Abstract
Gastric cancer remains a serious threat to public health with high incidence and mortality worldwide. Accumulating evidence demonstrates that long non-coding RNAs (lncRNAs) play important roles in regulating gene expression and are involved in various pathological processes, including gastric cancer. To investigate the possible role of dysregulated lncRNAs in gastric cancer development, we performed lncRNA microarray and identified 3141 significantly differentially expressed lncRNAs in gastric cancer tissues. Next, some of deregulated lncRNAs were validated among about 60 paired gastric cancer specimens such as Linc00261, DKFZP434K028, RPL34-AS1, H19, HOTAIR and Linc00152. Our results found that the decline of DKFZP434K028 and RPL34-AS1, and the increased expression of Linc00152 positively correlated with larger tumor size. The high expression levels of HOTAIR were associated with lymphatic metastasis and poor differentiation. Since the biological roles of Linc00152 are largely unknown in gastric cancer pathogenesis, we assessed its functions by silencing its up-regulation in gastric cancer cells. We found that Linc00152 knockdown could inhibit cell proliferation and colony formation, promote cell cycle arrest at G1 phase, trigger late apoptosis, reduce the epithelial to mesenchymal transition (EMT) program, and suppress cell migration and invasion. Taken together, we delineate the gastric cancer lncRNA signature and demonstrate the oncogenic functions of Linc00152. These findings may have implications for developing lncRNA-based biomarkers for diagnosis and therapeutics for gastric cancer.
Abbreviations
| AEG-1 | = | astrocyte elevated gene-1 |
| BACE1 | = | β-secretase 1 |
| EMT | = | epithelial-to-mesenchymal transition |
| GSEA | = | gene set enrichment analysis |
| LncRNA | = | long non-coding RNA |
Introduction
Gastric cancer is one of the most aggressive malignancies and represents the second frequent cause of cancer-related death worldwide.Citation1 The majority of patients suffering from gastric cancer are diagnosed at the advanced stages accompanied with malignant proliferation, extensive invasion, lymph node and distant metastasis.Citation2 Currently, surgical resection and chemotherapy are the main approaches for gastric cancer treatment, but recurrence and metastasis are serious events accounting for the poor prognostic outcomes.Citation2 Remarkable progress has been made in understanding the molecular mechanisms of cancer, such as oncogenes and tumor suppressor gene mutations,Citation3 aberrant genome alterations,Citation4 and identification of cancer stem cells.Citation5 However, successful clinical applications are limited and the mortality is still high in gastric cancer. Therefore, elucidating new mechanisms associated with gastric cancer pathogenesis are vital to identify useful biomarkers and develop effective targeted treatment for clinical benefits.
Recently, the large scale genome-wide sequencing projects revealed that up to 70% human genome is transcribed, whereas the coding-protein transcripts represent less than 2%, while the majority of transcriptome belong to non-protein-coding RNAs (ncRNA).Citation6 These ncRNAs were initially regarded as transcriptional “noise,” but accumulating evidence has demonstrated that ncRNAs play critical regulatory roles in gene expression, not only the small ncRNAs such as microRNAs, siRNAs and piRNAs but also the long ncRNAs (lncRNAs).Citation7 LncRNAs are collectively referred to the cellular RNA longer than 200 nucleotides and have the mRNA-like properties containing 5′ cap structure, exons, introns and 3′ polyadenylation, yet lack the potential of encoding proteins.Citation8 Unlike the mechanism of miRNA and siRNA silencing target genes via base-pairing the mRNA complementary sequences, lncRNAs have diversity and complex manners to control the local or global gene expression. LncRNA can function as scaffolds to affect complex assembly and chromatin remodeling, or as enhancers and decoys for promoting and inhibiting target gene transcriptions.Citation9-11 The dysregulated expression of lncRNAs has been implicated in many diseases, including cancers.Citation8 HOTAIR is one of the well-documented lncRNAs, and the high expression level of HOTAIR has become a powerful predictor of metastasis and death in breast cancer. Citation12 HOTAIR can serve as a scaffold with its 5′ domain interacting with polycomb repressive complex 2 (PRC2), whereas the 3′ domain binds to the LSD1/CoREST/REST complex. The ability of HOTAIR to assemble the 2 distinct histone modification enzymes enables the PRC2 and LSD1 complexes to re-target the genome-wide chromatin by regulating H3K27 trimethylation and H3K4 demethylation. The cancer epigenome alterations by HOTAIR lead to aberrant gene expression and induce cancer invasiveness and metastasis.Citation12,13 Several lncRNAs have been identified in specific cancer subtypes, such as the lncRNA PCA3 in prostate cancer, which has become the first approved diagnostic biomarker by FDA. However, in gastric cancer, dysregulated lncRNAs and their functional mechanisms have not yet been extensively studied.
Microarray is a reliable and low-cost approach to profile gene expression. The high technical stability and detection sensitivity make microarray popular in detecting lncRNA alterations in cancers.Citation14 The lncRNA expression signatures have been reported in multiple cancers including glioblastoma,Citation15 hepatocellular carcinoma,Citation16 renal cell carcinoma,Citation17,18 osteosarcoma,Citation19 lung adenocarcinoma,Citation20 esophageal squamous cell carcinomaCitation21 and gastric cancer.Citation22 These studies discovered a great deal of dysregulated lncRNAs in cancers, but the underlying mechanisms executed by these aberrant lncRNAs are largely unknown.
In the current study, we performed lncRNA microarray detection using 6 paired gastric cancer samples and delineated the lncRNAs expression signature in gastric cancer. After validating several up- and down-regulated lncRNAs in about 60 paired specimens, we found that some deregulated lncRNAs levels were associated with the clinical pathology features such as larger tumor size, poor differentiation, and lymphatic metastasis. Then, the functional involvements of one validated up-regulated lncRNA named Linc00152 were further evaluated. Our findings indicated that knockdown of Linc00152 led to cell proliferation repression, enhancing cell cycle G1 phase arrest, triggering late apoptosis, decreasing the epithelial-to-mesenchymal transition (EMT) program, and inhibiting cell migration and invasion. These results suggest that Linc00152 may serve as an oncogenic lncRNA that participates in gastric cancer development and aggressive progression. Linc00152 has the potential to become a new biomarker for gastric cancer diagnosis and therapy.
