Abstract
Viruses often exploit autophagy, a common cellular process of degradation of damaged proteins, organelles, and pathogens, to avoid destruction. HIV-1 dysregulates this process in several cell types by means of Nef protein. Nef is a small HIV-1 protein which is expressed abundantly in astrocytes of HIV-1-infected brains and has been suggested to have a role in the pathogenesis of HIV-Associated Neurocognitive Disorders (HAND). In order to explore its effect in the CNS with respect to autophagy, HIV-1 Nef was expressed in primary human fetal astrocytes (PHFA) using an adenovirus vector (Ad-Nef). We observed that Nef expression triggered the accumulation of autophagy markers, ATG8/LC3 and p62 (SQSMT1). Similar results were obtained with Bafilomycin A1, an autophagy inhibitor which blocks the fusion of autophagosome to lysosome. Furthermore co-expression of tandem LC3 vector (mRFP-EGFP-LC3) and Ad-Nef in these cells produced mainly yellow puncta (mRFP+, EGFP+) strongly suggesting that autophagosome fusion to lysosome is blocked in PHFA cells in the presence of Nef. Together these data indicate that HIV-1 Nef mimics Bafilomycin A1 and blocks the last step of autophagy thereby helping HIV-1 virus to avoid autophagic degradation in human astrocytes.
Introduction
A vast number of HIV-1-infected individuals suffer from HIV-associated neurocognitive disorders (HAND) despite very effective antiviral therapies such as Highly Active Antiretroviral Therapy (HAART).Citation1,2 HIV-1 infects predominantly T-cells and macrophages via cell surface receptor, CD4, a transmembrane glycoprotein. Viral infection starts when the HIV-1 envelope protein gp120 binds to CD4 and forms a complex enabling the virus to enter host cells.Citation1,3 Once the virus enters the cells, it reverse transcribes viral DNA from its RNA and proceeds to the nucleus where it eventually integrates into the host cell genome, replicates and either spreads to other cells or stays dormant in the form of a latent provirus. In addition to envelope protein, the HIV-1 genome encodes several structural and regulatory proteins such as Gag and reverse transcriptase. Furthermore, several small accessory proteins, Rev, Tat, Vpr, and Vpu are also generated by its genome as well as Nef.Citation4 Nef was originally thought to be a negative factor. However, in cases where the Nef gene is deleted, people infected with HIV-1 did not develop AIDS indicating that this small protein is involved in the replication of HIV-15.
Nef is a multifunctional small protein (25 to 34 kD), possessing no enzymatic activity and carrying a myristic acid residue at the N-terminus as a result of posttranslational modification. Myristylation is required for the functionality of the protein.Citation6 Nef is expressed in the early stages of HIV-1 infection and is known to interact with many cellular proteins and affects many cellular pathways including cytokine networks and signal transduction.Citation7,8 Nef primarily targets cell surface receptors such as CD4, which is responsible for HIV-1 entry and down-regulates these receptors to prevent superinfection.Citation9 Nef is also involved in endosome biogenesis to facilitate HIV-1 infectivity.Citation10 Nef is released in extracellular vesicles known as exosomes through which it can migrate to neighboring bystander cells where it inflicts most of its toxic effects. For example, release of Nef via exosomes from HIV-infected CD4+ T cells can induce apoptosis in neighboring cells.Citation11 Nef is one of the most abundantly expressed HIV-1 proteins and its effect in brain cells is detrimental due to its toxicity. Early studies by Ranki et al., 1995Citation12 analyzed postmortem brain tissues of HIV-infected individuals and found that patients with a history of moderate to severe dementia had Nef-positive astrocytes suggesting neurotoxic effects of this protein. AIDS patients with high viral loads show increased levels of Nef in their blood, which can be used to monitor progression of the disease. Intracellular Nef was also found in infected PBMCs from the AIDS patients.Citation13
HIV-1 can infect astrocytes and cause a latent infection suggesting that astrocytes can serve as viral reservoirs.Citation14,15,16 Nef is also known to affect neuronal cell viability.Citation17 Treatment of human glial cells and neurons with recombinant Nef showed 25% and 32% decrease in cell numbers, respectively.Citation17 Despite recent advancements in the HIV-1 field, the exact role of Nef in these cells is not completely understood.
