though in this case the milder effect on EXO1 stabilization might be the result of incomplete PIAS4 silencing (). Similar data were obtained using pools of siRNA oligonucleotides for PIAS1 and PIAS4 (data not shown).
Figure 4 (See previous page). EXO1 is target of sumoylation in vivo. (A) HEK-293T cells ectopically expressing GFP-EXO1 and Myc-SUMO1 or Myc-SUMO2 were treated with CPT (1 μM) and MG-132 (10 μM) for 4h. GFP-EXO1 was immunoprecipitated with a rabbit polyclonal antibody and visualized using a monoclonal antibody to the Myc-tag. The membrane was stripped and re-probed with a mouse monoclonal to GFP. IgG(H) were used as control for the quality of the IP. WCEs (inputs) were analyzed before IP. (B) PIAS1 or PIAS4 depletion increases EXO1 stability. HEK-293T cells ectopically expressing GFP-EXO1 were depleted for PIAS1 or PIAS4 and either left untreated or treated with CPT. WCEs were analyzed using the indicated antibodies. (C) Depletion of SENP6 affects EXO1 protein level. Western blot analysis of stable U2OS-GFP-EXO1 cells depleted for SENP5 or SENP6 and either left untreated or treated with CPT. WCEs were analyzed using the indicated antibodies. (D) EXO1 interacts with SENP6 in vivo. HEK-293T cells were transfected with Flag-SENP6 and GFP-EXO1 as indicated and either left untreated or treated with CPT. WCEs were immunoprecipitated with a mouse monoclonal antibody to the Flag and membranes were probed with a rabbit polyclonal antibody to GFP. The membrane was stripped and re-probed with a mouse anti-Flag monoclonal antibody. IgG(H) were used as control for the quality of the immunoprecipitation (IP). WCEs were analyzed before IP using the indicated antibodies. (E) EXO1 and SENP6 interact in vitro. Intein-tagged EXO1 was bound to chitin beads and used as prey to capture purified recombinant SENP6 protein (628–1112 aa). The silver-stained gel shows proteins remaining in the supernatant after capture (S) or elution from chitin beads (E) trapping intein-EXO1 (lanes 1 and 2) or beads alone (lanes 3 and 4). The position of EXO1 and SENP6 is indicated.
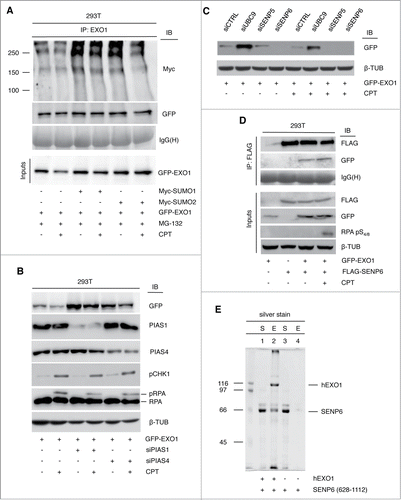
Since most PTMs are dynamic and reversible, de-sumoylation is equally important to sumoylation in the control of protein function. Therefore, we tested the effect of various SENP proteases on EXO1 stability. RNAi-mediated depletion of SENP proteases in stable U2OS cells revealed that SENP6 downregulation (Fig. S5) led to decrease of GFP-EXO1 protein to undetectable levels (). SENP6 does not participate in the maturation of SUMO1–3 precursors but can only deconjugate SUMO chains.Citation19 Hence, effects of SENP6 depletion on the pool of SUMO molecules could be ruled out as possible explanation for our results. Protein interaction studies in HEK-293T cells ectopically expressing Flag-SENP6 and GFP-EXO1 showed that the 2 proteins could be co-immunoprecipitated, in both untreated and CPT-treated conditions, indicative of constitutive physical interaction (). In addition, pull-down studies with recombinant proteins showed that intein-tagged EXO1, which was affinity-purified with chitin beads, was able to capture soluble SENP6 protein (), indicating direct protein interaction.
Taken together, these data point to a functional role for the E3-SUMO ligases PIAS1/PIAS4 and the de-sumoylating enzyme SENP6 in the control of EXO1 protein stability.