Results
Differential expression patterns of lncRNAs between gastric cancer tissues and the normal counterparts
To investigate the involvement of lncRNA in gastric cancer pathogenesis, we performed the lncRNA microarray detection with 6 paired gastric cancer tissues and their matched normal counterparts. The basic clinical information of the 6 patients is shown in Table S1 and the lncRNA profiling data are presented in Table S3. In , the scatter-plots above the top green line and below the bottom green line were those differentially expressed lncRNAs. Then, we performed the volcano-plot filtering, and the red plots in were the statistically significant lncRNAs between the gastric cancer and normal groups. From the overview of the scatter-plot and volcano-plot (), there were more downregulated lncRNAs than the upregulated ones. We also found that the down-regulated lncRNAs were more common in each paired gastric cancer tissues (), although the number of dysregulated lncRNAs varied in different patients. These significantly differentially expressed lncRNAs were defined as those with normalized intensities between gastric cancer tissues and the normal counterparts calculating the absolute fold change of ≥2. The differentially expressed lncRNA are shown in Table S4. In total, we identified 3141 differentially expressed lncRNAs, 1127 were up-regulated and 2014 were downregulated (Table S5). The most upregulated and down-regulated lncRNAs are listed in Table S6. The Hierarchical clustering result in showed a distinguishable lncRNA expression signature, which may help to discriminate gastric cancer tissues from normal counterparts.
Figure 1. Differential expression patterns of lncRNAs between gastric cancer tissues and the normal counterparts. (A) Scatter-plots presented the variations of lncRNA between the gastric cancer and matched normal tissues. The points deviated from the top and bottom green lines indicated the differentially expressed lncRNA in gastric cancer and the point color indicated the signal intensity from low (gray) to high (red). (B) Volcano plots of lncRNAs. The red plots were the differentially expressed lncRNAs with statistical significance (P < 0.05). The left and right red pots represented significantly down- and up-regulated lncRNAs in gastric cancer. (C) The number of significantly deregulated lncRNA in each paired gastric cancer specimens. (D) Heat map and hierarchical clustering analyzed the distinguishable lncRNA profiling in gastric cancer and normal samples. The relative expression from high to low level was indicated with red and blue color. (E) The number of significantly differentiate lncRNA in each lncRNA subgroups.
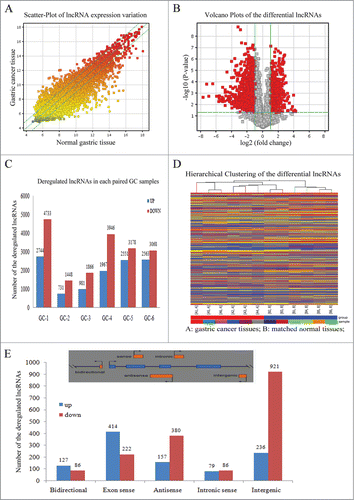
According to the lncRNA location with respect to the neighbor protein-coding transcripts, lncRNAs were stratified into bidirectional, sense, antisense, intronic and intergenic,Citation23 as shown in the upper panel of . This classification could provide some clues for exploring lncRNA functions. For examples, BACE1-AS was the conserved noncoding natural antisense transcript for β-secretase-1 (BACE1), which could form RNA duplex with each other to increase the stability of BACE1 mRNA, thereby aggravating the process of human Alzheimer disease.Citation24 In this study, we enumerated the number of altered lncRNAs in each subgroup including 213 bidirectional, 636 exon sense, 537 antisense, 165 intronic sense and 1157 intergenic lncRNAs as shown in .
Overall, the results from the lncRNA microarray analysis revealed that lncRNAs display aberrantly expressed patterns and these dysregulated lncRNAs may play important roles in the development and progression of gastric cancer.
Validation of the aberrantly expressed lncRNAs in paired gastric cancer tissues and cell lines
To validate the microarray findings, we randomly selected 3 downregulated lncRNAs Linc00261, DKFZP434K028 and RPL34-AS1 in the microarray for validation in about 60 paired gastric cancer specimens. The clinical characteristics of the cohort are summarized in . As shown in , Linc00261, DKFZP434K028 and RPL34-AS1 had lower expression levels in gastric cancer tissues than the normal counterparts. In gastric cell lines, the 3 lncRNAs were also down-regulated compared with the respective normal gastric epithelial cell line GES-1 (). Moreover, the low expression levels of DKFZP434K028 and RPL34-AS1 positively correlated with the larger tumor size as shown in .
Figure 2. Validation of the aberrantly downregulated lncRNAs in paired gastric cancer tissues and cell lines. (A) The expression levels of Linc00261 were validated in 61 paired gastric cancer specimens with qRT-PCR. (B) The levels of DKFZP434K028 were detected in 65 paired gastric cancer tissues. (C) RPL34-AS1 expression levels were evaluated in 61 paired gastric cancer cohort. GAPDH was the internal control. The-ΔCt value was calculated by GAPDH Ct value subtracting lncRNA Ct value. (D) The levels of Linc00261, DKFZP434K028 and RPL34-AS1 in gastric cell lines. HGC-27 and SGC-7901 were two gastric cancer cell lines, and GES-1 was the normal gastric epithelial cell. The data were presented as the mean ±SD (n = 3), *P < 0.05, and ** P < 0.001.
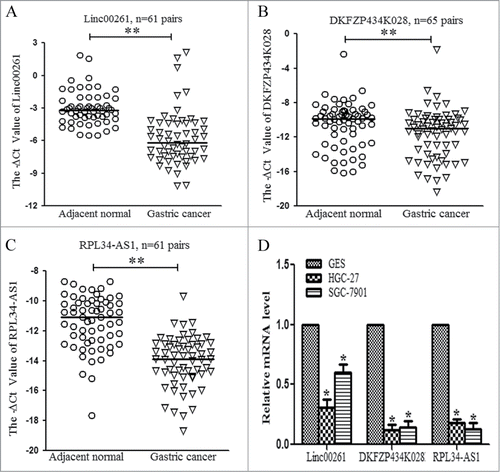
Table 1. The association of deregulated lncRNAs expression levels with clinical pathology features in gastric cancer
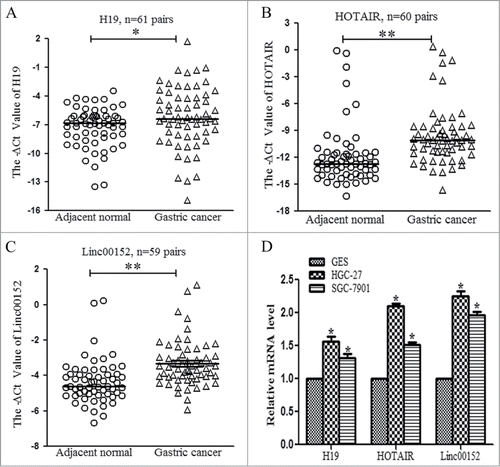
Next, we evaluated the up-regulated lncRNAs. H19 and HOTAIR are 2 well-known oncogenic lncRNAs in multiple cancers. In our validation cohort, H19 and HOTAIR also displayed significantly higher levels () and the upregulation of HOTAIR was associated with lymphatic metastasis and poor differentiation (). In addition, Linc00152 was validated to be increased in 59 paired gastric cancer specimens (), and the high levels of Linc00152 expression correlated with larger tumor size (). Moreover, H19, HOTAIR and Linc00152 were also overexpressed in gastric cancer cell lines ().