Macroautophagy (hereafter called autophagy) is a common cellular process whereby unwanted proteins, damaged organelles, viruses and pathogens are first captured as cargo, trapped in double-membraned vesicles, and then degraded and/or recycled back to the cell. These processes are regulated by more than 30 ATG (autophagy-related) proteins. Autophagy is composed of several steps: initiation, autophagosome formation, and fusion of lysosome with autophagosome to form autophagolysosome where degradation of the cargo takes place.Citation18,19 The relationship between HIV-1 and autophagy was first reported by Zhou and Spector,Citation20 demonstrating that the virus downregulates this cellular process during acute infection. Since then, regulation of autophagy by HIV-1 has increasingly received more attention.Citation21 Kyei et al., 2009Citation22 reported the involvement of Nef in autophagy by showing that Nef-GFP expression in HEK cells induced the autophagy marker LC3-II and further demonstrated that Nef binds to one of the autophagy proteins, ATG1/Beclin1 to block autophagy.Citation22 To further establish the role of Nef, macrophages infected with an HIV-1 strain carrying a Nef deletion did not block autophagy while wild-type virus with Nef did. These findings led to the discovery of a negative autophagy regulator GAPR-1 by exploiting the specific interactions between HIV-1 Nef and Beclin1.Citation23 HIV-1 Nef also induces autophagosome formation by interacting with IRGM an immunity associated GTPase family M, which is commonly targeted by RNA viruses.Citation24 All this evidence has suggested that Nef could be a multifaceted player in the dysregulation of autophagy by HIV-1 and exploit autophagy by interacting with several key regulatory cellular proteins thereby promoting productive HIV-1 infection.
The reports regarding Nef involvement in autophagy are limited to cells lines such as HEK, HeLa, and macrophages.Citation20,22 To date, there are no reports assessing the effects of HIV-1 Nef on autophagy in human astrocytes. Given the importance of astrocytes in various functions of the brain including secretion and absorption of neuronal transmitters, metabolism in the brain, neuronal circuitry and others, combined with the presence of Nef in these cells, it is essential to understand the role of Nef in pathways controlling cell survival and death. Here we show for the first time that expression of Nef in human astrocytes compromises the autophagic pathway by inducing autophagosome formation and blocking the assembly of autophagolysosomes.
Results
HIV-1 Nef was expressed in PHFA cells by transduction with Ad-Nef. Nef expression was detectable in most of the cells by immunocytochemistry. Interestingly, some of the HIV-1 Nef-expressing astrocytes showed morphological changes and shrunk drastically indicating formation of apoptotic bodies (). Western blot analysis of cell lysates from Ad-Nef transduced astrocytes shows a copious amount of Nef protein expression and a significant increase in the autophagy marker ATG8/LC3-II levels when compared to Ad-Null transduced control cell lysates at 48 hrs post-transductions (). In the presence of Nef, LC3-II levels increased nearly fold4- in PHFA cells. We also detected a substantial increase in p62 (SQSMT1) protein in cells expressing Nef (), suggesting that Nef dysregulates autophagy. These observations were reproducible in PHFA cells derived from 3 different fetal brain tissues. The Nef-dependent increase in the autophagy marker proteins appears to be restricted to ATG8/LC3-II and p62. Other autophagy-related proteins such as ATG3, ATG7 and Beclin1 showed no significant variations in their levels upon Nef production.
Figure 1. Effect of HIV-1 Nef expression on autophagy in Primary Human Astrocytes (PHFA). (A) Immunocytochemistry. PHFA cells were plated in 2 well chamber slides in astrocyte growth medium. Adenovirus transductions and fixing the cells were performed as described in the Materials and Methods. The fixed cells were probed with anti-GFAP antibody for astrocyte marker and anti-HIV-1 Nef (SF2) then mounted on slides using DAPI solution. The slides were visualized using a fluorescence microscopy. Green is GFAP; Red is HIV-1 Nef and blue is DAPI. (B) Immunoblotting of Cell lysates obtained from PHFA transduced with Ad-Nef or Ad-Null. PHFA cells were harvested and lysed using TNN buffer containing 1% NP40 supplemented with mammalian protease inhibitors at 4°C followed by SDS-PAGE and Western blots as described in Materials and Methods. Three different cultures of PHFA were used. The results are representative of at least 3 independent experiments. (C) MTT Assay. MTT proliferation assay was performed as described in Materials and Methods. The experiments were performed in duplicates in a 96 well plate and averages are shown as bar graphs. MTT activity is described differences in absorbances between 620 nm and 595 nm. (D) PHFA cells were transduced with Adnull and AdNef, harvested and lysed at 10, 24, and 48 hrs post-transductions (hpt) using TNN buffer containing 1% NP40 supplemented with mammalian protease inhibitors at 40C followed by SDS-PAGE and Western Blots as described in Materials and Methods.