Effect of EXO1 sumoylation on genome stability
In order to assess the physiological role of sumoylation, we set out to identify the SUMO conjugation sites in human EXO1. Analysis of EXO1 primary sequence revealed the presence of 4 potential sumoylation sites corresponding to the canonical consensus motif Φ-K-x-E/D, with 2 consecutive lysine residues located in the most C-terminal site (). Mass spectrometric analysis of in vitro sumoylated EXO1 revealed that lysine K655 and K801 were modified by SUMO (Fig. S6A and B), with the latter displaying conservation through evolution (). Hence, we generated the K655/K801/K802>R triple mutant (EXO1–3KR). In vitro assays conducted with purified recombinant EXO1–3KR showed that the sumoylation pattern of wt-EXO1 was essentially abolished in the mutant (). DNA resection activity of non-sumoylated EXO1–3KR mutant was similar to that of the WT protein (Fig. S7), indicating that Lysine to Arginine substitution at these sites did not alter EXO1 catalytic activity.
Figure 5 (See previous page). Identification of EXO1 sumoylation sites and effect of sumoylation on chromosomal aberrations caused by camptothecin. (A) Schematic representation of EXO1. The N- and I-catalytic domains are depicted in orange and 5 potential sumoylation sites are indicated in red. Amino acid sequences flanking sumoylation sites are shown. (B) Alignment of the human EXO1 region containing K801 with Exo1 orthologs. Fully and partially conserved amino acid residues are highlighted in black and gray boxes, respectively. The sumoylated lysine in human EXO1 is marked with an asterisk. (C) EXO1–3KR is not sumoylated in vitro. In vitro sumoylation assay of human EXO1-WT or EXO1–3KR (K655801802>R) was performed with yeast E1, E2, Smt3-KR and Siz1 in the presence or the absence of ATP. The asterisk indicates the sumoylated form of EXO1. (D) EXO1–3KR is not sumoylated in vivo. HEK-293 cells stably expressing GFP-EXO1-WT or GFP-EXO1–3KR were treated with CPT (1 μM, 4h) and extracted under denaturing conditions. Proteins were precipitated with a rabbit antibody to EXO1 and revealed with a monoclonal antibody to the Myc-tag. WCEs (inputs) were analyzed before IP using the indicated antibodies. (E) Sumoylation sites mutation rescues the chromosomal aberrations caused by EXO1. Chromosome breaks and fragments observed in metaphase spreads of HEK-293 cells stably transfected with the empty vector (EV) or expressing either GFP-EXO1-wt or GFP-EXO1–3KR and treated in the presence or the absence of CPT. Values are plotted as: [breaks or fragments / total metaphases counted]. At least 30 metaphases were examined for each condition. Inset: example of chromosome breaks and fragments used in the quantification.
![Figure 5 (See previous page). Identification of EXO1 sumoylation sites and effect of sumoylation on chromosomal aberrations caused by camptothecin. (A) Schematic representation of EXO1. The N- and I-catalytic domains are depicted in orange and 5 potential sumoylation sites are indicated in red. Amino acid sequences flanking sumoylation sites are shown. (B) Alignment of the human EXO1 region containing K801 with Exo1 orthologs. Fully and partially conserved amino acid residues are highlighted in black and gray boxes, respectively. The sumoylated lysine in human EXO1 is marked with an asterisk. (C) EXO1–3KR is not sumoylated in vitro. In vitro sumoylation assay of human EXO1-WT or EXO1–3KR (K655801802>R) was performed with yeast E1, E2, Smt3-KR and Siz1 in the presence or the absence of ATP. The asterisk indicates the sumoylated form of EXO1. (D) EXO1–3KR is not sumoylated in vivo. HEK-293 cells stably expressing GFP-EXO1-WT or GFP-EXO1–3KR were treated with CPT (1 μM, 4h) and extracted under denaturing conditions. Proteins were precipitated with a rabbit antibody to EXO1 and revealed with a monoclonal antibody to the Myc-tag. WCEs (inputs) were analyzed before IP using the indicated antibodies. (E) Sumoylation sites mutation rescues the chromosomal aberrations caused by EXO1. Chromosome breaks and fragments observed in metaphase spreads of HEK-293 cells stably transfected with the empty vector (EV) or expressing either GFP-EXO1-wt or GFP-EXO1–3KR and treated in the presence or the absence of CPT. Values are plotted as: [breaks or fragments / total metaphases counted]. At least 30 metaphases were examined for each condition. Inset: example of chromosome breaks and fragments used in the quantification.](/cms/asset/931188fd-ba34-45ee-b47b-8a09d94fa22f/kccy_a_1060381_f0005_c.gif)
To conduct studies in a cellular system, we generated HEK-293 cells stably expressing GFP-EXO1-WT or GFP-EXO1–3KR. Immunofluorescence experiments confirmed that both proteins were localized to the nucleus (data not shown). Treatment with CPT revealed that the sumoylation pattern of GFP-EXO1-WT was abolished in the mutant (), confirming the observations made in the studies conducted in vitro (). These data additionally indicated that mutation of the identified sumoylation sites did not effectively rescue EXO1 protein stability in response to CPT ().