Figure 3. Validation of the aberrantly upregulated lncRNAs in paired gastric cancer tissues and cell lines. (A) H19 expression levels were validated in 61 paired gastric cancer tissues with qRT-PCR. (B) The expression levels of HOTAIR were measured in 60 paired gastric cancer tissues. (C) Linc00152 expression levels were detected in 59 paired gastric cancer specimens. (D) The levels of H19, HOTAIR and Linc00152 in gastric cancer and normal gastric epithelial cell lines. The calculating method was the same as above described, *P < 0.05 and ** P < 0.001.
Taken together, our validation results further confirm that a set of lncRNAs is frequently dysregulated in gastric cancers, and their aberrant expression levels are associated with the clinical pathology features such as larger tumor size, poor differentiation and lymphatic metastasis.
Linc00152 knockdown inhibits cell viability and colony formation
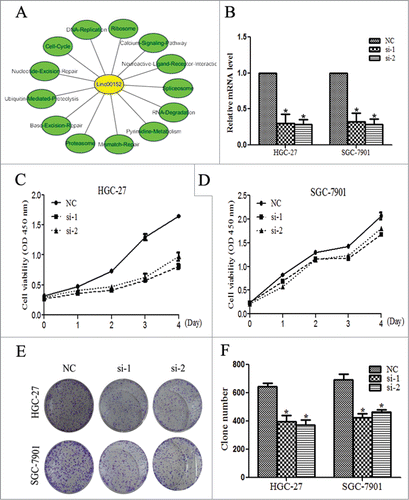
Our results indicated that the significant up-regulation of Linc00152 in gastric cancer specimens was associated with larger tumor size. Previous study found the high level of Linc00152 correlated with invasion in gastric cancer.Citation25 However, the underlying mechanisms of Linc00152 to promote gastric cancer development have not been elucidated, which prompted us to explore its biological functions. Gene Set Enrichment Analysis (GSEA) is a computational method to determine the possible biological processes and signaling pathways of defined gene sets. GSEA analysis suggested that Linc00152 might be involved in some critical biological processes including cell cycle, DNA replication, and mismatch-repair as shown in . Then, we investigated the Linc00152 function by employing 2 siRNA oligos to silence its upregulated expression in the 2 gastric cancer cell line HGC-27 and SGC-7901 (). Cell viability assay revealed that cell proliferation was significantly impaired with the Linc00152 knockdown, especially in HGC-27 cells (). The similar effects were also observed in the colony formation assay where the colony numbers were decreased following the Linc00152 knockdown (). Thus, these findings suggest that Linc00152 depletion may decrease malignant gastric cancer cell proliferation.
Figure 4. Linc00152 knockdown inhibits cell viability and colony formation. (A) The GESA results of Linc00152 with the absolute normalized enrichment score (|NES|) > 1 and false discovery rate (FDR) P < 0.001. (B) The knockdown result of Linc00152 by siRNAs in HGC-27 and SGC-7901 cell lines. The two cell lines were transfected with 40 nM siRNA oligos against Linc00152 for 48 h, qRT-PCR was performed to determine the Linc00152 mRNA level. (C) The cell viability was inhibited with Linc00152 knockdown by WST-1 assays in HGC-27. (D) The cell viability was detected with Linc00152 depletion in SGC-7901. (E) The representative picture of colony formation with Linc00152 reduction in HGC-27 and SGC-7901cells. (F) The histogram showed the average number of the survival clones. The data were presented as the mean ±SD (n = 3), and *P < 0.05.
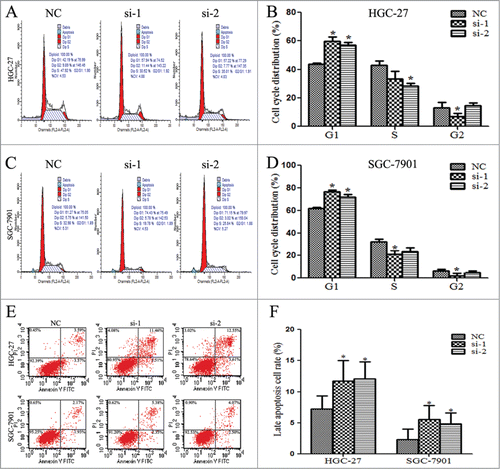
Linc00152 silence promotes cell cycle G1 phase arrest and apoptosis
To determine whether cell proliferation inhibition by Linc00152 silence resulted from the alteration of cell cycle or apoptosis, flow cytometry analysis was performed. The results indicated that the cell cycle of both HGC-27 and SGC-7901 were significantly arrested at G1 phase when Linc00152 was repressed. The percentage of G1 phase in HGC-27 was increased from 43.67% (NC) to 59.61% (si-1, P < 0.05) and 57.15% (si-2, P < 0.05) (). The G1 arrest percentage in SGC-7901 cell was elevated from 61.67% (NC) to 76.46% (si-1, P < 0.05) and 71.63 (si-2, P < 0.05) (). In addition, Linc00152 knockdown affected the late apoptosis of gastric cancer cells, but not the early apoptotic cells rates. The late apoptosis cell rates were increased from 7.23% (NC) to 11.75% (si-1, P < 0.05) and12.05% (si-2, P < 0.05) in HGC-27, and from 2.33% (NC) to 5.56% (si-1, P < 0.05) and 4.82% (si-2, P < 0.05) in SGC-7901 cells (). These findings indicate that Linc00152 knockdown can trigger cell cycle arrest at the G1 phase and drive late apoptosis, which may lead to inhibition of cell proliferation.