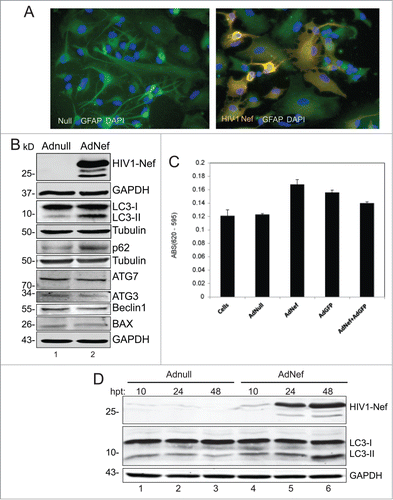
We also performed MTT assay to assess cell viability. Results showed that expression of Nef has no significant toxic effect on PHFA cells (). In fact, we found that MTT activity was modestly increased in the presence of Nef compared to Ad-Null. The increase in MTT activity may be a cellular response to expression of the foreign protein. An increase in MTT activity in the presence of Nef has also been reported by Trillo-Pazos et alCitation15 and does not correlate with cell viability. In fact, the same investigators found a decrease in cell viability using LDH release and trypan blue exclusion. In our experiments, we also detected dead cells after AdNef transductions.
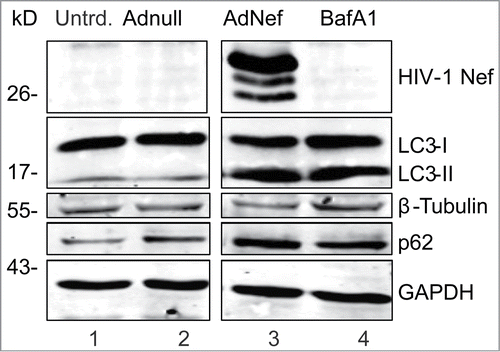
Bafilomycin A1 (BafA1), an inhibitor of V-ATPase, blocks autophagolysosome formation by preventing fusion between autophagosomes and lysosomes.Citation25 BafA1 leads to an accumulation of LC3-II and p62 proteins that are protected from the autolysomal protease degradation. The levels of LC3-II and p62 proteins were both increased similarly in Ad-Nef tranduced and BafA1 treated astrocytes suggesting that Nef mimics the effects of BafA1 in PHFA cells by blocking autophagolysosome formation ().
Figure 2. HIV-1 Nef mimics the BafA1 to block autophagy in primary human astrocytes. PFHA were plated in 6-well plates as described earlier and transduced with Ad-Nef or Ad-Null when the cells were confluent. In this experiment, 3 times more (3 × 106 U) Ad-Nef was used to maximize Nef expression (Lane 3) and BafA1 (10 nM) treatment in untransduced cells was shown in Lane 4. Controls are shown in lane 1, untransduced PHFA and lane 2, Ad-null transduced PHFA Lane 2. Cells were harvested after 24 hours and analyzed by SDS-PAGE/Western Blotting as described before. Immunoblotting was performed using anti-HIV-1 Nef, anti-LC3B and anti p62 antibodies along with loading controls β-tubulin and GAPDH similar to shown in . These results are representative of at least 3 different independent experiments.
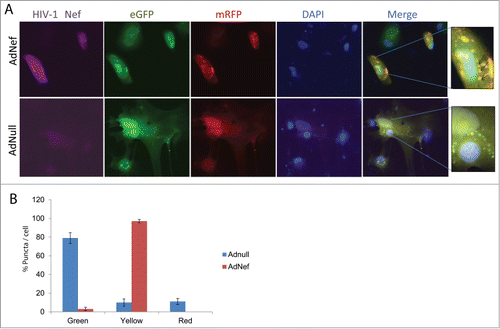
In order to demonstrate the impact of Nef on autophagy at the cellular level, we utilized a tandem expression vector producing mRFP-EGFP-LC3B fusion protein. Cells were first transfected with the tandem LC3 vector followed by transduction with Ad-Nef. mRFP-EGFP-LC3B expression was monitored by color puncta formation. Red puncta are an indicator of autophagolysosome formation where mRFP red fluorescence is visible while EGFP fluorescence is quenched due to acidic conditions. Yellow puncta indicates both mRFP and EGFP fluorescence are in autophagosomes demonstrating inhibition of autophagosome and lysosome fusion, while the green puncta indicate autophagosome formation. When Ad-Nef transduced astrocytes were analyzed by immunofluorescence, the majority of puncta observed in the cells were yellow indicating that fusion of autophagosome to lysosome was blocked. Ad-null transduced cells exhibited only few yellow and some red puncta, while the rest of the puncta remained green ().