To evaluate the biological effects of EXO1 sumoylation we examined DNA processing in vivo, using a flow cytometry-based method that couples quantification of DNA resection (chromatin-bound RPA) with analysis of DNA content (DAPI), according to an established protocol.Citation30 To this end, HEK-293 cells stably transfected with empty vector (EV), GFP-EXO1-WT or GFP-EXO1–3KR (Fig. S8A) were treated with thymidine to enrich the early S-phase population. Under these basal conditions we observed a ∼10-fold of chromatin-bound RPA in cells expressing GFP-EXO1-WT over empty vector-transfected cells, with GFP-EXO1–3KR cells displaying a more modest increase of the RPA signal (Fig. S8B). Next, we examined chromosomal abnormalities. CPT-treatment of GFP-EXO1-WT expressing cells resulted in a significant amount of aberrations, mostly consisting of breaks and fragments, a pattern that was clearly reduced in cells expressing the GFP-EXO1–3KR mutant ( and Fig. S9).
In summary, these data show that interfering with sumoylation rescues the deleterious effect of supernumerary EXO1 molecules on DNA, contributing to maintain genome integrity.
Discussion
Genetic conditions characterized by dysfunction of the machinery that signals DNA damage and/or addresses its repair are associated with a predisposition to the development of cancer and resistance to therapy,Citation1,Citation31 providing a direct demonstration of the importance of surveillance pathways in genome stability.Citation32 DSBs, which are among the most dangerous DNA lesions, are estimated to occur at a rate of 10 per cell per day in primary human or mouse fibroblasts.Citation33 These naturally occurring lesions are generated upon collapse of stalled DNA replication forks, replication across nicks, reactive oxygen species of endogenous origin or the untimely action of DNA endonucleases (topoisomerases or RAG and AID).Citation33 Additionally, DSBs can occur as a result of lesions caused by external agents, such as ionizing radiation and mutagenic chemicals.Citation6,Citation34
Despite the intense effort currently being exerted to identify proteins and pathways involved in recognition and repair of the various forms of DNA damage, we are only beginning to appreciate the function of DNA repair mechanisms, their cooperation and regulation. In particular, the hierarchy and coordination of post-translational modifications (PTMs) on recruitment, function and stability of DNA repair proteins at sites of damage represent new challenges in the field.Citation19,Citation21,Citation35 To advance our understanding, and based on previous evidence from our laboratory,Citation5,Citation22–24 in this study we characterize the molecular mechanism controlling Exonuclease-1 (EXO1), a common component of machineries processing stalled replication forks, DNA base mismatches and DSBs.Citation18,Citation36–39
Given the low level of EXO1 expression in mammalian cellsCitation5,Citation23,Citation24 and legitimated by studies that used over-expressed EXO1 to assess its role in nucleotide-excision repair pathways,Citation40 we first demonstrated that endogenous () and exogenous EXO1 () are controlled in a similar manner upon DNA damage, and then used GFP-EXO1 for our study. In search of the pathway that controls EXO1 protein stability, we found that EXO1 is a target of sumoylation (), extending observations made in proteome-wide studies.Citation41,42 The finding that stabilization of EXO1 occurred in non-damaged cells depleted for UBC9 () is consistent with the fact that EXO1 undergoes constitutive degradation, as we previously demonstrated by chemical inhibition of the proteasome with MG-132 or in ATR-deficient Seckle cells.Citation23,24 In vitro () and in vivo studies () confirmed that EXO1 is sumoylated and allowed establishing a role for UBC9-PIAS1/PIAS4 in the regulation of EXO1 protein stability in vivo (). Furthermore, EXO1 constitutively interacts with the SUMO-protease SENP6 () that, in turn, regulates EXO1 protein level in the cell ().
SUMO-Targeted Ubiquitin E3 Ligases (STUbL) recognize sumoylated proteins targeting them to degradation. Hence, we examined whether RNF4, a STUbL involved in DNA damage response,Citation43,44 might be a component of the pathway controlling EXO1 degradation. The E2 siRNA-library screening ruled out that enzymes cooperating with RNF4, such as UBE2D and UBE2E – human homologues of yeast Ubc4/Ubc5 – could affect EXO1 stability (), and RNF4 depletion with a specific oligonucleotideCitation44 had no effect on EXO1 protein level (data not shown), ruling out RNF4 as candidate Ubiquitin E3 Ligase for EXO1.
