Epigenetics controls cell dynamics in the absence of DNA sequence modifications, adjusting cell metabolism to better adapt cells to the changing microenvironment. However, mutations cannot be entirely excluded from the equation, especially if we interrogate the pillars of carcinogenesis. In healthy cells these rearrangements are not usually accompanied by a mutational load, but in cancer cells the transformation process is simultaneously facilitated by driver and passenger mutations. In this context, it is worth noting not only the scientific success in revealing how singular events contribute to tumorigenesis, but also the limitations we face in identifying which events precede others. At present, the difficulty of understanding the tumorigenic process lies in the way of our unifying epigenetics and genetics – a goal similar to that faced by physicists of unifying general relativity and quantum mechanics.
As is the case with general relativity, Epigenetics is highly plastic and ductile, but during development, thisplasticity becomes diminished once cellular differentiation begins in order to configure the various tissues from stem cells. So, although adult cells retain some adaptive properties, we consider them to be epigenetically fairly rigid, with the tissue associated-pluripotent cells being those that, for the most part, control the repair, maintenance and homeostatic processes of the whole organism. Unlike differentiated cells, however, cells developing a tumor demand the return of epigenetic plasticity. Tumorigenesis can therefore be interpreted as the dysregulation of the epigenomeCitation1 (). Normal tissues are characterized by a degree of epigenomic stability. The lack of DNA methylation at CG-enriched regions (known as CpG islands or CGIs) of promoters of housekeeping genes, tissue-specific genes and developmental regulator genes is a typical hallmark of normal cells. By contrast, transposable elements are highly methylated in order to avoid unexpected and undesirable DNA arrangements. Moreover, euchromatin (transcribed open regions) is differentiated from heterochromatin (transcribed repressed regions) on the basis of the deposition of large organized chromatin lysine post-translational modifications (LOCKs) and its organization in laminar-associated domains (LADs). Conversely, during tumorigenesis a widespread hypomethylation cascade occurs, which affects the aforementioned LOCK and LAD regions, repetitive elements, and single-copy genes. This phenomenon has been related to the gain of cell invasive properties among other phenotypes. Along with this generalized hypomethylation event, cancer is also driven by the hypermethylation of specific CGIs located at tumor suppressor gene promoters. In addition, Timp and co-workersCitation1 recently found a dramatic DNA methylation shift affecting the boundaries of promoter-associated CGIs. On occasion, there are shifts in the CGIs promoting CGI hypermethylation in cancer, and in other circumstances, when the boundary shifts in an opposite manner the CGI and the associated shores become hypomethylated. This discovery had tremendous implications for tumor biology, given that CGI shores are known to activate alternative transcription start sites (TSS) of cancer-specific differentially methylated regions (cDMRs). Finally, we should highlight the role of Polycomb machinery that leads to the blocking of transcription at methylated regions and the release of pluripotency loci from DNA methylation repression.
Figure 1. Epigenetic plasticity throughout the genome drives tumor initiation, promotion and progression. Panel illustrating tumorigenesis, from normal cells expressing common cell adhesion molecules and miRNA profiles to proliferative and invasive cells with an uncountable number of epigenetic disruptions. The picture highlights the importance of epigenetic plasticity to the invasive capacity of tumor cells and the EMT/MET processes in tumorigenesis. Head arrows indicates upregulation or downregulation in gene expression; green denotes hypomethylation events while red hypermethylation events.
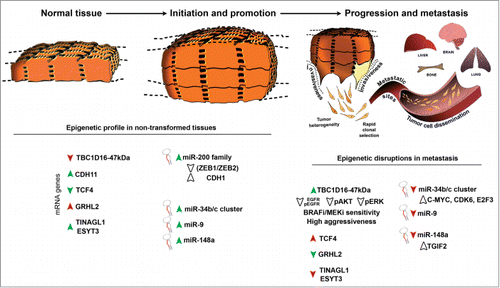
To a large extent, this epigenetic knowledge has been gained from the development of novel comprehensive strategies and improved technologies. For instance, Lujambio and colleagues,Citation2 using a pharmacological and genomic approach, identified an aberrant epigenetic silencing program of miRNAs tightly linked to the metastasis process by hybridization to an expression microarray. The authors reported the cancer-associated epigenetic silencing of 3 of these small molecules (miR-34b/c cluster, miR-9, and miR-148a) that are specialized in abrogating the expression of well-known anti-apoptotic and cell cycle oncogenes in normal cells. In a similar fashion, Carmona and colleaguesCitation3 described the epigenetic silencing of a relevant cell adhesion molecule ascribed to the metastatic cascade by using GoldenGate DNA methylation-based technology. The authors found that CDH11 DNA methylation-associated transcriptional silencing occurred in the corresponding lymph node metastases but not in the primary tumors. Taking advantage of new advanced platforms, Fang and colleaguesCitation4 were confident of discriminating breast tumor subtypes on the basis of the presence or absence of hypermethylation events in a large number of genes that define the breast CG island methylator phenotype (B-CIMP). Finally, Vizoso and colleagues,Citation5 using a next-generation platform, the HumanMethylation450 array, deciphered one of the primordial activator mechanisms of metastasis in melanoma, the epigenetic reactivation of a cryptic isoform of the TBC1D16 gene. The authors linked this local hypomethylation event to a dynamic rewiring between MAPK/RAS/RAF/ERK and PI3K/AKT pathways led by the downregulation of the EGFR protein.
Here, we have briefly summarized how DNA methylation plasticity may contribute to the natural history of metastasis. However, epigenetic dynamics is also a critical determinant of natural processes such as organogenesis and wound-healing. Curiously, most of these phenomena are commonly controlled by a disturbance called epithelial-to-mesenchymal transition (EMT), which plays a significant role not only in development but also in the initiation of metastases. However, there is a reverse process, the mesenchymal-to-epithelial transition (MET), which contributes to normal development and to the establishment and stabilization of distant metastases. Working in this field, Davalos and colleaguesCitation6 reported how epigenetic plasticity, based on the DNA methylation of a microRNA family, can shift between MET and EMT states during tumor progression. Another study, led by Carmona and colleagues,Citation7 examined the conservation of EMT and MET processes among species, and found that common methylation events act as commanders of the metastasizing cells. Intriguingly, these findings also imply that DNA methylation affects unfamiliar parts of the genome during tumor progression, thereby establishing a milestone in the exciting race to untangle the complexity of the genome and epigenetic plasticity.
References
- Timp W, Feinberg AP. Nat Rev Cancer 2013; 13:497-510; PMID:23760024; http://dx.doi.org/10.1038/nrc3486
- Lujambio A, et al. Proc Natl Acad Sci U S A 2008; 105:13556-61; PMID:18768788; http://dx.doi.org/10.1073/pnas.0803055105
- Carmona FJ, et al. J Pathol 2012; 228:230-40; PMID:22374749; http://dx.doi.org/10.1002/path.4011
- Fang F, et al. Sci Transl Med 2011; 3:75ra25; PMID:21430268; http://dx.doi.org/10.1126/scitranslmed.3001875
- Vizoso M, et al. Nat Med 2015; 21(7):741-50; PMID:26030178; http://dx.doi.org/10.1038/nm.3863
- Davalos V, et al. Oncogene 2012; 31:2062-74; PMID:21874049; http://dx.doi.org/10.1038/onc.2011.383
- Carmona FJ, et al. Cancer Res 2014; 74:5608-19; PMID:25106427; http://dx.doi.org/10.1158/0008-5472.CAN-13-3659