Figure 3. Effect of HIV-1 Nef expression on mRFP-EGFP-LC3 puncta in PHFA cells by Immunocytochemistry (ICC). (A) PHFA cells were plated in 2 well chamber slides the day before the transfection. The cells were first transfected with ptfLC3 plasmid using Lipofectamine™ 3000 according to the manufacturer's guidelines followed by transduction with Ad-null or Ad-Nef and immunocytochemistry was performed as described in Materials and Methods. The upper panel shows the ICC of the Ad-Nef transduced primary astrocytes. From left to right purple for HIV-1 Nef, green for EGFP, red for mRFP, blue for DAPI, and merging EGFP-mRFP-DAPI where puncta were shown. The bottom panel is control with Ad-null transduced PHFA and the slides followed the same order. The far right panels depict the zoom-in where yellow puncta are shown in detail as indicated by arrows. (B) Percentage of Green-red-yellow puncta signal over total puncta per cell in merged images was calculated and shown as a bar graph. Data are shown as mean ± SD and are representative of at least 10 different cells from 10 different fields.
Discussion
Besides HIV-1, other viruses such as human T-cell leukemia virus type 1 (HTLV-1) and influenza A are also known to dysregulate autophagy in order to avoid degradation by autolysosomes.Citation26,27 Influenza A virus and HTLV-1 utilize small auxiliary proteins to facilitate control of autophagy. Influenza A virus matrix protein 2 inhibits the fusion of autophagosome with lysosome to ensure the survival of virus-infected cells without affecting viral replication.Citation26 Likewise, HTLV-1 Tax protein also inhibits fusion of autophagosome and lysosome. The Tax protein not only helps to increase HTLV-1 infection, but also prevents its degradation by autophagy.Citation27
The relationship between autophagy and HIV-induced cell damage is not clear. While astrocytes have been shown to harbor HIV-1, they have yet to robustly support viral replication and thereby may serve as a reservoir for viral latency site in the brain.Citation28 Nef, a HIV-1 protein, has been shown to cause neurological impairments directly or indirectly. Earlier studies have shown that HIV-1 Nef expression in astrocytes leads to deficiencies in spatial memory.Citation29 Astrocytes also release inflammatory proteins such as IP-10 known to be neurotoxic.Citation30 The amount of Nef present in the CNS as a result of HIV-1 infection could be a determining factor in neurotoxicity. Dysregulation of autophagy is a common feature of several neurological diseases.Citation31 It has been shown that HIV-1 can suppress autophagy in neuronal cells as evidenced by the accumulation of p62/SQSMT1.Citation32
In the era of HAART, while the replication of HIV-1 is controlled, neurological disorders resulting from neurotoxic impact of viral regulatory proteins remain a major clinical problem.Citation33 At the clinical level it has become clear that the effect of HIV-1 on various biological events including autophagy plays a significant role in pathophysiology of HIV-1/CNS disorders. Although it is known that HIV-1 dysregulates autophagy by promoting and blocking its early and late steps respectively, the details of the viral role in this cellular process are not completely understood. Recently the effect of HIV-1 Tat protein on neuronal autophagy and the implications of this effect for neurological disorders have been reported.Citation34 Interestingly, unlike Nef, HIV-1 Tat increases autophagic degradation in neurons.Citation34 It is important to understand the details of how HIV-1 controls and exploits autophagy for its own advantage with respect to HIV-1-associated neurocognitive disorders. In conclusion, our results suggest that HIV-1 Nef dysregulates autophagy by blocking the assembly of autophagolysosomes, and may help HIV-1 to escape degradation by autophagy.