To address the biological impact of sumoylation, we identified SUMO modified residues in human EXO1 (, and Figs. S6A and B). Sequence alignment showed that K655 was not conserved, whereas K801 displayed conservation in M. musculus, D. rerio, X. laevis, D. melanogaster and S. cerevisiae (). Mutation of the identified lysines to arginine led to suppression of EXO1 sumoylation in vitro () and in vivo (), although this was not mirrored by a significant rescue of EXO1 degradation in response to CPT (). These data likely indicate that either additional minor sites of sumoylation are present in EXO1 and play a role in the regulation of protein stability or that UBC9 affects components of the EXO1 ubiquitylation pathway that in turn control EXO1 protein level.
Next we examined the phenotype of cells expressing a SUMO-mutant EXO1. We observed that over-expression of EXO1-WT led to increased DNA resection (Fig. S8B) and chromosome breaks in comparison to control cells. On the other hand, overexpression of a comparable amount of SUMO-mutant EXO1 displayed reduced rates of chromosomal aberrations in cells experiencing replication stress ( and Fig. S9). These data suggest that sumoylation may have multiple roles, with the control of EXO1 stability representing one aspect of the coin, and the regulation of EXO1 function at sites of DNA damage likely being the most relevant facet.
Hierarchy and reciprocal influence of PTMs are of key importance in determining protein function.Citation19 For instance, it has been shown that sequential PTMs of FEN1 set the stage for its recruitment to DNA and the subsequent degradation of this important nuclease, thus ensuring precise cell cycle progression and preventing transformation.Citation45 We have previously shown that, in response to replication stress, ATR-dependent phosphorylation of EXO1 at 3 sites promotes its degradation through ubiquitin-mediated proteasomal pathways.Citation23,24 Our early data indicated the presence of 12 sites of phosphorylation in EXO1,24 while the most recent estimate is approaching 25 sites (Eid W, El-Shemerly M, Hess D and Ferrari S, manuscript in preparation). Studies focused on the effect of PTMs on EXO1 activity showed that ATM-dependent phosphorylation at a single site possibly occurs after recruitment of EXO1 to DSBs and serves to modulate its activity during repair of DSBs by homologous recombination.Citation25 Along this line, a recent study showed that CDK-dependent phosphorylation of EXO1 at 4 sites in S- and G2-phase increases its recruitment to DNA DSBs, possibly through interactions with BRCA1. This, in turn, dictates the choice for error-free repair of DNA DSBs via homologous recombination as opposed to an error-prone non-homologous end-joining mode of repair.Citation27 On the other hand, phosphorylation of yeast Exo1 in a checkpoint-dependent manner, at a set of sites that are not conserved in the human protein and that do not conform to the recognition by CDKs, was proposed to limit Exo1 activityCitation26,46 The data presented in our study add a further layer of complexity to this picture, showing that sumoylation limits pathological resection of DNA by human EXO1, thus contributing to prevent genomic instability. Eventual hierarchy and reciprocal influence of cell cycle-dependent EXO1 phosphorylation and sumoylation in modulating recruitment to DNA and resection activity, as well the participation of checkpoint-dependent EXO1 phosphorylation to this scenario, are interesting issues awaiting exploration.
With regard to possible translation of our findings, based on the data presented here we envisage exploiting pathways of EXO1 post-translational modification in cancer cells to induce apoptosis as result of uncontrolled DNA damage processing. Specifically, we speculate that further characterization of the effect of EXO1 sumoylation on interaction with its partners and recruitment to DNA will provide the knowledge to modulate both EXO1 protein stability and permanence at sites of DNA damage. This, in turn, would favor pathological resection of DNA ends and result in apoptosis.
Sumoylation, as part of the mechanism controlling EXO1 function, may also help explaining the fitness of cancer cells characterized by up-regulated EXO1 gene expression (Citation47 and www.nextbio.com). These cells possibly exploit inefficient sumoylation of supernumerary EXO1 molecules to limit pathological resection of DNA, thus suppressing apoptotic signals that result from the accumulation of chromosomal aberrations of the type that we report.
In light of the multifaceted role of EXO1 in various DNA repair processes, the hypotheses discussed above will require in-depth studies.
Materials and Methods
Cell culture and transfections
Low passage, mycoplasma-free HEK-293, HEK-293T and U2OS cells, either wild-type or stably expressing GFP-EXO1, were maintained as described.Citation5,23 Transient transfections were performed using Truefect-Lipo (United Biosystems Inc..). Single siRNA oligonucleotides (Microsynth, Table S1) were transfected at 40 nM concentration using Truefect-Lipo (United BioSystems Inc.. USA) in 2 consecutive rounds. Experiments were typically performed 48–72 h.