Figure 5. Linc00152 silence promotes cell cycle G1 phase arrest and apoptosis. (A and B). Cell cycle was arrested in G1 phase with Linc00152 silence in HGC-27. (C and D) Cell cycle arrest in G1 phase was increased in SGC-7901 with Linc00152 depletion. Flow cytometry was used to analyze the effects of Linc00152 on cell cycle when cells were treated with 40 nM siRNA targeting Linc00152 for 36 h in HGC-27 and SGC-7901 cells, respectively. (E and F) The late apoptosis cell rates were increased with Linc00152 knockdown in gastric cancer cells. The apoptotic cells were detected by flow cytometry after staining with Annexin V and PI after cells were transfected with 40 nM siRNA against Linc00152 for 36 h. The data were presented as the mean ± SD (n = 3), and *P < 0.05.
Linc00152 depletion represses epithelial-to-mesenchymal transition (EMT) program
Given that the epithelial–mesenchymal transition (EMT) plays important roles in cancer dissemination and metastatic spread, we assessed if Linc00152 had any impact on EMT program via examining the protein and mRNA levels of some EMT related markers. As shown in , when the Linc00152 was knocked down, the mesenchymal markers N-cadherin and Vimentin were downregulated, and the epithelial marker E-cadherin protein were up-regulated, whereas the EMT related transcriptional factors Snail and Slug were not altered remarkably. Additionally, we examined the level of AEG-1 expression since AEG-1 was established as the oncogenic proteins associating with cancer metastasis and invasion, and our previous studies found AEG-1 reduction could inhibit invasion, EMT in cervical cancerCitation26 and suppress cell migration in hepatocellular carcinoma.Citation27 We found that AEG-1 protein decreased with Linc00152 depletion.
Figure 6. Linc00152 depletion represses epithelial-to-mesenchymal transition (EMT) program. (A) Western Blot analyzed the expression of EMT related protein factors with Linc00152 knockdown in HGC-27 cells. (B) The relative mRNA levels of these EMT related factors were detected by qRT-PCR in HGC-27 with Linc00152 silence. (C) Western Blot detected EMT related protein expression with Linc00152 reduction in SGC-7901 cells. (D) qRT-PCR results of these EMT factors in SGC-7901 with Linc00152 depletion. The data were presented as the mean ± SD (n = 3), and *P < 0.05.
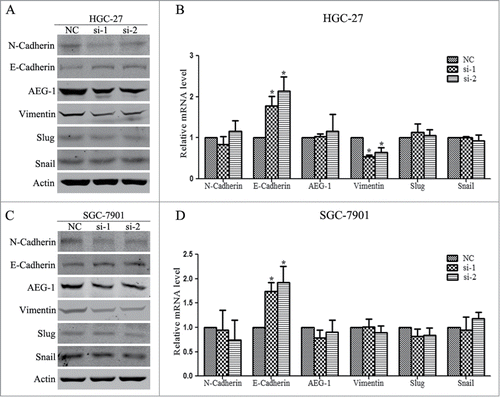
The qRT-PCR results of EMT markers indicated that the E-Cadherin mRNA levels were increased significantly in both HGC-27 and SGC-7901 cells, while the mRNA level of Vimentin was down-regulated remarkably in HGC-27 but not in SGC-7901, the transcriptional levels of N-cadherin, AEG-1, Snail and Slug had not been affected by Linc00152 knockdown (). Taken together, these findings indicate that depletion of Linc00152 inhibits the EMT progression in gastric cancer cells.
Linc00152 knockdown decreases migration and invasion of gastric cancer cells
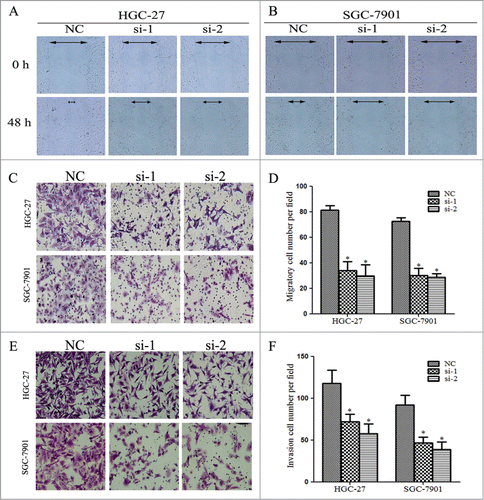
The EMT programs may cause dissociated epithelial cells to acquire migration and invasive capacities, which confer cancer cells the ability to pass through the basement membrane and migrate to distant tissues. Since we found that Linc00152 knockdown led to decreased EMT, we evaluated the effects of Linc00152 on cell migration and invasion. The wound-healing assay revealed that cells with Linc00152 knockdown showed a notably slower scratch closure rate than control cells (), which suggests mobility inhibition. The two-chamber transwell assay further confirmed that silencing Linc00152 remarkably decreased the migration ability in both gastric cancer cell lines HGC-27 and SGC-7901 (). Furthermore, invasion assay demonstrated that loss of Linc00152 displayed significantly lower invasion potential in gastric cancer cells compared with control cells (). Collectively, these results suggest that Linc00152 knockdown suppressed the migration and invasion of gastric cancer cells.
Figure 7. Linc00152 knockdown decreases migration and invasion of gastric cancer cells. (A) Linc00152 knockdown exhibited slower scratch closure rate by the wound-healing detection in HGC-27. (B) Wound-healing assay in SGC-7901 with Linc00152 silence. (C) The representative pictures of cell migration across the membrane in HGC-27 and SGC-7901 cells with Linc00152 reduction. (D) The histograms showed the average number of migrated cells per visual field calculated from 10 representative fields. (E) The representative pictures of transwell invasion assay in HGC-27 and SGC-7901 cells with Linc00152 depletion. (F) The average number of invasion cells per visual field from 10 representative fields. Data were presented as mean ± SD. (n = 3), and *P < 0.05.
Discussion
Although several studies have carried out lncRNA microarray profiling among gastric cancer tissues and cell lines, Citation22,28-31 their functional roles in gastric cancer pathogenesis remain largely unknown. Compared with the previously reported lncRNA profiles, our samples for lncRNA microarray could be divided into 2 subgroups (Table S1): 3 patients with tumor size <3 cm accompanying lymph metastasis; and three specimens with tumor size >4 cm without lymph node metastasis. Hence, our lncRNA signature may be better able to identify the dysregulated lncRNAs associated with gastric cancer proliferation and metastasis. Additionally, the availability of all these published lncRNA microarray data together would provide more information to interrogate lncRNAs mechanisms and clinical applications in gastric cancer. Taken the glioblastoma multiforme (GBM) as example, by mining previous published lncRNA expression profiling in 213 GBM tumors from The Cancer Genome Atlas (TCGA), a recent study identified a 6-lncRNA signature with prognostic value in GBM.Citation15 The 6 GBM prognostic lncRNAs contain one risky KIAA0495 and 5 protective PART1, MGC21881, MIAT, GAS5 and PAR5.Citation15 In esophageal squamous cell carcinoma (OSCC), a 3-lncRNA signature was found to associate with the survival of OSCC patients via detecting and analyzing 119 paired OSCC samples lncRNA microarray profiling. The dysregulation of 3 lncRNAs ENST00000435885.1, XLOC_013014 and ENST00000547963.1 in OSCC could serve as a new biomarker to predict the overall survival rate. Citation21 In gastric cancer, more lncRNA microarray analysis may help to identify valuable and effective lncRNA-based diagnosis, prognosis and therapeutic biomarkers.
