Abstract
Retinoblastoma-1 (RB1), and the RB1-related proteins p107 and p130, are key regulators of the cell cycle. Although RB1 is required for normal erythroid development in vitro, it is largely dispensable for erythropoiesis in vivo. The modest phenotype caused by RB1 deficiency in mice raises questions about redundancy within the RB1 family, and the role of RB1 in erythroid differentiation. Here we show that RB1 is the major pocket protein that regulates terminal erythroid differentiation. Erythroid cells lacking all pocket proteins exhibit the same cell cycle defects as those deficient for RB1 alone. RB1 has broad repressive effects on gene transcription in erythroid cells. As a group, RB1-repressed genes are generally well expressed but downregulated at the final stage of erythroid development. Repression correlates with E2F binding, implicating E2Fs in the recruitment of RB1 to repressed genes. Merging differential and time-dependent changes in expression, we define a group of approximately 800 RB1-repressed genes. Bioinformatics analysis shows that this list is enriched for terms related to the cell cycle, but also for terms related to terminal differentiation. Some of these have not been previously linked to RB1. These results expand the range of processes potentially regulated by RB1, and suggest that a principal role of RB1 in development is coordinating the events required for terminal differentiation.
Introduction
RB1 is the first tumor suppressor gene discovered and founding member of the pocket protein family. RB1 and the related proteins, p107 and p130, are adaptors that interact with other proteins through a conserved protein fold. Of particular significance, pocket proteins bind E2F transcription factors and repress their activity (for a review see refs. 1,2). E2Fs are categorized as activators (E2F1, E2F2, and E2F3a) or repressors (E2F3b, E2F4, E2F5, E2F6, E2F7, and E2F8). E2F4 and E2F5 repress genes by recruiting pocket proteins, whereas E2F6-8 are pocket protein-independent repressors. Activator E2Fs can also function as repressors, by recruiting RB1.
E2Fs operate downstream of a complex regulatory network. Cyclins, cyclin-dependent kinases, and pocket proteins integrate extracellular signals with the cell cycle machinery. Elegant models based on in vitro studies have assigned specific activities to cyclins and Cdks; however, genetic experiments in mice have shown that there is redundancy in the network. Mice deficient for all D-type cyclins survive until midgestation but die of severe anemia and heart defects.Citation3 Single D-type cyclin mice exhibit tissue specific defects; cyclins D2 and D3, but not D1, support erythropoiesis,Citation4 and cyclin D3 regulates the number of terminal erythroid cell divisions.Citation5 Mice with combined deficiencies of cyclins E1 and E2 survive at least until birth.Citation6
D- and E-type cyclins form complexes with Cdk2, Cdk4, and Cdk6. Mice without Cdk4 and Cdk6 die midgestation of severe anemia.Citation7 Embryos lacking Cdk2 and Cdk4 die of heart defects.Citation8 Embryos lacking all 3 Cdks (Cdk2, Cdk4, and Cdk6) exhibit haematopoietic and heart defects but still survive to mid-gestation.Citation9 By contrast, Cdk1 deficient embryos fail to reach the blastocyst stage; further, Cdk1 is required for S phase entry and RB1 phosphorylation in cells lacking other Cdks. Thus, Cdk1 has an integral role in cell cycle progression, whereas the interphase cyclin-Cdk complexes act in specific developmental contexts. Cyclin D-Cdk4/6 complexes are RB1-specific kinases.Citation10 Given the role these complexes have in erythropoiesis, it follows that one or more of the pocket proteins is active in erythroid cells, and must be inhibited at some phase of erythroid development.
The role of RB1 in development has been assessed in mice. Germline RB1 deficiency causes developmental defects,Citation11-13 due to placental insufficiency.Citation14 Erythropoiesis can be restored, although some developmental defects persist.Citation14-20 Erythropoiesis is partially rescued by concurrent E2F2 deficiency, which establishes E2F2 as a target of RB1-mediated repression.Citation21 However, even as a consensus emerges regarding the phenotypic effects of RB1 deficiency in the erythroid lineage, questions remain. Among these is whether there is redundancy between the pocket proteins in erythroid cells, and whether the role of the pocket proteins is limited to cell cycle regulation. In the present study, we address these questions through the use of mouse genetics and expression profiling. Our results suggest that RB1-mediated repression of E2F target genes has a central role in erythroid development: regulating the transition of erythroid cells from precursors to a fully differentiated state.
Materials and Methods
Analysis of mice
Conditional Rb1fl/fl mice,Citation22 p107−/− and p130−/− mice,Citation23,24 and Epor-cre miceCitation25 were bred to obtain embryos with all 8 possible combinations of homozygous mutations. Timed mated pregnant females were anesthetized with inhaled isoflurane and euthanized by cervical dislocation. Embryos were harvested and fetal livers obtained by dissection. Animal studies were performed under protocol in accordance with the guidelines established by the Institutional Animal Care and Use Committees of St. Jude Children's Research Hospital and the New York Blood Center.
Genotyping
Embryos were genotyped by PCR (Qiagen Taq, Q-solution, dNTPs, and 10X coral load buffer), with the following primers and conditions.
Rb1: (94°C × 5 min) 1 cycle; (94°C × 30 sec, 58°C × 30 sec, 72°C × 40 sec) 35 cycles; (72°C × 8 min) 1 cycle.
5′-ggcgtgtgccatcaatg-3′; 5′-ctcaagagctcagactcatgg-3′
Wild type = 235 bp. Unrecombined floxed allele = 283 bp.
p107: (94°C × 5 min) 1 cycle; (94°C × 10 sec, 60°C × 1 min, 68°C × 2 min) 32 cycles; (72°C × 10 min) 1 cycle.
5′-tgtcctgagcatgaacagac-3′; 5′-tcgctggcagtctgagtcag-3′; 5′-acgagactagtgagacgtgc-3′
Wild type = 280 bp. Mutated allele = 330 bp.
p130: (94°C × 5 min) 1 cycle; (94°C × 10 sec, 60°C × 1 min, 68°C × 2 min) 32 cycles; (72°C × 10 min) 1 cycle.