High-content single-cell imaging of cell populations
96-well plates were imaged with a wide-field 20x objective microscope (ImageExpress Micro, Molecular Devices). Five focal planes per image and 49 sites per well were acquired. The maximum intensity projection of the 5 focal planes was saved for each site and used for further analysis. Images were stored as 16-bit uncompressed TIFFs.
Image analysis pipeline, single-cell feature
For each single cell, nuclei were detected based on the DAPI signal using the open-source software, Cell Profiler.Citation48 For every nucleus, the intensity for DAPI (blue) and EXO1 (green) (11 features per object and channel), shape (30 features per object) and texture features (15 features per object and channel) were extracted. For measurements of the population context of each single cell a point-spread function was used to measure the local cell density, the position of the cell in an islet (being on the edge or inside) and distance to cell islet edge (minimal distance between a cell and the edge of an islet).Citation49 In total ∼50 features per cell were extracted. For the single-cell EXO1 readout we z-scored the mean intensity per cell (subtracting the average value over all single cells in the plate from each single-cell value and dividing this with the standard deviation of all single cells of the plate).
In vitro sumoylation assay
The in vitro sumoylation assay using yeast proteins was performed as previously described.Citation28 Briefly, 150 nM Aos1/Uba2, 250 nM Ubc9, 4.3 μM Smt3-KR, 10 nM Siz1 or Siz2, 1 mM ATP, buffer S1 (100 mM Tris-HCl pH 7.5, 10 mM MgCl2), and 1 μM human or yeast Exo1 were incubated in a 10 μl volume at 30°C for 45 min. Reactions were stopped by addition of SDS Laemmli buffer and analyzed by SDS-PAGE.
The in vitro sumoylation assay using human proteins was performed in a 10 μl volume containing 100 nM GST-SAE2/SAE1, 2.8 μM UBC9, 4.3 μM SUMO1, 4.3 μM SUMO2, 1 mM ATP, buffer S (50 mM HEPES, 10 mM MgCl2, 0.1 mM DTT), and 1 μM EXO1. Reactions were incubated at 30°C for 1h, stopped by addition of SDS Laemmli buffer and analyzed by SDS-PAGE.
Pull-down, immunoprecipitation and Western blotting
Purified SENP6 protein was incubated with 25 μl of intein-tagged EXO1 pre-bound to chitin beads (New England Biolabs) for 30 min at 4°C with gentle rocking in 25 μl of buffer T (25 mM Tris-HCl pH 7.5, 0.5 mM EDTA, 10% glycerol) containing 100 mM KCl. After incubation, the supernatants (S) were collected and 20 μl SDS Laemmli buffer was added. Beads were washed with 100 μl of buffer T and bound proteins were eluted with 30 μl of SDS Laemmli buffer (eluate, E). S and E fractions were analyzed by SDS-PAGE.
To assess ubiquitylation or SUMOylation in vivo, cells expressing HA-ubiquitin or Myc-SUMO1/Myc-SUMO2 were lysed in buffer B (50 mM Tris-HCl pH 7.5, 5 mM dithiothreitol, 0.5 mM N-ethylmaleimide (NEM), 1% SDS) and incubated for 10 min at 95°C. Samples were sonicated and clarified by centrifugation for 10 min at maximum speed in an Eppendorf centrifuge. Finally, samples were diluted with 4 volumes of buffer A (50 mM Tris-HCl pH 7.5, 120 mM NaCl, 20 mM NaF, 1 mM EDTA, 6 mM EGTA, 15 mM Na-pyrophosphate, 0.5 mM Na-orthovanadate, 1 mM benzamidine, 0.1 mM phenylmethylsulfonyl fluoride, 1% Nonidet P-40) prior to immunoprecipitation.
Protein co-immunoprecipitation studies were performed in buffer A in the presence of ethidium bromide to rule out DNA-mediated interactions.
Immunoblot analysis was performed as previously describedCitation24 using the FUSION SOLO® chemiluminescence imaging system (Vilber).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
1060381_supplemental_files.zip
Download Zip (1.3 MB)Acknowledgments
We would like to thank S.P. Jackson, University of Cambridge, UK, for kindly providing U2OS-GFP-EXO1 cells and for support to SB. We are also indebted to A. Sisakova and R. Aithal, Masaryk University, Brno, Czech Republic, for providing proteins. L. Penengo (IMCR) and H.P. Naegeli (Institute of Pharmacology and Toxicology, University of Zurich, Switzerland) are greatly acknowledged for critical reading of the manuscript and useful suggestions.