In this study, we validated 3 downregulated lncRNAs Linc00261, DKFZP434K028 and RPL34-AS1 () in the microarray data, and found that the low expression levels of DKFZP434K028 and RPL34-AS1 positively correlated with the larger tumor size as shown in . The three lncRNAs have not been reported in previous studies and their functions in cancer are largely unknown. LncRNAs act mainly through regulating nearby or global gene expression in cis or in trans. The relative locations of lncRNAs with neighbor coding genes may provide some implications for their biological roles, such as the BACE1-AS could increase the stability of BACE1 to aggravate Alzheimer disease.Citation24 Among our validated 3 down-regulated lncRNAs, Linc00261 is intergenic and transcribed from the minus strand of chr20; DKFZP434K028 is located at the intronic antisense of MYRF gene, and MYRF encodes a membrane-bound transcription factor that autoproteolytically cleaves to activate myelin genes.Citation32 RPL34-AS1 is a bidirectional lncRNA of RPL34, and RPL34 is a ribosomal protein as a component of the 60S subunit which may catalyze protein synthesis. Whether the low levels of DKFZP434K028 and RPL34-AS1 have effect on MYRF and RPL34 expression needs to be investigated in future.
H19 has been found to be upregulated in multiple cancers including gastric cancer. H19 has the capacity to produce miR-675 precursor. The H19-derived miR-675 could target tumor suppressor RB in colorectal cancerCitation33 and silence RUNX1 and CALN1 to drive gastric cancer development. Citation34,35 Additionally, H19 could interact with tumor suppressor P53 to decrease Bax expression.Citation36 ISM1 was another binding protein of H19 in gastric cancer cells, and ISM1 level positively correlated with that of H19.Citation35 Via directly associating with proteins and indirectly silencing targets by producing miR-675, H19 overexpression led to cell proliferation, anti-apoptosis, invasion and metastasis in gastric cancer. Gastric cancer patients with high H19 levels had shorter survival times.Citation35 The activation of oncogene c-Myc was found to increase H19 expression.Citation36 Our result also demonstrated that H19 was overexpressed in gastric cancer tissues, despite lacking correlations with clinical characteristics, partly owing to the limited sample size and clinical information.
HOTAIR is another well-studied up-regulated lncRNAs in cancers, which affect cell proliferation, the EMT program, migration and invasion of gastric cancer. The well-established mechanism for HOTAIR was to bind PRC2 and LSD1/CoREST/REST complexes and silence genes in trans via H3K27 methylation and H3K4 demethylation.Citation12,13 In addition, HOTAIR could interact with miR-331–3p and acted as the endogenous decoy to protect HER2 from degradation by miR-331–3p.Citation37 The increased HER2 expression led to more aggressive properties and poor survival outcome in gastric cancer patients.Citation37 Our findings demonstrated that HOTAIR was upregulated in gastric cancer tissues (). Moreover, the high level of HOTAIR expression was associated with lymphatic metastasis and poor differentiation (). The multiple mechanisms of oncogenic HOTAIR in cancer development may account for the association.
Recently, Linc00152 was found to be increased in gastric cancer LncRNA profiles and correlated with invasion.Citation25,29 Our results demonstrated that Linc00152 was indeed up-regulated in gastric cancer specimens and its high levels of expression were associated with larger tumor size. However, the Linc00152 functions have not been elucidated so far. We performed the bioinformatic Linc00152 GSEA, which suggested that Linc00152 may participate in cell cycle, DNA replication, and mismatch-repair (). These findings provided clues about how to evaluate Linc00152 functions in gastric cancer development. Our results showed that Linc00152 depletion could inhibit gastric cancer cell proliferation (), colony formation (), promote cell cycle arrest at the G1 phase () and trigger late apoptosis (). Moreover, we found that Linc00152 knockdown could suppress EMT program by decreasing mesenchymal markers N-cadherin, Vimentin and oncogenic AEG-1 protein levels, and increasing epithelial marker E-cadherin expression (). The wound-healing, trans-well migration and invasion assays further demonstrated that depleting Linc00152 could reduce the migration and invasion ability of gastric cancer cells (). These results provide an explanation for Linc00152 upregulation associating with larger gastric tumor size in our study and gastric cancer invasion reported in other study.Citation25 Linc00152 may be a useful biomarker in gastric cancer.
In this study, we found that the mRNA alterations of EMT factors were not completely consistent with their protein levels. When Linc00152 was depleted, E-cadherin mRNA and protein expression were increased. But N-cadherin, Vimentin and AEG-1 proteins were reduced while their transcriptional levels were not altered significantly except the reduction of Vimentin mRNA in HGC-27 cells (). These inconsistencies may reflect the versatile mechanisms of lncRNAs to manipulate gene expression in gastric cancer, that lncRNAs can control expression at transcriptional, post-transcriptional and epigenetic levels as we previously reviewed.Citation38 For example, our recent study found that lncRNA GAS5 interacted with YBX1 protein, and GAS5 could increase YBX1 protein stability without affecting YBX1 mRNA level. The down-regulation of GAS5 in gastric cancer reduced YBX1 protein level, which subsequently decreased YBX1-transactivated p21 expression and abolished G1 phase cell cycle arrest.Citation39 Another example is LncRNA ANRIL. ANRIL could recruit and associate with PRC2 complex to epigenetically repress miR-99a/miR-449a expression in trans, which activated the miR-99a/miR-449a targets: mTOR and CDK6/E2F1 pathways, and led to aberrant cell proliferation.Citation40 The high level of ANRIL could serve as an independent predictor for poor overall survival, and markedly correlated with aggressive TNM stage and larger tumor size in gastric cancer.Citation40
In summary, we performed lncRNA microarray analysis and validated the dysregulated lncRNAs in about 60 paired gastric cancer specimens. The low expression of DKFZP434K028 and RPL34-AS1, and the high levels of Linc00152 positively correlated with larger tumor size (). The up-regulation of HOTAIR associated with lymphatic metastasis and poor differentiation (). Furthermore, we found that Linc00152 knockdown could inhibit cell proliferation and colony formation, promote cell cycle G1 phase arrest, trigger late apoptosis, reduce EMT, and suppress cell migration and invasion. Collectively, these results suggest that Linc00152 may function as an oncogenic LncRNA to drive cancer development and have great potential to become a promising biomarker and therapeutic target in gastric cancer.