Materials and Methods
Ethics statement
Fetal human brain tissues were obtained and utilized in accordance with Temple University human subject protocols and the approval of the Temple University Institutional Review Board.
Cell lines
PHFA were obtained from the Comprehensive NeuroAIDS Center (CNAC) at Temple University School of Medicine. Cells were maintained in astrocyte growth media, Dulbecco's Modified Eagle Medium (DMEM): F12 medium supplemented with 10% fetal bovine serum (FBS), 10% Glutamax, insulin and gentamycin (10 μg/ml) at 37°C under 5% CO2 atmosphere. The growth media were changed every 3–4 d The cells were plated at densities of 500,000 cells/well in 6 well plates or at densities of 100,000 cells/chamber in 2 chamber slides.
Plasmids, adenovirus constructs and reagents
ptfLC3, tandem LC3 plasmid (mRFP-EGFP-LC3B, plasmid # 21074, characterized by Kimura et al.Citation35) was purchased from Addgene. pCDNA-3.1-sf2-Nef was obtained from the NIH-AIDS Reagent program (Germantown, MD). The HIV-1 Nef gene from this vector was subcloned into Adenovirus-5 (Ad5) and propagated in HEK292A cells and purified using OptiPrep™ Gradient to its pure form. The final titer of the purified AdNef is 1 × 109 U/ml. AdGFP was also cloned in Ad5 also prepared from the same cell line and purified using CsCl gradient (courtesy Dr. Alexey Bogush). The final AdGFP titer was 3 × 1010. AdNull (1.5 × 1010 U/ml) was a gift from Dr. Satish Deshmane.Citation36 Monoclonal antibodies were obtained from the following sources; anti-Nef (SF2), EH1 from NIH-AIDS Reagent Program, anti-GAPDH from Santa Cruz (Dallas, TX). Polyclonal antibodies were obtained from the following sources; anti-LC3B from Sigma-Aldrich (St. Loius, MO), β-tubulin from LI-COR, Odyssey (Lincoln, NE) and anti-p62(SQSTM1), anti-ATG7, anti-ATG3 anti-Beclin1 and anti-BAX from Cell Signaling (Beverly, MA). Mammalian protease inhibitors and Bafilomycin A1 were obtained from Sigma-Aldrich (St Louis, MO). Bradford reagent was from BioWorld (Dublin, OH). Vybrant MTT proliferation assay kit was purchased from Life Technologies (Carlbad, CA).
Transduction of human primary astrocytes (PHFA)
PHFA cultures in 6-well plates were transduced using 1–3 × 106 units of AdNef or AdNull as a control. For the Bafilomycin A1 (BafA1) experiment, un-transduced PHFA cells were treated with 10 nM BafA1 3 hrs prior to harvesting. The cells were harvested after 10–24–48 hrs and lysed using TNN buffer with 1% NP40 supplemented with protease inhibitors. The clear cell lysates were obtained after centrifugation at max speed at 4°C for 10 min (Eppendorf Microfuge). Protein concentrations were determined by Bradford Assay.
Western blotting
Cell lysates (40–50 μg) were prepared in 1 x SDS loading buffer after heating at 95°C for 5 min. The lysates were loaded onto 10 or 12% SDS-polyacrylamide gels and electrophoresis was performed. After the electrophoresis, the gels were transferred onto nitrocellulose membranes at 250 mA for 30 min for 0.22 μm membranes (LI-COR Odyssey) or 2 hrs at 250 mA or overnight at 40 mA for 0.45 μm membranes (Santa Cruz Technologies) in 1 x cold transfer Buffer (100 mM TrisHCl pH 7.4, 150 mM NaCl, 20% methanol). After transfer, membranes were blocked in either 1x PBST buffer containing 10% dried fat free milk or 1x Li-Cor blocking buffer at least 1 hr at RT. The primary antibodies; monoclonal anti-Nef sf2 (EH1), monoclonal anti-GAPDH, polyclonal anti-LC3B, polyclonal anti-p62 (SQSTM1), polyclonal β-tubulin, polyclonal anti-ATG7, polyclonal anti-ATG3, polyclonal anti-Beclin1 and polyclonal anti-BAX were diluted 1/1000 in PBST buffer containing 5% dried fat free milk and the nitrocellulose membranes were incubated with these antibodies where needed overnight at 4°C with gentle shaking. After three 5 minute washes with PBST buffer, the secondary antibodies [Li-Cor goat against rabbit 680 for polyclonal, Li-Cor goat against mouse 800 for monoclonal] were added to the membranes followed by 45 min incubation at RT. The final step was 3 5-minute wash using PBST buffer and 5 min final wash using PBS buffer. Washed membranes were stored in 1x PBS before scanning the image. The Western blot images were obtained using the ODYSSEY® CLx Imaging system. The intensity of the protein bands was determined using ImageJ software (NIH) to quantitate relative protein levels.