5′-tacatagtttccttcagcgg-3′; 5′-gaagaacgagatcagcagc-3′; 5′-acggatgtcagtgtcacg-3′
Wild type = 230 bp. Mutated allele = 330 bp.
Epor-cre: (94°C × 5 min) 1 cycle; (94°C × 10 sec, 60°C × 1 min, 68°C × 2 min) 32 cycles; (72°C × 10 min) 1 cycle.
5′-gtgtggctgccccttctgcca-3′; 5′-ggcagcctgggcaccttcac-3′; 5′-caggaattcaagctcaacctca-3′
Wild type = 431 bp. Mutated allele = 679 bp.
Embryonic blood smears
Embryos were bled by puncturing the carotid artery with a 2 microliter glass microcapillary tube and the blood expelled to prepare a peripheral blood smear. Blood smears were stained with Wright-Giemsa, and the erythrocyte morphology examined by light microscopy. Blood smears were viewed with a Nikon Eclipse E600 microscope and Nikon Plan Apo 40× objective, NA 0.95. Images were acquired with a Nikon DXM1200 digital camera and ACT-1 software.
Protein assays and antibodies
For Western blots, proteins were separated with NuPAGE Bis-Tris gels (Life Technologies) and transferred to 0.45 μm PVDF membrane. We used the following primary antibodies: RB1 (554136), Ter119 (553670), N-terminal p130 (610262) from BD Transduction Labs; p107 and C-terminal p130 (C-20) from Santa Cruz; β-actin (A5441) from Sigma. Secondary antibodies: Donkey anti-rabbit IgG HRP (NA934V) and Sheep anti-mouse IgG HRP (NA931V) from GE Healthcare Life Sciences.
Cell culture
Fetal liver cells (FLC) were disaggregated in 1 ml of phosphate-buffered saline by passage through a 21 gauge needle. FLC were cultured at a final concentration of 5 × 105 cells/ml in complete medium (30% fetal bovine serum, 1% deionized bovine serum albumin, 0.001% monothioglycerol, 2 mM glutamine, and penicillin-streptomycin in Iscove's modified Dulbecco medium). Cells were maintained in a humidified incubator at 37°C, 5% CO2 and harvested by pipeting at the indicated times. Erythroid colonies were cultured in methylcellulose (M3234 for CFU-E; M3534 for BFU-E) from Stem Cell Technologies. M3534 contains rmSCF, rmIL3, rhIL6, rh-insulin, and human transferrin (iron saturated). M3234 contains no cytokines. We added rhEPO to both to a final concentration of 4 units/ml. CFU-E and BFU-E were scored at 2 days and 7 days, respectively.
Flow cytometry
For cell cycle analysis, we centrifuged 1 × 106 FLC at 400g for 5 min and aspirated the supernatant. The cells were resuspended in 1 ml of PI solution (0.05 mg/ml propidium, 0.1% sodium citrate, 0.1% Triton X-100) and stored at 4°C in the dark. Samples were treated with 4 μg/ml RNase for 30 min at room temperature. Samples were filtered through 40 micron mesh just prior to analysis. Samples were run on a FACSCalibur analyzer (BD Biosciences) and the data processed with ModFit software (Verity Software House).
For Brdu incorporation studies, 10 μM Brdu was added to FLC in culture. For time course experiments, Brdu was added 45 min before each time point. For pulse chase, Brdu was added for 45 min initially, then washed out, and the cells harvested at hourly intervals thereafter. Cells were stained with the FITC Brdu flow kit (BD Transduction Labs) in accordance with the manufacturer's instructions. Samples were run on an LSR analyzer (BD Biosciences) and the data processed with FACSDiva software (BD Biosciences).
For routine flow cytometry, 2 × 106 FLC were blocked with γ-globulin and stained with CD71-PE and Ter119-APC (BD PharMingen). Samples were run on an LSR analyzer (BD Biosciences) and the data processed with FACSDiva software. Sorting was performed on FACSVantage and FACS Aria cell sorters (BD Biosciences).
Microarray procedure
FLC were obtained from E13.5-14.5 embryos. 3-4 × 106 FLC were washed once in phosphate-buffered saline and total RNA extracted with 1 ml of RNA-Bee (Tel-Test) and 0.1 ml of chloroform, in accordance with the manufacturer's instructions. RNA quality was verified with an Agilent 2100 Bioanalyzer. Total RNA (1-10 μg) was processed in the Hartwell Center microarray core facility following the Affymetrix eukaryote one-cycle target labeling protocol (701025, Rev. 6). Biotin-labeled cRNA (15 μg) was hybridized overnight at 45°C to Mouse Genome 430 2.0 GeneChip arrays which correspond to more than 39,000 transcripts. After staining and washing, the arrays were scanned, and expression values were summarized using the MAS5 algorithm as implemented in the GCOS v1.4 software (Affymetrix). Signals were normalized for each array by scaling to a 2% trimmed mean of 500. Detection calls (present, absent, and marginal) were determined using the default parameters.
Microarray data analysis
In the first experiment, we performed expression profiling on 34 fetal liver samples. All Rb1 genotypes were floxed/floxed. There were 12 samples without Epor-cre (RB1 replete) and 22 samples with Epor-cre (RB1 deficient). To maximize the number of samples available for the RB1 comparison, we used all 34 samples, without regard to the genotypes of the other pocket proteins. Because RB1 had a greater effect, for p107 and p130 we excluded RB1-deficient samples, leaving 12 samples for the comparison. The second experiment was a time course (0, 24, and 48 hours). In this experiment, there were 12 RB1-replete samples and 15 RB1-deficient samples. All raw data files (cel) are submitted to the GEO database (GSE67285). These experiments comply with MIAME (minimal information about a microarray experiment) standards.