Materials and Methods
Patient samples
The gastric cancer samples and matched adjacent normal tissues were obtained from surgical operation patients at Huashan Hospital, Fudan University, from November 2012 to December 2013. Informed consent was written by each enrolled subject and the Human Research Ethics Committee of Huashan Hospital granted the ethics approval for this study. All our methods and experimental protocols were according to the guidelines approved by the Human Research Ethics Committee of Huashan Hospital. The gastric cancer specimens were diagnosed by 2 individual pathologists, and the corresponding non-tumorous tissues were 5 cm away from the edge of tumor without obvious tumor cells existence. All the enrolled subjects had no radiotherapy and chemotherapy history prior to the surgical operation. Tumor staging was performed according to the tumor-node-metastasis (TNM) grading system. All samples were immediately snap-frozen in liquid nitrogen after surgical resection and then stored at −80°C until RNA extraction. Of these paired gastric cancer tissues, 6 paired samples were used for lncRNAs microarray analysis and their detailed information is summarized in Table S1.
Microarray and computational analysis
For microarray analysis, Agilent Array platform was employed. Sample preparation and microarray hybridization were performed based on the manufacturer's standard protocols with minor modifications. Briefly, mRNA was purified from total RNA after removal of rRNA (mRNA-ONLY™ Eukaryotic mRNA Isolation Kit, Epicentre). Then, each sample was amplified and transcribed into fluorescent cRNA along the entire length of the transcripts without 3′ bias utilizing a random priming method. The labeled cRNAs were hybridized onto the Human LncRNA Array v3.0 (8 × 60 K, Arraystar). After washing, the arrays were scanned by the Agilent Scanner G2505C. Agilent Feature Extraction software (version 11.0.1.1) was used to analyze acquired array images. Quantile normalization and subsequent data processing were performed using the GeneSpring GX v11.5 software package (Agilent Technologies). After quantile normalization of the raw data, the 12 lncRNAs samples with flags in Present or Marginal (“All Targets Value”) were chosen for further data analysis. Differentially expressed lncRNAs with statistical significance between the gastric cancer tissues and the corresponding normal tissues were identified through Volcano Plot filtering. Finally, Hierarchical Clustering was performed to show the distinguishable lncRNAs expression pattern among samples. The microarray work was performed by KangChen Bio-tech, Shanghai P.R. China.
Gene set enrichment analysis (GSEA)
GSEA was performed by the JAVA program (http://www.broadinstitute.org/gsea) with MSigDB C2 CP: Canonical pathways gene set collection. Gene sets with the absolute normalized enrichment score (|NES|) > 1, and the false discovery rate (FDR) P < 0.001 were considered to be significantly enriched. Cytoscape was used for visualization of the GSEA results.
Cell culture and siRNA transfection
Human gastric epithelial cell line (GES-1), gastric cancer cell lines (HCG-27 and SGC-7901) were obtained from the Shanghai Institute of Biochemistry and Cell Biology. The cells were cultured in 1640 medium with 10% fetal bovine serum (GIBCO, USA) at 37°C in 5% CO2-humidified atmosphere. Transfection was conducted with Lipofectamine 2000 reagent (Invitrogen, USA) according to the manufacturer's instructions. When cell densities were about 60%, 40 nM siRNA oligos were introduced into cells. The siRNA oligos sequences targeting Linc00152 were:
Linc00152-si-1: 5′ GGGAAAUAAAUGACUGGAUdTdT 3′;
Linc00152-si-2: 5′ GGAGAUGAAACAGGAAGCUdTdT 3′;
Negative control (NC): 5′ UUCUCCGAACGUGUCACGUdTdT 3′;
RNA extraction and qRT-PCR detection
Total RNA was extracted from gastric cancer samples using TRIzol reagent (Invitrogen, CA, USA) according to the manufacturer's protocol. The RNA quantity and quality was evaluated by Nano Drop ND-2000 spectrophotometer. To validate the microarray data and detect the target gene transcriptional level, 1 μg RNA was reverse-transcribed to cDNA with ReverTra Ace kit (Toyobo, Japan) following the manufactures' protocols. qRT-PCR was performed with standard SYBR-Green PCR kit protocol (Takara, Japan) in ABI 7900 Real Time PCR System. All samples were done in triplicate and normalized by the housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The qRT-PCR primers are listed in Table S2.
Cell proliferation assays
Cell proliferation assays were performed with Cell Counting Kit-8 (Dojindo, Japan) according to the manufacturer's protocol. About 2000 cells per well were seeded into 96-well plates after Linc00152 siRNAs and negative control (NC) transfection for 24 h. The number of proliferating cells per well was evaluated by the 450 nm absorbance of reduced WST-8 at the indicated time points. Three independent experiments were carried out for each assay.
Colony formation assays
Cells were trypsinized into a single-cell suspension after transfection with siRNA oligos targeting Linc00152 for 48 h. About 1000 cells were plated in each well of the 12-well plate and maintained for 2 weeks to form colony. Then, the colonies were fixed using methanol and stained with 0.1% crystal violet. All samples were done in triplicate.
Flow cytometry analysis for cell cycle and apoptosis
For cell cycle analysis, 1 × 106 cells were seeded into 6-well plates. At 36 h after transfection with Linc00152 siRNAs duplexes, cells were collected and fixed in chilled 70% ethanol at −20°C for 2 h, followed by washing with phosphate-buffered saline (PBS), and the fixed cells were stained with 50 μg/ml propidium iodide (PI) at room temperature for 20 min before analysis.
For apoptosis assay, cells were harvested after transfection with Linc00152 siRNAs for 36 h, and then processed to stain with Annexin V-fluorescein isothiocyanate/ Propidium Iodide kit (KeyGen Biotech, China) according to the manufacturer's instructions. The flow cytometry analysis was performed by FACSCalibur (BD Biosciences, USA). Results were the representative of 3 independent experiments with triplicate samples for each assay.