Immunocytochemistry
PHFA cultures were grown in 2 well chamber slides until 80–90% confluency. Transfection of ptfLC3 was performed using Lipofectamine™ 3000 (Life Technologies, Carlsbad, CA) according to the manufacturer's guidelines. The following day, growth media in slides was replaced with fresh media and the transductions were performed using 1 × 106 U AdNef or AdNull. One day after transduction, the slides were fixed using 4% paraformaldehyde, 0.25% Triton X-100 for 10 min followed by 3 washes using 1x PBS. The fixed slides were first incubated in 400 μl blocking solution 1 x PBST containing 1% BSA for 1 hr at RT, then anti-Nef antibodies (EH1) were added at 1/200 dilution in the same blocking solution and incubated overnight at RT. The next day the slides were washed 3 times (10 min) using 1x PBST and treated with Li-Cor donkey anti mouse 680 secondary antibodies (1/400 dilution) in dark for 45 min. The slides were washed 3 times (10 min each) with 1 x PBST and mounted using Vectashield™ mounting solution containing DAPI. The slides were allowed to dry overnight and fluorescence images were obtained using a Leica Fluorescence Microscope.
MTT assay
PHFA cells were plated in 96-well plates in astrocyte growth medium. Adenovirus titers were scaled down to 96 wells. After 24 hrs transduction, MTT assay was performed using Vybrant ® MTT cell proliferation assay from Life Technologies according to the manufacturer's guidelines. Absorbances were measured in a Multiscan FC plate reader (Thermo Fisher Scientific) and the differences between 620 nm and 595 nm were calculated at least from 2 independent experiments and plotted using Microsoft Excel.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors thank the past and present members of the Department of Neuroscience/Center for Neurovirology for sharing their ideas and reagents. We thank Drs. Satish Deshmane and Alexey Bogush for the Adenoviral constructs. We also thank Drs. Martyn K. White and Prasun K Datta for critical reading and editing of this manuscript. This study utilized services offered by core facilities of the Comprehensive NeuroAIDS Center (CNAC NIMH Grant Number P30MH092177) at Temple University School of Medicine.
Funding
Research reported in this publication was supported by the National Institutes of Health under Award number R01MH086358. The funding organization played no role in the design of the study, in the collection, analysis, and interpretation of the data and in the decision to submit the manuscript for publication.
References
- Moir S, Chun TW, Fauci AS. Pathogenic mechanisms of HIV disease. Annu Rev Pathol Mech Dis 2011; 6:223-48; http://dx.doi.org/10.1146/annurev-pathol-011110-130254
- Fraser C, Lythgoe K, Leventhal GE, Shirreff G, Hollingsworth TD, Alizon S, Bonhoeffer S. Virulence and pathogenesis of HIV-1 infection: an evolutionary perspective. Science 2014; 343(6177):1243727; PMID:24653038
- Pantaleo G, Fauci AS, Immunopathogenesis of HIV infection. Ann Rev Microbiol 1996; 50:825-54; http://dx.doi.org/10.1146/annurev.micro.50.1.825
- Malim, MH, Emerman, M. HIV-1 accessory proteins-ensuring viral survival in a hostile environment. Cell Host Microbe 2008; 3:388-98; PMID:18541215
- Gorry PR, McPhee DA, Verity E, Dyer WB, Wesselingh SL, Learmont J, Sullivan JS, Roche M, Zaunders JJ, Gabuzda D, et al. Pathogenicity and immunogenicity of attenuated, nef-deleted HIV-1 strains in vivo. Retrovirology 2007; 4:66; PMID:17888184; http://dx.doi.org/10.1186/1742-4690-4-66
- Geyer M, Fackler OT, Peterlin BM. Structure-function relationships in HIV-1 Nef. EMBO Rep 2001; 2: 580-5; http://dx.doi.org/10.1093/embo-reports/kve141
- Percario ZA, Ali M, Mangino G, Affabris E. Nef, the shuttling molecular adaptor of HIV, influences the cytokine network. Cytokine Growth Factor Rev 2014; 26(2):159-73.