Butterfly plots were generated on the Gene Set Enrichment Analysis server.Citation26 The data was permuted 1,000 times with randomized column headings to establish 1% and 5% significance thresholds. Hierarchical clustering was performed on the GenePattern server.Citation27 Next, the data were ranked employing the Signal-to-Noise (S2N) metric. The genes were divided into 16 bins (1,000 genes/bin), based on rank order, with the genes most derepressed in the absence of RB1 in bin 1, and those most repressed in bin 16. Finally, the data were resorted according to their original hierarchical clustering position. The S2N metric is the difference in the mean of the 2 groups in a comparison, divided by the sum of the standard deviations. K-means clustering of human expression dataCitation28,29 was performed on the GenePattern server. Microarray data and external data sets were merged with Microsoft Access and Excel, 2010. False discovery rates (FDR) were determined by comparing gene sets corresponding to a group of interest (e.g., E2F1 bound genes) against the rank order of the expression data, based on the K-S statistic (GSEA server). Functional annotation was performed on the DAVID server, employing the default settings.Citation30
Results
p107 and p130 do not compensate for RB1 in erythroid cells
A plausible explanation for the modest effect of RB1 deficiency on erythropoiesis in vivo is that the other pocket proteins supply redundant activity in erythroid cells. Pocket protein redundancy exists during development,Citation23,31-33 and in retinoblastomaCitation34,35 and other cancers.Citation36 To address this, we generated mice lacking pocket proteins in the erythroid lineage. We bred mice with a conditional Rb1 allele with mice that express Cre in an erythroid-restricted pattern.Citation22,25 RB1 accumulates in erythroid cells in its hypophosphorylated state during terminal differentiation (Figure S1A).Citation37 Rb1fl/fl;Epor-cre FLC did not express RB1 (Figure S1B); further, they lacked RB1 from the CD71+Ter119- stage onward (Figure S1C). Rb1-floxed mice were bred with p107 and p130 mice.Citation23,24 FLC from p130−/− embryos expressed a faster migrating protein that reacts with carboxyl-terminal p130 antibody (Figure S1B). The mutant p130 allele used in our studies has a similar effect on fibroblast immortalization as another null allele;Citation38,39 however, we cannot exclude the possibility that this protein has retained some p130 function.
Because combined p107 and p130 deficiency is associated with neonatal lethality,Citation24 we limited our studies to midgestational (E13.5-14.5) embryos. Genetic crosses produced 250 embryos representing all possible combinations of RB, p107, and p130 deficiency (). All combinations, including the triple knockout (TKO), were obtained in the expected proportions. Differences in the number of FLC and erythroid colony-forming units were not significant (data not shown). Thus, regarding erythroid develop-ment, p107 and p130 do not provide redundant activity.
Table 1. Midgestational Rb1 family erythroid TKO embryos are viable
Cell cycle withdrawal is a feature of terminally differentiating cells. Isolated FLC showed an increase in S phase cells in the absence of RB1, which was unchanged in the absence of all 3 pocket proteins (Figure S2A, TKO). FLC in S phase are poised to exit the cell cycle, so to monitor this process we cultured the cells for 48 hours. RB1-replete cells exited the cell cycle efficiently, regardless of the status of the other pocket proteins, whereas RB1-deficient cells arrested in S phase (Figure S2B). The TKO defect was equivalent to that of cells lacking RB1 alone. Similar results were obtained with FLC sorted for maturity (Figure S2C). Thus, p107 and p130 do not contribute to cell cycle withdrawal during terminal erythroid differentiation.
We confirmed the existence of an enucleation defect in the absence of RB1,Citation19 and noted that it affects the definitive lineage based on morphology and is no worse in the absence of RB1 and p107 (Figure S3). Given the increase in cells with S phase DNA content, we examined the proliferation of RB1-deficient FLC in culture. RB1-deficient cells incorporated more Brdu initially than controls (54.4 ± 3.8% versus 48.0 ± 1.6%), which reversed itself by the end of the time course (18.9 ± 3.3% vs. 22.7 ± 2.5%). At the same time, RB1-deficient cells with >2N DNA content accumulated in the Brdu negative gate (Figure S4, arrow). These results suggest that deregulation of the G1-S transition causes an initial increase in Brdu incorporation, which is followed by the activation of S phase checkpoints, and decreased Brdu incorporation at later time points. Consistent with this interpretation, Brdu pulse-chase showed impaired S phase progression in the absence of RB1 (Figure S5).
RB1 deficiency is associated with large-scale changes in gene expression in erythroid cells
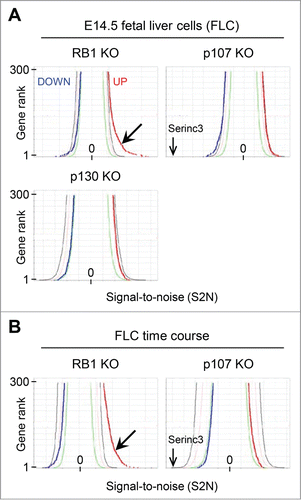
To extend existing concepts of RB1 function, we performed expression profiling of all genotypes. We did not observe major differences in erythroid subpopulations by surface marker expression (data not shown); therefore, we used unfractionated cells. We compared 34 fetal livers, employing the Signal-to-Noise (S2N) metric.Citation26 Significance levels were set by random permutation of columnar data (GSEA server), and the results presented in butterfly plots.Citation26 About 300 genes were upregulated in RB1-deficient fetal livers above a 1% threshold (, barbed arrow). Expression of these genes increased a mean 1.4 ± 0.3 fold in the absence of RB1. By contrast, no genes were downregulated below the 1% threshold. Genes in p107- and p130-deficient fetal livers did not deviate significantly above or below the 1% thresholds. Furthermore, no effect was observed in TKO fetal livers, beyond that attributable to RB1 deficiency. We conclude that RB1 deficiency significantly upregulates a large number of genes, and that p107 and p130 do not have significant genome-scale effects on expression. However, loss of p107 did decrease expression of Serinc3, which was independent of RB1.