Western blot and antibodies
Cell lysates were separated by 10% SDS-PAGE and transferred to nitrocellulose membranes (GE Healthcare, USA). The membrane was blocked with 5% milk in TBS (20 mM Tris-HCl, 137mM NaCl, pH 7.6) for 1 h at room temperature, and incubated with specific primary antibodies at 4°C overnight, followed by IRDye secondary antibodies at room temperature for 1 h. Then the protein bands of interest were visualized with the Odyssey system (LI-COR, USA). The specific antibodies used for Western blot were as follows: anti-AEG-1 (ab45338, Abcam, British), anti-N-Cadherin (#4061, CST, USA), anti-E-Cadherin (#3195, CST, USA), anti-Vimentin (#5741, CST, USA), anti-Snail (#3879, CST, USA), anti-Slug (#9585, CST, USA) and anti-β-actin (#12620, CST, USA).
Cell migration and invasion assay
The migration assay was performed by using transwell insert chambers (8 μm pore size, Corning, USA). About 2 × 104 cells were seeded into the upper chamber in serum free medium in triplicate. For the invasion assay, Matrigel Invasion Chambers in the 24-well plates of 8 μm (BD Bioscience, USA) were used and 1 × 105 cells were plated into the upper chamber without serum in triplicate. The lower chamber was filled with 600 μl 1640 medium containing 10% FBS as chemo-attractant. After incubation for 24 h, non-migrating or non-invasion cells in upper chambers were removed by cotton swab, and cells migrating or invading to the lower surface of membrane were fixed using methanol and stained with 0.1% crystal violet. The migrating and invasion cells were counted at least 10 visual fields per membrane under the light microscope.
Statistical analysis
All statistical data were analyzed by Statistical Program for Social Sciences 18.0 software (SPSS, Chicago, IL) and GraphPad Prism 6.0 (GraphPad Software, La
Jolla, CA). Two-tailed Student's t-test and Wilcoxon signed rank-sum test were used as appropriate. The association between lncRNAs levels and clinical pathology features were analyzed with Mann-Whitney U test. P < 0.05 was considered statistically significant.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
1078034_supplemental_files.zip
Download Zip (6.3 MB)Funding
This study was supported by the Natural Science Foundation of China (81201521, 81400007), the China Postdoctoral Science Foundation Funded Project (2013M541467), and the Foundation from Science and Technology Commission of Shanghai (13XD1401200).
References
- Ferro A, Peleteiro B, Malvezzi M, Bosetti C, Bertuccio P, Levi F, Negri E, La Vecchia C, Lunet N. Worldwide trends in gastric cancer mortality (1980–2011), with predictions to 2015, and incidence by subtype. Eur J Cancer 2014; 50:1330-44; PMID:24650579; http://dx.doi.org/10.1016/j.ejca.2014.01.029
- Karimi P, Islami F, Anandasabapathy S, Freedman ND, Kamangar F. Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev 2014; 23:700-13; PMID:24618998; http://dx.doi.org/10.1158/1055-9965.EPI-13-1057
- Nagarajan N, Bertrand D, Hillmer AM, Zang ZJ, Yao F, Jacques PE, Teo AS, Cutcutache I, Zhang Z, Lee WH, et al. Whole-genome reconstruction and mutational signatures in gastric cancer. Genome Biol 2012; 13:R115; PMID:23237666; http://dx.doi.org/10.1186/gb-2012-13-12-r115
- Zang ZJ, Cutcutache I, Poon SL, Zhang SL, McPherson JR, Tao J, Rajasegaran V, Heng HL, Deng N, Gan A, et al. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat Genet 2012; 44:570-4; PMID:22484628; http://dx.doi.org/10.1038/ng.2246
- Zhou J, Zhang Y. Cancer stem cells: Models, mechanisms and implications for improved treatment. Cell Cycle 2008; 7:1360-70; PMID:18418062; http://dx.doi.org/10.4161/cc.7.10.5953
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 2007; 447:799-816; PMID:17571346; http://dx.doi.org/10.1038/nature05874
- Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell 2011; 145:178-81; PMID:21496640; http://dx.doi.org/10.1016/j.cell.2011.03.014
- Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol 2012; 9:703-19; PMID:22664915; http://dx.doi.org/10.4161/rna.20481
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet 2009; 10:155-9; PMID:19188922; http://dx.doi.org/10.1038/nrg2521
- Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell 2011; 43:904-14; PMID:21925379; http://dx.doi.org/10.1016/j.molcel.2011.08.018
- Au PC, Zhu QH, Dennis ES, Wang MB. Long non-coding RNA-mediated mechanisms independent of the RNAi pathway in animals and plants. RNA Biol 2011; 8:404-14; PMID:21525783; http://dx.doi.org/10.4161/rna.8.3.14382
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010; 464:1071-6; PMID:20393566; http://dx.doi.org/10.1038/nature08975
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010; 329:689-93; PMID:20616235; http://dx.doi.org/10.1126/science.1192002
- Du Z, Fei T, Verhaak RG, Su Z, Zhang Y, Brown M, Chen Y, Liu XS. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat Struct Mol Biol 2013; 20:908-13; PMID:23728290; http://dx.doi.org/10.1038/nsmb.2591
- Zhang XQ, Sun S, Lam KF, Kiang KM, Pu JK, Ho AS, Lui WM, Fung CF, Wong TS, Leung GK. A long non-coding RNA signature in glioblastoma multiforme predicts survival. Neurobiol Dis 2013; 58:123-31; PMID:23726844
- Yang F, Zhang L, Huo XS, Yuan JH, Xu D, Yuan SX, Zhu N, Zhou WP, Yang GS, Wang YZ, et al. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology 2011; 54:1679-89; PMID:21769904; http://dx.doi.org/10.1002/hep.24563
- Yu G, Yao W, Wang J, Ma X, Xiao W, Li H, Xia D, Yang Y, Deng K, Xiao H, et al. LncRNAs expression signatures of renal clear cell carcinoma revealed by microarray. PLoS One 2012; 7:e42377; PMID:22879955; http://dx.doi.org/10.1371/journal.pone.0042377
