Exonuclease 1 (EXO1) is one of several nucleic acid-trimming enzymes required for the repair of double strand breaks that constitute the most toxic form of DNA damage. By resecting DNA ends, this processive 5′ to 3′ exonuclease helps to produce recombinogenic 3′ overhangs, which are than captured by the homologous recombination machinery to repair double strand breaks, i.e., to reestablish an intact DNA double helix.Citation1 This mechanism raises the question of how EXO1 is prevented from carrying out excessive DNA digestions at strand breaks, possibly generating even more deleterious intermediates responsible for chromosomal aberrations. Clearly, further research is needed to understand the regulation of EXO1 in the maintenance of genome stability but, in the current issue of Cell Cycle, Bologna et al.Citation2 provide important new insights into this challenging problem. They report that, in conjunction, 2 related peptide modifiers control the level and enzymatic activity of EXO1 in human cells. A first hint for this newly discovered regulatory circuit came from the finding that EXO1 undergoes ubiquitin-dependent proteasomal degradation in response to agents like hydroxyurea, aphidicolin or camptothecin that cause replication stress by stalling DNA replication forks. Interestingly, the authors discovered that, in such genome-threatening situations like replication stress, both the ubiquitination of EXO1 and its degradation are triggered by a preceding conjugation with SUMO (for Small Ubiquitin-like MOdifier).
The SUMO cascade is an intensively studied protein modification system with key roles in diverse cellular processes. Like for posttranscriptional ubiquitination, the toolbox for protein sumoylation comprises a hierarchy of E1, E2 and E3 enzymes. The SUMO moiety is activated in an ATP-dependent manner by the E1 activating enzyme and then transferred to an E2 conjugating enzyme. This E2 conjugates SUMO to target lysine residues either by direct substrate recognition or with the assistance of E3 ligases acting as substrate adapters. Recent proteomic screens revealed a high prevalence of SUMO substrates in DNA-metabolic pathways.Citation3,4 By demonstrating sumoylation of EXO1, Bologna and colleagues extend this list of sumoylated DNA-resecting enzymes (already including other nucleases like Rad1, FEN1, Sae2/CtIP and the Mre11 complex) by one additional entry and confirm the paradigm of SUMO-dependent processing of DNA double strand breaks. In fact, the E2 and E3 enzymes (UBC9 and PIAS1, respectively) involved in EXO1 sumoylation had been shown previously to co-localize with sites of DNA double strand break repair.Citation5,6 The finding that EXO1 is de-sumoylated by the SENP6 protease completes this regulatory circuit.
To address the biological relevance of EXO1 sumoylation, Bologna and colleagues identified the lysine acceptor sites and generated an EXO1 triple mutant that is catalytically active, but refractory to sumoylation. Since a central arena for SUMO actions relates to pathways promoting genome stability, the authors next tested the impact of EXO1 overexpression on chromosomal aberrations in camptothecin-treated cells. Overexpression of wild-type EXO1 increases the prevalence of chromosomal breaks, presumably due to a disproportionate DNA-resecting activity. However, this negative effect of EXO1 on genome stability is attenuated when, instead of the wild-type protein, the SUMO-refractory triple mutant was overexpressed at the same protein level and under the same conditions. Evidently, interference with sumoylation protects cells experiencing replication stress from the deleterious consequences of an excess level of EXO1
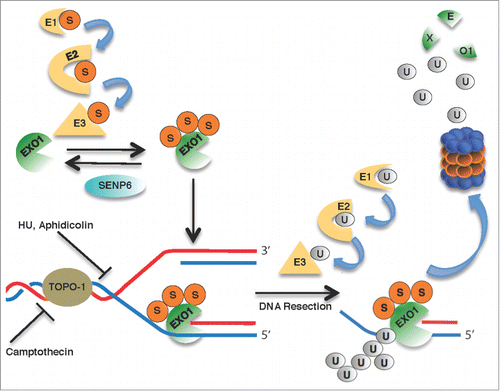
Based on these findings, Bologna and colleagues.Citation2 proposed a model of how the conjugation of EXO1 with peptide modifiers is linked to its function in handling replication stress (). Their scheme involves recruitment of constitutively sumoylated EXO1 to DNA ends at stalled replication forks. SUMO stimulates the catalytic activity of EXO1 during DNA resection in the context of stalled replication forks and, upon full substrate engagement, induces its ubiquitination. Thus, this combination of constitutive sumoylation and on-site ubiquitination ensures fit-to-purpose resection but prevents excessive digestion of free DNA ends by EXO1. A challenge for the future will be to understand how SUMO is able to fine-tune the enzymatic activity of EXO1 independently of ubiquitin-dependent degradation. In general terms, SUMO can have effects either as a molecular “glue” or as an “anti-glue” in modulating protein-protein or protein-DNA interactions.Citation7 Thus, one testable possibility is that SUMO moieties mediate the association of EXO1 to binding partners displaying SUMO-interacting motifs in structures assembled at stalled replication forks. Another possibility is that SUMO residues might facilitate the displacement of DNA-binding proteins that sterically hinder accessibility and DNA resection by EXO1.
Figure 1. Dual control of EXO1 activity by conjugation with SUMO (S) and ubiquitin (U) reported by Bologna et al.Citation2 Constitutively sumoylated EXO1 is recruited to stalled replication forks induced by treatment with hydroxyurea (HU; a ribonucleotide reductase inhibitor), aphidicolin (a DNA polymerase inhibitor) or camptothecin (a topoisomerase-1 inhibitor). Upon substrate engagement and resection, EXO1 is ubiquitinated and delivered to proteasomal destruction.
References
- Mimitou EP, Symington LS. DNA Rep 2009; 8:983-95; PMID:19473888; http://dx.doi.org/10.1016/j.dnarep.2009.04.017
- Bologna S, et al. Cell Cycle 2015; 14(15):2473-84; http://dx.doi.org/10.1080/15384101.2015.1060381
- Tatham MH, et al. Sci Signal 2011; 4:rs4; PMID:21693764; http://dx.doi.org/10.1126/scisignal.2001484
- Hendriks IA, et al. Nat Struct Mol Biol 2014; 21:927-36; PMID:25218447; http://dx.doi.org/10.1038/nsmb.2890
- Galanty Y, et al. Nature 2009; 462:935-39; PMID:20016603; http://dx.doi.org/10.1038/nature08657
- Psakhye I, et al. Cell 2012; 151:807-20; PMID:23122649; http://dx.doi.org/10.1016/j.cell.2012.10.021
- Sarangi P, Zhao X. Trends Biochem Sci 2015; 40:233-42; PMID:25778614; http://dx.doi.org/10.1016/j.tibs.2015.02.006