Figure 1. Genome-wide effects of RB1 deficiency. (A) Butterfly plots comparing microarray results from pocket protein deficient FLC to controls employing the Signal-to-noise (S2N) metric. The blue line corresponds to downregulated genes (the 300 most negative S2N values), the red line to upregulated genes (the 300 most positive S2N values), in rank order. The green, tan, and black lines represent 50%, 5%, and 1% thresholds, respectively, generated by permutation of randomized columnar data. The barbed arrow points a shoulder of several hundred genes expressed above the 1% threshold in the absence of RB1. A second arrow points to the position of the gene most repressed in the absence of p107, Serinc3. (B) Butterfly plots comparing microarray results from pocket protein deficient FLC to controls, in culture for 0, 24, and 48 hours. All time points were combined in calculating the S2N metric. The labels and scale are the same as in panel A.
RB1 repressed genes are well expressed but downregulated late in development
RB1 activity increases during erythroid differentiation; therefore, to examine the relationship between RB1-mediated repression and differentiation, we performed expression profiling on FLC in culture (0, 24, and 48 hours), with an additional 27 samples. Combining the time points, we confirmed genome-scale changes in expression, except, perhaps reflecting greater RB1 activity at later times, more genes were upregulated in the absence of RB1 (, barbed arrow). Once again, p107 had little effect, except on Serinc3. Thus, our subsequent analysis was limited to RB1.
To characterize RB1 repressed genes we organized the microarray data by 2 independent criteria and merged the results graphically. First, the genes (32,170 probe sets reduced to 16,047 unique genes) were organized by hierarchical clustering, based on the expression pattern of the control samples at 0, 24, and 48 hours (5, 4, and 3 lanes of data, respectively), employing the Broad Institute algorithm (Gene Pattern).Citation27 Six clusters were assigned colors (gray for no pattern) with lighter shades representing regions of transition between clusters. Second, the data were divided into 16 bins (1,000 genes per bin), based on rank order, employing the S2N metric, with the genes most upregulated by loss of RB1 in bin 1 and those most downregulated in bin 16. Third, the genes in each bin were resorted according to their original hierarchical clustering position. Consequently, the genes are clustered by their native expression pattern along the vertical axis and by their response to the loss of RB1 along the horizontal axis ( and Supplemental File S1). This two-dimensional representation reveals several distinctive gene clusters. Of these, the genes in cluster C are most enriched in bin 1 (upregulated following loss of RB1), followed by those in clusters A and D.
Figure 2. Clustering analysis. (Top) Heat map of microarray results comparing RB1-deficient FLC in culture to controls. The results show the expression patterns of genes in control cells cultured for 0 hours (5 lanes), 24 hours (4 lanes), and 48 hours (3 lanes). Z-scores are gene normalized over the time course, using only the control samples. The data is divided into 16 bins. The genes are binned according to their regulation by RB1, employing the S2N metric (combining all time points), with those most derepressed in the absence of RB1 in bin 1 and those most repressed in bin 16. There are 1,000 unique genes per bin, except for bin 16, which has 1,047 genes. See supplemental file S1 for greater detail. (Bottom) Color coding and labeling of the major expression pattern clusters.
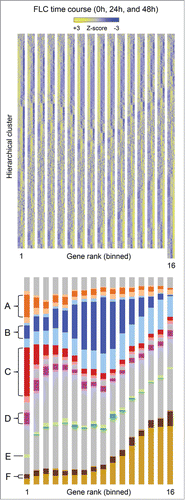
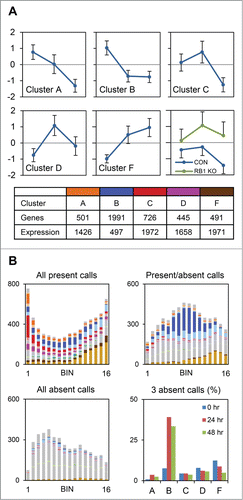
Next, we examined the expression patterns of the clusters in detail. The mean Z-scores of the genes in cluster C, and to a lesser extent clusters A and D, are downregulated between 24 and 48 hours ( and S6). By contrast, the genes in cluster B are downregulated in the initial 24 hours, and those in cluster F are upregulated. Bin 1 genes (clusters A, C, and D) were derepressed in the absence of RB1, although they retained a biphasic expression pattern. The expression ratio of this group of genes (sans gene normalization) at 48 hours compared to 24 hours was 0.61 ± 0.15 in the presence of RB1 and 0.87 ± 0.20 in the absence of RB1; thus, approximately two-thirds of the decrease in expression at 48 hours is due to RB1. Cluster E, a small group enriched in ribosomal genes, was not considered further. The median expression of the genes in clusters A, C, D, and F was similar, whereas that of cluster B was one-third as much. Present and absent calls provide an independent assessment of expression. Most of the genes in bin 1 were flagged present in all 27 microarrays (). Consistent with their lower median expression, the genes in cluster B exhibited a mix of present and absent calls. The latter, however, occurred mostly at 24 and 48 hours, suggesting that the genes in this cluster were undergoing RB1-independent silencing. Based on these analyses, we conclude that RB1 downregulates hundreds of well-expressed genes late in erythroid development.
Figure 3. RB1-repressed genes are well expressed but downregulated late in development. (A) Charts show the mean Z-scores of genes in each of the major clusters in the control samples at 0 hours, 24 hours, and 48 hours. The chart at the lower right shows the Z-scores of the genes in bin 1 (clusters A, C, and D), in the control and RB1 deleted (KO) samples, at the same time points. Beneath the charts, the table shows the number of genes and relative median expression of all the genes in each cluster. (B) Bar graphs show the number of genes in each bin and cluster that satisfy the indicated condition. Present and absent calls are determined by Affymetrix software. The color schemes are consistent throughout. The bar graph at the lower right shows the percentage of genes in each cluster with 3 absent calls, at each time point.
Comparisons with external data sets
To assess the correlation between RB1-mediated repression, transcription factor binding, and histone modifications, we compared our results to several external ChIP-Seq and ChIP-chip data sets (). Alignment of our results with the top 2,000 E2F1 peaks in a breast cancer cell line,Citation40 showed that E2F1-bound genes are significantly enriched in bin 1. For E2F4-bound genes in primary murine tissues,Citation41 or genes bound by DREAM complex components (of which E2F4 is one) in quiescent human glioblastoma cells,Citation42 the degree of enrichment was even greater. KLF1 regulates cell cycle related and lineage marker genes in erythroid cells.Citation43,44 Consistent with this dual role, KLF1-bound genes were modestly enriched in bin 1; however, there was no such enrichment for genes that exhibited stage-specific KLF1 binding (data not shown). GATA1-bound genes,Citation45 by contrast were not significantly enriched.