Funding
This work was supported by grants from the Swiss National Science Foundation (PDFMP3_127523), the Hartmann-Müller Foundation, the Swiss Foundation for Fight Against Cancer, the Hermann Foundation, the Huggenberger-Bischoff Foundation, the University of Zurich Research Funds, the Czech Science Foundation (GACR 13- 26629S and 207/12/2323), the European Regional Development Fund (Project FNUSA-ICRC) (No. CZ.1.05/1.1.00/02.0123) (to LK), and “Employment of Newly Graduated Doctors of Science for Scientific Excellence” (CZ.1.07/2.3.00/30.0009) co-financed from European Social Fund.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- Curtin NJ. DNA repair dysregulation from cancer driver to therapeutic target. Nat Rev Cancer 2012; 12:801-17; PMID:23175119; http://dx.doi.org/10.1038/nrc3399
- Whitby MC. Making crossovers during meiosis. Biochem Soc Trans 2005; 33:1451-5; PMID:16246144; http://dx.doi.org/10.1042/BST20051451
- Bassing CH, Swat W, Alt FW. The mechanism and regulation of chromosomal V(D)J recombination. Cell 2002; 109:S45-55; PMID:11983152; http://dx.doi.org/10.1016/S0092-8674(02)00675-X
- Errico A, Costanzo V. Mechanisms of replication fork protection: a safeguard for genome stability. Critical Rev Biochem Mol Biol 2012; 47:222-35; PMID:22324461; http://dx.doi.org/10.3109/10409238.2012.655374
- Eid W, Steger M, El-Shemerly M, Ferretti LP, Pena-Diaz J, Konig C, Valtorta E, Sartori AA, Ferrari S. DNA end resection by CtIP and exonuclease 1 prevents genomic instability. EMBO Rep 2010; 11:962-8; PMID:21052091; http://dx.doi.org/10.1038/embor.2010.157
- Thompson LH. Recognition, signaling, and repair of DNA double-strand breaks produced by ionizing radiation in mammalian cells: the molecular choreography. Mutation Res 2012; 751:158-246; PMID:22743550; http://dx.doi.org/10.1016/j.mrrev.2012.06.002
- Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, Modrich P, Kowalczykowski SC. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Devel 2011; 25:350-62; PMID:21325134; http://dx.doi.org/10.1101/gad.2003811
- Karanja KK, Cox SW, Duxin JP, Stewart SA, Campbell JL. DNA2 and EXO1 in replication-coupled, homology-directed repair and in the interplay between HDR and the FA/BRCA network. Cell Cycle 2012; 11:3983-96; PMID:22987153; http://dx.doi.org/10.4161/cc.22215
- Szankasi P, Smith GR. A DNA exonuclease induced during meiosis of Schizosaccharomyces pombe. J Biol Chem 1992; 267:3014-23; PMID:1737756
- Tishkoff DX, Amin NS, Viars CS, Arden KC, Kolodner RD. Identification of a human gene encoding a homologue of Saccharomyces cerevisiae EXO1, an exonuclease implicated in mismatch repair and recombination. Cancer Res 1998; 58:5027-31; PMID:9823303
- Lee BI, Wilson DM 3rd. The RAD2 domain of human exonuclease 1 exhibits 5′ to 3′ exonuclease and flap structure-specific endonuclease activities. J Biol Chem 1999; 274:37763-9; PMID:10608837; http://dx.doi.org/10.1074/jbc.274.53.37763
- Szankasi P, Smith GR. A role for exonuclease I from S. pombe in mutation avoidance and mismatch correction. Science 1995; 267:1166-9; PMID:7855597; http://dx.doi.org/10.1126/science.7855597
- Fiorentini P, Huang KN, Tishkoff DX, Kolodner RD, Symington LS. Exonuclease I of Saccharomyces cerevisiae functions in mitotic recombination in vivo and in vitro. Mol Cell Biol 1997; 17:2764-73; PMID:9111347
- Kirkpatrick DT, Ferguson JR, Petes TD, Symington LS. Decreased meiotic intergenic recombination and increased meiosis I nondisjunction in exo1 mutants of Saccharomyces cerevisiae. Genetics 2000; 156:1549-57; PMID:11102356
- Tsubouchi H, Ogawa H. Exo1 roles for repair of DNA double-strand breaks and meiotic crossing over in Saccharomyces cerevisiae. Mol Biol Cell 2000; 11:2221-33; PMID:10888664; http://dx.doi.org/10.1091/mbc.11.7.2221