- Mukerji J, Olivieri KC, Misra V, Agopian KA, Gabuzda D. Proteomic analysis of HIV-1 Nef cellular binding partners reveals a role for exocyst complex proteins in mediating enhancement of intercellular nanotube formation. Retrovirology 2012; 9:33; http://dx.doi.org/10.1186/1742-4690-9-33
- Venzke S, Michel N, Allespach I, Fackler OT, Keppler OT. Expression of Nef downregulates CXCR4, the major coreceptor of human immunodeficiency virus, from the surfaces of target cells and thereby enhances resistance to superinfection. J Virol 2006; 80:11141-52.
- Madrid R, Janvier K, Hitchin D, Day J, Coleman S, Noviello C, Bouchet J, Benmerah A, Guatelli J, Benichou S. Nef-induced alteration of the early/recycling endosomal compartment correlates with enhancement of HIV-1 infectivity. J Biol Chem 2005; 280: 5032-44.
- Lenassi M, Cagney G, Liao M, Vaupotic T, Bartholomeeusen K, Cheng Y, Krogan NJ, Plemenitas A, Peterlin BM. HIV Nef is secreted in exosomes and triggers apoptosis in bystander CD4+ T cells. Traffic 2010; 1:110-22; http://dx.doi.org/10.1111/j.1600-0854.2009.01006.x
- Ranki A, Nyberg M, Ovod V, Haltia M, Elovaara I, Raininko R, Haapasalo H, Krohn K. Abundant expression of HIV Nef and Rev proteins in brain astrocytes in vivo is associated with dementia. AIDS 1995; 9:1001-8; PMID:8527071; http://dx.doi.org/10.1097/00002030-199509000-00004
- Wang T, Green LA, Gupta SK, Amet T, Byrd DJ, Yu Q, Twigg HL 3rd, Clauss M. Intracellular Nef detected in peripheral blood mononuclear cells from HIV patients. AIDS Res and Hum Retrovir 2015; 31:217-20; PMID:25062432
- Kramer-Hämmerle S, Rothenaigner I, Wolff H, Bell JE, Brack-Werner R. Cells of the central nervous system as targets and reservoirs of the human immunodeficiency virus. Virus Res 2005; 111(2):194-213; http://dx.doi.org/10.1016/j.virusres.2005.04.009
- Eugenin EA, Clements JE, Zink MC, Berman JW. Human immunodeficiency virus infection of human astrocytes disrupts blood-brain barrier integrity by a gap junction-dependent mechanism. J Neurosci 2011; 31(26):9456-65; PMID:21715610; http://dx.doi.org/10.1523/JNEUROSCI.1460-11.2011
- Meulendyke KA, Croteau JD, Zink MC. HIV life cycle, innate immunity and autophagy in the central nervous system. Curr Opin HIV AIDS 2014; 6:565-71; http://dx.doi.org/10.1097/COH.0000000000000106
- Trillo-Pazos G, McFarlane-Abdulla E, Campbell IC, Pilkington GJ, Everall IP. Recombinant nef HIV-IIIB protein is toxic to human neurons in culture. Brain Res 2000; 864: 315-26; PMID:10802040; http://dx.doi.org/10.1016/S0006-8993(00)02213-7
- Deretic V. Autophagy in infection. Curr Opin Cell Biol 2010; 22:252-62; PMID:20116986; http://dx.doi.org/10.1016/j.ceb.2009.12.009
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature, 2008; 451:1069-75; http://dx.doi.org/10.1038/nature06639
- Zhou D, Spector SA. Human immunodeficiency virus type-1 infection inhibits autophagy. AIDS 2008; 22: 695-9; PMID:18356598; http://dx.doi.org/10.1097/QAD.0b013e3282f4a836
- Dinkins C, Pilli M, Kehrl JH. Roles of autophagy in HIV infection. Immunol Cell Biol 2015; 93:11-7; PMID:25385065; http://dx.doi.org/10.1038/icb.2014.88
- Kyei GB, Dinkins C, Davis AS, Roberts E, Singh SB, Dong C, Wu L, Kominami E, Ueno T, Yamamoto A, et al. Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. J Cell Biol 2009; 186: 255-68; http://dx.doi.org/10.1083/jcb.200903070