Figure 4. Repression by RB1 correlates with E2F binding. Comparison to external ChIP-chip, ChIP-Seq, and alternative splicing data sets. The bar graphs show the number of genes in each bin and cluster bound by the protein indicated at the top. H3K27me3 (down) represents genes exhibiting downregulation of this histone mark at the Ter119- to Ter119+ transition. Color schemes are consistent with previous figures.
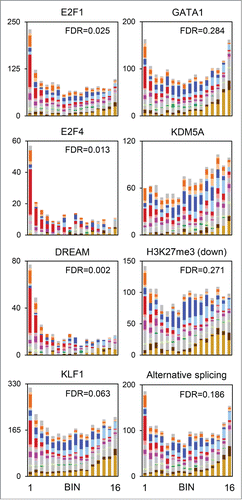
KDM5A is an RB1-interacting protein and histone lysine demethylase,Citation46-48 which inhibits differentiation.Citation49 KDM5A has been linked to the regulation of mitochondrial and cell cycle genes.Citation50 In our data set, genes belonging to these categories co-clustered by temporal expression pattern but differed in their response to RB1. Genes bound by KDM5A at the transcription start siteCitation50 were not enriched in bin 1, suggesting that the response to RB1 may be dictated in part by this factor. A panel of histone marks has been examined for changes during erythroid development at the Ter119- to Ter119+ transition.Citation51 Of these, only genes exhibiting downregulation of H3K27me3 showed evidence of enrichment ( and S7). However, RB1 recruitment may elicit histone modifications after the Ter119 transition, coincident with repression.
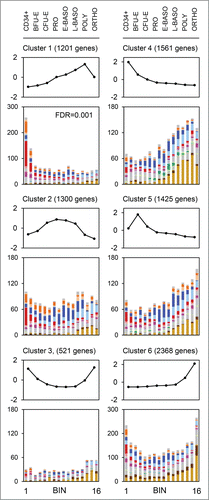
We also compared our results to 2 expression data sets. First, terminal erythroid differentiation is characterized by genome-scale changes in mRNA splicing,Citation52 and we observed modest enrichment for alternatively-spliced genes in bin 1. Second, we compared our results to those from human CD34-derived erythroid cells, cultured and sorted by developmental stage.Citation28,29 We sorted the latter into 6 groups by k-means clustering (GenePattern). The genes in k-means cluster 1 were highly significantly enriched in bin 1 (). The expression of these genes increases progressively until the polychromatophilic erythroblast stage, then decreases. Of the 1201 human genes exhibiting this expression pattern, the orthologs of about one-third are RB1-repressed in mice. Whether RB1 is responsible for this expression pattern in human cells remains to be determined.
Figure 5. Murine orthologs of human genes downregulated in orthochromatophilic erythroblasts are repressed by RB1. The line charts show human RNA sequencing data gene normalized by Z-score and organized by k-means clustering into 6 clusters. Eight sorted human populations are shown: CD34+, BFU-E, CFU-E, proerythroblast (PRO), early basophilic erythroblast (E-BASO), late basophilic erythroblast (L-BASO), polychromatophilic erythroblast (POLY), and orthochromatophilic erythroblast (ORTHO). The bar graphs show the distribution of the murine orthologs of the genes in each cluster by bin. The color scheme is consistent with other figures.
Bioinformatics predictions of RB1 regulated processes
To gain insight into the role of RB1 in development, we assembled a list of RB1 repressed genes. To increase the stringency of our analysis we merged differential and time-dependent changes in gene expression; thus, we took the intersection of the genes in clusters A, C, or D, and bins 1 or 2. This approach yielded 801 RB1-repressed genes (group ACD (1-2)) (Supplemental File S2). Functional annotation clustering was performed on the DAVID bioinformatics server ().Citation30 RB1 repressed genes were enriched for terms involved in the cell cycle, DNA replication, and DNA repair. However, the terms could also be related to processes involved in terminal differentiation, such as chromatin condensation, mRNA splicing, nucleocytoplasmic transport, and modification-dependent protein catabolism. By comparison, the genes in cluster B were enriched for terms related to the extracellular matrix and cell adhesion, and those in cluster F were enriched for terms related to vesicular trafficking and apoptosis.
Table 2. DAVID functional annotation clustering
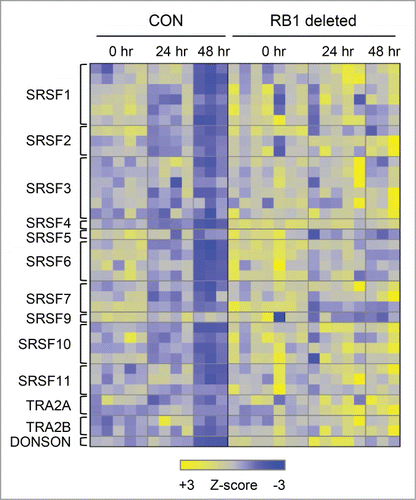
Included in the mRNA processing term are members of the Ser/Arg-rich splicing factor family; collectively, these genes are repressed by RB1 (). These proteins regulate alternative splicing,Citation53 a process which is active during erythroid differentiation,Citation52 and is implicated in hematological disease.Citation54,55 Other genes shared the biphasic expression pattern, but were unaffected or even activated by RB1 (ACD (4-16)). These were enriched for terms related to noncoding- and tRNA processing, translation, mitochondrial ribosomes, and the electron transport chain (). Within this group, NADH:ubiquinone oxidoreductase genes were downregulated in the absence of RB1 (Figure S8).
Figure 6. Ser/Arg-rich splicing factors are repressed by RB1 during terminal erythroid differentiation. Heat map of Ser/Arg-rich splicing factor expression in control and RB1-deficient FLC in culture at 0 hours, 24 hours, and 48 hours. Each row corresponds to a unique probe set. Z-scores are gene normalized over the time course (using control and RB1 deleted samples).