- Qin C, Han Z, Qian J, Bao M, Li P, Ju X, Zhang S, Zhang L, Li S, Cao Q, et al. Expression pattern of long non-coding RNAs in renal cell carcinoma revealed by microarray. PLoS One 2014; 9:e99372; PMID:24905231; http://dx.doi.org/10.1371/journal.pone.0099372
- Li JP, Liu LH, Li J, Chen Y, Jiang XW, Ouyang YR, Liu YQ, Zhong H, Li H, Xiao T. Microarray expression profile of long noncoding RNAs in human osteosarcoma. Biochem Biophys Res Commun 2013; 433:200-6; PMID:23466354; http://dx.doi.org/10.1016/j.bbrc.2013.02.083
- Xu G, Chen J, Pan Q, Huang K, Pan J, Zhang W, Yu F, Zhou T, Wang Y. Long noncoding RNA expression profiles of lung adenocarcinoma ascertained by microarray analysis. PLoS One 2014; 9:e104044; PMID:25089627; http://dx.doi.org/10.1371/journal.pone.0104044
- Li J, Chen Z, Tian L, Zhou C, He MY, Gao Y, Wang S, Zhou F, Shi S, Feng X, et al. LncRNA profile study reveals a three-lncRNA signature associated with the survival of patients with oesophageal squamous cell carcinoma. Gut 2014; 63(11):1700-10; PMID:24522499
- Lin XC, Zhu Y, Chen WB, Lin LW, Chen DH, Huang JR, Pan K, Lin Y, Wu BT, Dai Y, et al. Integrated analysis of long non-coding RNAs and mRNA expression profiles reveals the potential role of lncRNAs in gastric cancer pathogenesis. Int J Oncol 2014; 45:619-28; PMID:24819045
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell 2009; 136:629-41; PMID:19239885; http://dx.doi.org/10.1016/j.cell.2009.02.006
- Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G, 3rd, Kenny PJ, Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of beta-secretase. Nat Med 2008; 14:723-30; PMID:18587408; http://dx.doi.org/10.1038/nm1784
- Pang Q, Ge J, Shao Y, Sun W, Song H, Xia T, Xiao B, Guo J. Increased expression of long intergenic non-coding RNA LINC00152 in gastric cancer and its clinical significance. Tumour Biol 2014; 35:5441-7; PMID:24523021; http://dx.doi.org/10.1007/s13277-014-1709-3
- Liu X, Wang D, Liu H, Feng Y, Zhu T, Zhang L, Zhu B, Zhang Y. Knockdown of astrocyte elevated gene-1 (AEG-1) in cervical cancer cells decreases their invasiveness, epithelial to mesenchymal transition, and chemoresistance. Cell Cycle 2014; 13:1702-7; PMID:24675891; http://dx.doi.org/10.4161/cc.28607
- Zhao J, Wang W, Huang Y, Wu J, Chen M, Cui P, Zhang W, Zhang Y. HBx elevates oncoprotein AEG-1 expression to promote cell migration by downregulating miR-375 and miR-136 in malignant hepatocytes. DNA Cell Biol 2014; 33(10):715-22; PMID:25050974
- Song H, Sun W, Ye G, Ding X, Liu Z, Zhang S, Xia T, Xiao B, Xi Y, Guo J. Long non-coding RNA expression profile in human gastric cancer and its clinical significances. J Transl Med 2013; 11:225; PMID:24063685; http://dx.doi.org/10.1186/1479-5876-11-225
- Cao WJ, Wu HL, He BS, Zhang YS, Zhang ZY. Analysis of long non-coding RNA expression profiles in gastric cancer. World J Gastroenterol 2013; 19:3658-64; PMID:23801869; http://dx.doi.org/10.3748/wjg.v19.i23.3658
- Ding J, Li D, Gong M, Wang J, Huang X, Wu T, Wang C. Expression and clinical significance of the long non-coding RNA PVT1 in human gastric cancer. Onco Targets Ther 2014; 7:1625-30; PMID:25258543; http://dx.doi.org/10.2147/OTT.S68854
- Han Y, Ye J, Wu D, Wu P, Chen Z, Chen J, Gao S, Huang J. LEIGC long non-coding RNA acts as a tumor suppressor in gastric carcinoma by inhibiting the epithelial-to-mesenchymal transition. BMC Cancer 2014; 14:932; PMID:25496320; http://dx.doi.org/10.1186/1471-2407-14-932
- Bujalka H, Koenning M, Jackson S, Perreau VM, Pope B, Hay CM, Mitew S, Hill AF, Lu QR, Wegner M, et al. MYRF is a membrane-associated transcription factor that autoproteolytically cleaves to directly activate myelin genes. PLoS Biol 2013; 11:e1001625; PMID:23966833; http://dx.doi.org/10.1371/journal.pbio.1001625
- Tsang WP, Ng EK, Ng SS, Jin H, Yu J, Sung JJ, Kwok TT. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis 2010; 31:350-8; PMID:19926638; http://dx.doi.org/10.1093/carcin/bgp181
- Zhuang M, Gao W, Xu J, Wang P, Shu Y. The long non-coding RNA H19-derived miR-675 modulates human gastric cancer cell proliferation by targeting tumor suppressor RUNX1. Biochem Biophys Res Commun 2014; 448:315-22; PMID:24388988; http://dx.doi.org/10.1016/j.bbrc.2013.12.126
- Li H, Yu B, Li J, Su L, Yan M, Zhu Z, Liu B. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget 2014; 5:2318-29; PMID:24810858
- Yang F, Bi J, Xue X, Zheng L, Zhi K, Hua J, Fang G. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J 2012; 279:3159-65; PMID:22776265; http://dx.doi.org/10.1111/j.1742-4658.2012.08694.x
- Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB, Yin DD, Kong R, Xia R, Lu KH, Li JH, et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer 2014; 13:92; PMID:24775712; http://dx.doi.org/10.1186/1476-4598-13-92
- Zhao J, Liu Y, Huang G, Cui P, Zhang W, Zhang Y. Long non-coding RNAs in gastric cancer: versatile mechanisms and potential for clinical translation. Am J Cancer Res 2015; 5:907-27; PMID:26045977
- Liu Y, Zhao J, Zhang W, Gan J, Hu C, Huang G, Zhang Y. lncRNA GAS5 enhances G1 cell cycle arrest via binding to YBX1 to regulate p21 expression in stomach cancer. Sci Rep 2015; 5:10159; PMID:25959498; http://dx.doi.org/10.1038/srep10159
- Zhang EB, Kong R, Yin DD, You LH, Sun M, Han L, Xu TP, Xia R, Yang JS, De W, et al. Long noncoding RNA ANRIL indicates a poor prognosis of gastric cancer and promotes tumor growth by epigenetically silencing of miR-99a/miR-449a. Oncotarget 2014; 5:2276-92; PMID:24810364