- Mimitou EP, Symington LS. DNA end resection: many nucleases make light work. DNA Repair (Amst) 2009; 8:983-95; PMID:19473888; http://dx.doi.org/10.1016/j.dnarep.2009.04.017
- Qiu J, Qian Y, Chen V, Guan MX, Shen B. Human exonuclease 1 functionally complements its yeast homologues in DNA recombination, RNA primer removal, and mutation avoidance. J Biol Chem 1999; 274:17893-900; PMID:10364235; http://dx.doi.org/10.1074/jbc.274.25.17893
- Cotta-Ramusino C, Fachinetti D, Lucca C, Doksani Y, Lopes M, Sogo J, Foiani M. Exo1 processes stalled replication forks and counteracts fork reversal in checkpoint-defective cells. Mol Cell 2005; 17:153-9; PMID:15629726; http://dx.doi.org/10.1016/j.molcel.2004.11.032
- Bologna S, Ferrari S. It takes two to tango: Ubiquitin and SUMO in the DNA damage response. Front Genet 2013; 4:106; PMID:23781231; http://dx.doi.org/10.3389/fgene.2013.00106
- Bennetzen MV, Larsen DH, Dinant C, Watanabe S, Bartek J, Lukas J, Andersen JS. Acetylation dynamics of human nuclear proteins during the ionizing radiation-induced DNA damage response. Cell Cycle 2013; 12:1688-95; PMID:23656789; http://dx.doi.org/10.4161/cc.24758
- Jackson SP, Durocher D. Regulation of DNA damage responses by ubiquitin and SUMO. Mol Cell 2013; 49:795-807; PMID:23416108; http://dx.doi.org/10.1016/j.molcel.2013.01.017
- Engels K, Giannattasio M, Muzi-Falconi M, Lopes M, Ferrari S. Fourteen-3-3 proteins regulate exonuclease 1-dependent processing of stalled replication forks. PLoS Genet 2011; 7:e1001367; PMID:21533173; http://dx.doi.org/10.1371/journal.pgen.1001367
- El-Shemerly M, Janscak P, Hess D, Jiricny J, Ferrari S. Degradation of human exonuclease 1b upon DNA synthesis inhibition. Cancer Res 2005; 65:3604-9; PMID:15867354; http://dx.doi.org/10.1158/0008-5472.CAN-04-4069
- El-Shemerly M, Hess D, Pyakurel AK, Moselhy S, Ferrari S. ATR-dependent pathways control hEXO1 stability in response to stalled forks. Nucleic Acids Res 2008; 36:511-9; PMID:18048416; http://dx.doi.org/10.1093/nar/gkm1052
- Bolderson E, Tomimatsu N, Richard DJ, Boucher D, Kumar R, Pandita TK, Burma S, Khanna KK. Phosphorylation of Exo1 modulates homologous recombination repair of DNA double-strand breaks. Nucleic Acids Res 2010; 38:1821-31; PMID:20019063; http://dx.doi.org/10.1093/nar/gkp1164
- Morin I, Ngo HP, Greenall A, Zubko MK, Morrice N, Lydall D. Checkpoint-dependent phosphorylation of Exo1 modulates the DNA damage response. EMBO J 2008; 27:2400-10; PMID:18756267; http://dx.doi.org/10.1038/emboj.2008.171
- Tomimatsu N, Mukherjee B, Catherine Hardebeck M, Ilcheva M, Vanessa Camacho C, Louise Harris J, Porteus M, Llorente B, Khanna KK, Burma S. Phosphorylation of EXO1 by CDKs 1 and 2 regulates DNA end resection and repair pathway choice. Nature Commun 2014; 5:3561; PMID:24705021; http://dx.doi.org/10.1038/ncomms4561
- Altmannova V, Eckert-Boulet N, Arneric M, Kolesar P, Chaloupkova R, Damborsky J, Sung P, Zhao X, Lisby M, Krejci L. Rad52 SUMOylation affects the efficiency of the DNA repair. Nucleic Acids Res 2010; 38:4708-21; PMID:20371517; http://dx.doi.org/10.1093/nar/gkq195
- Galanty Y, Belotserkovskaya R, Coates J, Polo S, Miller KM, Jackson SP. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature 2009; 462:935-9; PMID:20016603; http://dx.doi.org/10.1038/nature08657
- Forment JV, Walker RV, Jackson SP. A high-throughput, flow cytometry-based method to quantify DNA-end resection in mammalian cells. Cytometry Part A 2012; 81:922-8; PMID:22893507; http://dx.doi.org/10.1002/cyto.a.22155