Discussion
In mice, germline mutation of Rb1 is associated with cell death in the nervous system, anemia, and embryonic lethality,Citation11-13 secondary to placental insufficiency.Citation14 RB1 deficiency limited to the embryo proper causes unscheduled DNA replication in the brain, lens defects, and pituitary adenomas, but does not generally affect development.Citation15,16,56, RB1-deficient neonates have defects in skeletal muscle development,Citation57 due at least in part to a mitochondrial defect.Citation58 In the retina, RB1 is required for rod photoreceptor development;Citation59 retinoblastoma arises from cells that develop in the absence of RB1.Citation60 RB1 is also required for Purkinje cellCitation15 and lens fiber development.Citation61 In sum, RB1 deficiency causes specific developmental defects; however, RB1 is not generally required for the implementation of lineage-specific genetic programs.
The limited developmental defects caused by RB1 deficiency may be attributable to the existence of RB1-related proteins. The RB1 family is differentially expressed during development,Citation62 and in several tissues p107 and p130 supply redundant function. These include skin,Citation31 heart,Citation32 muscle,Citation63 and embryonic central nervous system, lens, and blood vessels.Citation33 p107 and p130 can prevent immortalization,Citation38,39 and supply tumor suppressor activity in the retina,Citation34,35 and other tissues.Citation36 In contrast, p107 and p130 are not redundant with RB1 during terminal erythroid differentiation.
Regarding the lack of redundancy, it is instructive to consider how the functional role of the pocket proteins has evolved. Drosophila dE2F2-RBF complexes maintain genes in a repressed state during S phase; dE2F1 reverses this effect at cell cycle- but not developmentally-regulated genes.Citation64,65 This repressive effect is mediated through the dREAM complex (Drosophila, RBF, E2F, Myb-interacting proteins).Citation66 Human DREAM has a similar composition, with the role of RBF filled by p130.Citation42 In contrast to the Drosophila complex, human DREAM is principally associated with cell cycle regulated genes. In mammals, the developmental role of dREAM could be met by RB1, the principal repressor of activator E2Fs, and most rapidly evolving member of the pocket protein family.Citation67 Regarding the unique requirement for RB1 in some tissues but not others, the answer may lie in the divergent activities and tissue-specific expression patterns of the E2F family.
The requirement for RB1 in erythroid cells could be related to increased E2F2 expression in late stage erythroblasts.Citation21 E2F2 expression is preceded by E2F4; paradoxically, since E2F4 is considered a repressor, E2F4-deficient embryos are transiently anemic due to impaired FLC proliferation.Citation68-70 Once E2F2 is expressed, it becomes the principal RB1-associated E2F.Citation21 E2F2 deficiency causes hypoproliferation of erythroid cells and erythropoietic abnormalities.Citation21,71 By contrast, E2F2 deficiency causes hyperproliferation of T cells and autoimmunity.Citation72 Whether E2F2 activates or represses cell cycle regulators depends on the absence or presence of RB1, respectively.Citation73 Consistent with this interpretation, genetic experiments in mice indicate that the erythroid defects caused by RB1 deficiency are diminished by the loss of E2F2.Citation21 This relationship may explain the lack of redundancy with p107 and p130, because they do not interact with E2F2 and therefore are unable to suppress its activity.
Depending on the spectrum of E2F proteins present, the other pocket proteins may play a role earlier in hematopoiesis. Indeed, deficiency of all 3 pocket proteins causes a myeloproliferative disorder.Citation74 It is interesting that all of the pocket proteins are able to establish quiescence, but only RB1 is able to establish senescence, in human fibroblasts.Citation75 Perhaps all the pocket proteins are capable of maintaining haematopoietic progenitor quiescence, whereas RB1 is uniquely required during terminal erythroid differentiation, which like senescence is characterized by changes in chromatin structure, and is normally an irreversible transition.
If RB1 supplies the required pocket protein activity during terminal erythroid differentiation, as our studies suggest, then why is RB1 deficiency associated with a mild erythroid defect in mice? One possibility is that differentiation does not require repression of E2F activity; however, E2F targets are still partially suppressed in RB1-deficient cells, suggesting that there is compensation. One mechanism of RB1-independent E2F repression is competition with repressor E2Fs for binding to E2F sites. E2F8 is an RB1-independent repressor; mice with combined deficiency of RB1 and E2F8 have hemolytic anemia,Citation76 which is caused at least in part by unrestrained E2F2 activity.Citation77 In Drosophila, S phase entry triggers ubiquitin-dependent degradation of dE2F1.Citation78 In mammals, phosphorylation of E2F-bound DP by cyclin A-Cdk2 prevents E2F-DP binding to DNA.Citation79 Thus, redundant mechanisms exist that mitigate the effect of RB1 deficiency on E2F target gene expression.
Loss of p107 and p130 did not have a broad effect on gene transcription. However, p107 deficiency did affect expression of the gene Serinc3. Serinc3 is a member of a family of proteins that incorporate serine into membranes for phosphatidylserine and sphingolipid synthesis.Citation80 Notably, RB1 had no effect on Serinc3 expression. Although p107 selectively represses certain genes,Citation81 this is an example of p107-specific gene activation, which has not been previously described. Interestingly, the Serinc3 locus (Tde1) is a target of insertional mutagenesis in Arf-null mice.Citation82 Whether there is crosstalk between p107 and the p53 pathway, as this observation implies, is unknown.
By contrast, RB1 deficiency has a major effect on erythroid gene transcription, causing derepression of hundreds of genes. At the single gene level, the magnitude of the effect is modest but statistically significant. Compounded over hundreds of genes, there are likely to be cellular consequences; this point is illustrated by the cell cycle defects present in RB1-deficient erythroid cells. From a network perspective, it is logical to think of RB1 as a horizontally-applied brake on E2F-driven biological processes; along this line, an accurate compilation of RB1-repressed genes should improve our understanding of the genes and processes involved in terminal differentiation.