- Shoji-Kawata S1, Sumpter R, Leveno M, Campbell GR, Zou Z, Kinch L, Wilkins AD, Sun Q, Pallauf K, MacDuff D, et al. Identification of a candidate therapeutic autophagy-inducing peptide. Nature 2013; 494:201-9; PMID:23364696; http://dx.doi.org/10.1038/nature11866
- Grégoire IP, Richetta C, Meyniel-Schicklin L, Borel S, Pradezynski F, Diaz O, Deloire A, Azocar O, Baguet J, Le Breton M, et al. IRGM is a common target of RNA viruses that subvert the autophagy network. Plos Pathog 2011;Dec; 7(12):e1002422; http://dx.doi.org/10.1371/journal.ppat.1002422
- Yamamoto A, Tagawa Y, Yoshimori T, Moriyama Y, Masaki R, Tashiro Y. Bafilomycin A prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct Funct 1998; 23:33-42; PMID:9639028; http://dx.doi.org/10.1247/csf.23.33
- Gannagé M, Dormann D, Albrecht R, Dengjel J, Torossi T, Rämer PC, Lee M, Strowig T, Arrey F, Conenello G, et al. Matrix protein 2 of influenza A virus blocks autophagosome fusion with lysosomes. Cell Host & Microbe 2009; 6: 367-80; http://dx.doi.org/10.1016/j.chom.2009.09.005
- Tang SW, Chen CY, Klase Z, Zane L, Jeang KT. The cellular autophagy pathway modulates human T-cell leukemia virus type 1 replication. J Virol 2013; 87: 1699-707; http://dx.doi.org/10.1128/JVI.02147-12
- Narasipura SD, Kim S, Al-Harthi L. Epigenetic regulation of HIV-1 latency in astrocytes. J Virol 2014; 88(5):3031-8; http://dx.doi.org/10.1128/JVI.03333-13
- Chompre G, Cruz E, Maldonado L, Rivera-Amill V, Porter JT, Noel RJ Jr. Astrocytic expression of HIV-1 Nef impairs spatial and recognition memory. Neurobiol Dis 2013; 49:128-36; PMID:22926191; http://dx.doi.org/10.1016/j.nbd.2012.08.007
- van Marle G, Henry S, Todoruk T, Sullivan A, Silva C, Rourke SB, Holden J, McArthur JC, Gill MJ, Power C. Human immunodeficiency virus type 1 Nef protein mediates neural cell death: a neurotoxic role for IP-10. Virology 2004; 3292:302-18; http://dx.doi.org/10.1016/j.virol.2004.08.024
- Martinez-Vicente M, Cuervo AM. Autophagy and neurodegeneration: when the cleaning crew goes on strike. Lancet Neurol 2007; 4:352-61; http://dx.doi.org/10.1016/S1474-4422(07)70076-5
- Alirezaei M, Kiosses WB, Fox HS. Decreased neuronal autophagy in HIV dementia: a mechanism of indirect neurotoxicity. Autophagy 2008; 7:963-6; http://dx.doi.org/10.4161/auto.6805
- Kramer-Hämmerle S, Rothenaigner I, Wolff H, Bell JE, Brack-Werner R. Cells of the central nervous system as targets and reservoirs of the human immunodeficiency virus. Virus Res 2005; 111:194-213; http://dx.doi.org/10.1016/j.virusres.2005.04.009
- Fields J, Dumaop W, Elueteri S, Campos S, Serger E, Trejo M, Kosberg K, Adame A, Spencer B, Rockenstein E, et al. HIV-1 Tat alters neuronal autophagy by modulating autophagosome fusion to the lysosome: implications for HIV-associated neurocognitive disorders. J Neurosci 2015; 35:1921-38; http://dx.doi.org/10.1523/JNEUROSCI.3207-14.2015
- Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 2007; 3(5):452-60; http://dx.doi.org/10.4161/auto.4451
- Deshmane SL, Mukerjee R, Fan S, Del Valle L, Michiels C, Sweet T, Rom I, Khalili K, Rappaport J, Amini S, et al. Activation of the oxidative stress pathway by HIV-1 Vpr leads to induction of hypoxia-inducible factor 1alpha expression. J Biol Chem 2009; 284:11364-73; PMID:19204000; http://dx.doi.org/10.1074/jbc.M809266200