- Smits VA, Gillespie DA. Cancer therapy. Targeting the poison within. Cell Cycle 2014; 13:2330-3; PMID:25483183; http://dx.doi.org/10.4161/cc.29756
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144:646-74; PMID:21376230; http://dx.doi.org/10.1016/j.cell.2011.02.013
- Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem 2010; 79:181-211; PMID:20192759; http://dx.doi.org/10.1146/annurev.biochem.052308.093131
- Wogan GN, Hecht SS, Felton JS, Conney AH, Loeb LA. Environmental and chemical carcinogenesis. Semin Cancer Biol 2004; 14:473-86; PMID:15489140; http://dx.doi.org/10.1016/j.semcancer.2004.06.010
- Altmannova V, Kolesar P, Krejci L. SUMO Wrestles with Recombination. Biomolecules 2012; 2:350-75; PMID:24970142; http://dx.doi.org/10.3390/biom2030350
- Genschel J, Modrich P. Mechanism of 5′-directed excision in human mismatch repair. Mol Cell 2003; 12:1077-86; PMID:14636568; http://dx.doi.org/10.1016/S1097-2765(03)00428-3
- Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 2008; 134:981-94; PMID:18805091; http://dx.doi.org/10.1016/j.cell.2008.08.037
- Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 2008; 455:770-4; PMID:18806779; http://dx.doi.org/10.1038/nature07312
- Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev 2008; 22:2767-72; PMID:18923075; http://dx.doi.org/10.1101/gad.503108
- Sertic S, Pizzi S, Cloney R, Lehmann AR, Marini F, Plevani P, Muzi-Falconi M. Human exonuclease 1 connects nucleotide excision repair (NER) processing with checkpoint activation in response to UV irradiation. Proc Natl Acad Sci U S A 2011; 108:13647-52; PMID:21808022; http://dx.doi.org/10.1073/pnas.1108547108
- Tatham MH, Matic I, Mann M, Hay RT. Comparative proteomic analysis identifies a role for SUMO in protein quality control. Science Signal 2011; 4:rs4; PMID:21693764; http://dx.doi.org/10.1126/scisignal.2001484
- Hendriks IA, D'Souza RC, Yang B, Verlaan-de Vries M, Mann M, Vertegaal AC. Uncovering global SUMOylation signaling networks in a site-specific manner. Nat Struct Mol Biol 2014; 21:927-36; PMID:25218447
- Galanty Y, Belotserkovskaya R, Coates J, Jackson SP. RNF4, a SUMO-targeted ubiquitin E3 ligase, promotes DNA double-strand break repair. Genes Dev 2012; 26:1179-95; PMID:22661229; http://dx.doi.org/10.1101/gad.188284.112
- Yin Y, Seifert A, Chua JS, Maure JF, Golebiowski F, Hay RT. SUMO-targeted ubiquitin E3 ligase RNF4 is required for the response of human cells to DNA damage. Genes Dev 2012; 26:1196-208; PMID:22661230; http://dx.doi.org/10.1101/gad.189274.112
- Guo Z, Kanjanapangka J, Liu N, Liu S, Liu C, Wu Z, Wang Y, Loh T, Kowolik C, Jamsen J, et al. Sequential posttranslational modifications program FEN1 degradation during cell-cycle progression. Molecular cell 2012; 47:444-56; PMID:22749529; http://dx.doi.org/10.1016/j.molcel.2012.05.042
- Doerfler Z, Schmidt KH. Exo1 phosphorylation status controls the hydroxyurea sensitivity of cells lacking the Pol32 subunit of DNA polymerases delta and zeta. DNA Repair (Amst) 2014; 24:26-36; PMID:25457771; http://dx.doi.org/10.1016/j.dnarep.2014.10.004
- Muthuswami M, Ramesh V, Banerjee S, Viveka Thangaraj S, Periasamy J, Bhaskar Rao D, Barnabas GD, Raghavan S, Ganesan K. Breast tumors with elevated expression of 1q candidate genes confer poor clinical outcome and sensitivity to Ras/PI3K inhibition. PloS one 2013; 8:e77553; PMID:24147022; http://dx.doi.org/10.1371/journal.pone.0077553
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol 2006; 7:R100; PMID:17076895; http://dx.doi.org/10.1186/gb-2006-7-10-r100
- Liberali P, Snijder B, Pelkmans L. A hierarchical map of regulatory genetic interactions in membrane trafficking. Cell 2014; 157:1473-87; PMID:24906158; http://dx.doi.org/10.1016/j.cell.2014.04.029