RB1 recruits chromatin modifying enzymes to genes through its interaction with E2Fs.Citation83 In the absence of RB1, E2Fs are effectively converted from repressors to activators, derepressing E2F targets. In this regard, E2Fs and RB1 may act as a switch during development, first activating then repressing E2F targets.Citation73 During terminal erythroid differentiation, Cdk4/6-associated RB1 kinase activity decreases, and hypophosphorylated RB1 increases,Citation37 suggesting the switch is linked to a change in the phosphorylation state of RB1. Comparison of our results with those from human erythroid cells Citation28,29 further suggests that an RB1-regulated switch may be a conserved aspect of development.
Repression by RB1 correlates with the presence of bound E2F1, E2F4, and DREAM complex components. In Drosophila, dE2F-dDP complexes are required for RBF recruitment.Citation84 In human senescent fibroblasts, RB1 recruitment correlates with the presence of E2F binding sites.Citation75 In murine FLC, DP2 knockdown interferes with downregulation of E2F2 target genes.Citation85 Together, these findings support the idea that E2Fs mediate RB1 recruitment and gene repression in erythroid cells.
KLF1 appears to have a dedicated role regulating the expression of RB1-repressed genes, which is not limited to direct targets, since E2F2 itself is regulated by KLF1.Citation86,87 GATA1-bound genes were not significantly enriched for RB1-repressed genes, but GATA1 interacts with RB1 and positively regulates its activity;Citation88 thus, depending on the relative abundance of E2Fs and RB1, KLF1 and GATA1 could have reinforcing or opposing effects on the expression of E2F target genes.
By merging differential and temporal gene expression patterns, we identified 801 RB1-repressed genes in erythroid cells. Approximately one-third of the genes we identified are bound by E2Fs in other cell types.Citation40-42 Cell cycle related genes and confirmed E2F targets are enriched in the list;Citation89 however, so are genes and processes not known to be regulated by E2F and RB1. Because they share differential and temporal gene expression patterns, we anticipate that many of the novel genes will also prove to be direct targets of RB1-mediated repression in erythroid cells, but this will require validation. In the interim, we expect that this gene list will be a useful resource for studies of the mechanisms of terminal erythroid differentiation.
The large number of RB1-repressed genes is consistent with a developmental role for RB1, possibly one that involves coordinating terminal differentiation with cell cycle withdrawal. A recent survey found that 2,250 genes are alternative spliced during terminal erythroid differentiation.Citation52 Splicing events occurred in late stage erythroblasts, concurrent with extensive cellular remodeling. RB1 repressed Ser/Arg-rich splicing factors, which are key regulators of alternative splicing.Citation53 Ser/Arg-rich splicing factors are themselves inactivated by a separate mechanism, alternative splicing-nonsense mediated decay, which further argues that their downregulation is important for terminal differentiation.Citation52
RB1 repressed genes involved in transcription and chromatin structure, such as GFI1B, EZH2, and SUZ12. The PRC2 subunits EZH1 and EZH2 undergo an expression switch during terminal erythroid differentiation, which is regulated by GATA2 and GATA1.Citation90 Repression by RB1 adds another dimension to their regulation. Ubiquitin-dependent catabolism is another process represented on the list; KCTD6 is one of several Cullin 3 adaptors identified and is implicated in ankyrin protein turnover and hereditary spherocytosis.Citation91
Genes involved in mitochondrial biogenesis and respiration are downregulated in the absence of RB1.Citation20 We showed this effect persists throughout terminal differentiation, and extended it to mitochondrial ribosomes, noncoding RNA metabolism, and translation. Regarding the role of RB1 in oxidative phosphorylation, the finding that mitochondrial biogenesis is diminished in RB1-deficient muscle cells is concordant with our results.Citation58 Conversely, findings that oxidative phosphorylation is repressed by RB1,Citation92 and degradation-defective cyclin E increases mitochondrial biogenesis are not.Citation93 Potentially, RB1 activates these genes by reversing KDM5A-mediated repression.Citation50 Additional experiments are needed, since the metabolic derangements associated with RB1 deficiency are related to those present in cancer cells.Citation94
RB1 regulates the expression of genes involved in the cell cycle, chromatin organization, alternative splicing, transcription, and ubiquitination. These processes set the stage for the final phase of erythroid development, which includes cellular remodeling, enucleation, and the expression of erythroid-specific protein isotypes. We anticipate that processes like these will be regulated by pocket proteins in a wide range of tissue types, but that the specific genes may differ. For some combinations of RB1 targets and tissues, down-regulation may be essential for terminal differentiation to proceed to completion. Identification of these genes will yield insights into development, and possibly into the causes of cancer associated with mutations in the RB1 pathway.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
1090067_Supplemental_Material.zip
Download Zip (853.2 KB)Acknowledgments
The authors acknowledge Drs. Anton Berns, Tyler Jacks, Ursula Klingmüller, and Michael Dyer for the conditional Rb1, p107 and p130 null, and Epor-cre mouse strains. The authors thank Drs. Mohandas Narla, Xiuli An, and David Bodine for sharing data. We thank the Laboratory Animal Research Service of the New York Blood Center. We also thank the Animal Resource Center, and the Flow Cytometry and Cell Sorting Shared Resource of St. Jude Children's Research Hospital.
Funding
This work was supported by grants from the American Society of Hematology and the F.M. Kirby Foundation (PAN), the New York Blood Center, and the American, Lebanese, and Syrian Associated Charities (ALSAC).
References
- Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev 1998; 12:2245-62; PMID:9694791; http://dx.doi.org/10.1101/gad.12.15.2245
- Dimova DK, Dyson NJ. The E2F transcriptional network: old acquaintances with new faces. Oncogene 2005; 24:2810-26; PMID:15838517; http://dx.doi.org/10.1038/sj.onc.1208612
- Kozar K, Ciemerych MA, Rebel VI, Shigematsu H, Zagozdzon A, Sicinska E, Geng Y, Yu Q, Bhattacharya S, Bronson RT, et al. Mouse development and cell proliferation in the absence of D-cyclins. Cell 2004; 118:477-91; PMID:15315760; http://dx.doi.org/10.1016/j.cell.2004.07.025
- Ciemerych MA, Kenney AM, Sicinska E, Kalaszczynska I, Bronson RT, Rowitch DH, Gardner H, Sicinski P. Development of mice expressing a single D-type cyclin. Genes Dev 2002; 16:3277-89; PMID:12502747; http://dx.doi.org/10.1101/gad.1023602
- Sankaran VG, Ludwig LS, Sicinska E, Xu J, Bauer DE, Eng JC, Patterson HC, Metcalf RA, Natkunam Y, Orkin SH, et al. Cyclin D3 coordinates the cell cycle during differentiation to regulate erythrocyte size and number. Genes Dev 2012; 26:2075-87; PMID:22929040; http://dx.doi.org/10.1101/gad.197020.112
- Geng Y, Yu Q, Sicinska E, Das M, Schneider JE, Bhattacharya S, Rideout WM, Bronson RT, Gardner H, Sicinski P. Cyclin E ablation in the mouse. Cell 2003; 114:431-43; PMID:12941272; http://dx.doi.org/10.1016/S0092-8674(03)00645-7
- Malumbres M, Sotillo R, Santamaria D, Galán J, Cerezo A, Ortega S, Dubus P, Barbacid M. Mammalian cells cycle without the D-type cyclin-dependent kinases Cdk4 and Cdk6. Cell 2004; 118:493-504; PMID:15315761; http://dx.doi.org/10.1016/j.cell.2004.08.002
- Berthet C, Klarmann KD, Hilton MB, Suh HC, Keller JR, Kiyokawa H, Kaldis P. Combined loss of Cdk2 and Cdk4 results in embryonic lethality and Rb hypophosphorylation. Dev Cell 2006; 10:563-73
- Santamaria D, Barriere C, Cerqueira A, Hunt S, Tardy C, Newton K, Cáceres JF, Dubus P, Malumbres M, Barbacid M. Cdk1 is sufficient to drive the mammalian cell cycle. Nature 2007; 448:811-815; PMID:17700700; http://dx.doi.org/10.1038/nature06046
- Matsushime H, Ewen ME, Strom DK, Kato JY, Hanks SK, Roussel MF, Sherr CJ. Identification and properties of an atypical catalytic subunit (p34PSK-J3/cdk4) for mammalian D type G1 cyclins. Cell 1992; 71:323-34; PMID:1423597; http://dx.doi.org/10.1016/0092-8674(92)90360-O
- Lee EY, Chang CY, Hu N, Wang YC, Lai CC, Herrup K, Lee WH, Bradley A. Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature 1992; 359:288-94; PMID:1406932; http://dx.doi.org/10.1038/359288a0
- Jacks T, Fazeli A, Schmitt EM, Bronson RT, Goodell MA, Weinberg RA. Effects of an Rb mutation in the mouse. Nature 1992; 359:295-300; PMID:1406933; http://dx.doi.org/10.1038/359295a0
- Clarke AR, Maandag ER, van RM, van der Lugt NM, van der Valk M, Hooper ML, Berns A, te Riele H. Requirement for a functional Rb-1 gene in murine development. Nature 1992; 359:328-30; PMID:1406937; http://dx.doi.org/10.1038/359328a0
- Wu L, de BA, Saavedra HI, Starovic M, Trimboli A, Yang Y, Opavska J, Wilson P, Thompson JC, Ostrowski MC, et al. Extra-embryonic function of Rb is essential for embryonic development and viability. Nature 2003; 421:942-7; PMID:12607001; http://dx.doi.org/10.1038/nature01417
- Williams BO, Schmitt EM, Remington L, Bronson RT, Albert DM, Weinberg RA, Jacks T. Extensive contribution of Rb-deficient cells to adult chimeric mice with limited histopathological consequences. EMBO J 1994; 13:4251-9; PMID:7925270
- Maandag EC, van d V, Vlaar M, Feltkamp C, O'Brien J, van Roon M, van der Lugt N, Berns A, te Riele H. Developmental rescue of an embryonic-lethal mutation in the retinoblastoma gene in chimeric mice. EMBO J 1994; 13:4260-8; PMID:7925271
- Hu N, Gulley ML, Kung JT, Lee EY. Retinoblastoma gene deficiency has mitogenic but not tumorigenic effects on erythropoiesis. Cancer Res 1997; 57:4123-9; PMID:9307303
- Clark AJ, Doyle KM, Humbert PO. Cell-intrinsic requirement for pRb in erythropoiesis. Blood 2004; 104:1324-6; PMID:15155463; http://dx.doi.org/10.1182/blood-2004-02-0618
- Spike BT, Dirlam A, Dibling BC, Marvin J, Williams BO, Jacks T, Macleod KF. The Rb tumor suppressor is required for stress erythropoiesis. EMBO J 2004; 23:4319-29; PMID:15457215; http://dx.doi.org/10.1038/sj.emboj.7600432
- Sankaran VG, Orkin SH, Walkley CR. Rb intrinsically promotes erythropoiesis by coupling cell cycle exit with mitochondrial biogenesis. Genes Dev 2008; 22:463-75; PMID:18258751; http://dx.doi.org/10.1101/gad.1627208
- Dirlam A, Spike BT, Macleod KF. Deregulated E2f-2 underlies cell cycle and maturation defects in retinoblastoma null erythroblasts. Mol Cell Biol. 2007; 27:8713-28; PMID:17923680; http://dx.doi.org/10.1128/MCB.01118-07
- Marino S, Vooijs M, van Der GH, Jonkers J, Berns A. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev 2000; 14:994-1004; PMID:10783170
- Lee MH, Williams BO, Mulligan G, Mukai S, Bronson RT, Dyson N, Harlow E, Jacks T. Targeted disruption of p107: functional overlap between p107 and Rb. Genes Dev 1996; 10:1621-32; PMID:8682293; http://dx.doi.org/10.1101/gad.10.13.1621
- Cobrinik D, Lee MH, Hannon G, Mulligan G, Bronson RT, Dyson N, Harlow E, Beach D, Weinberg RA, Jacks T. Shared role of the pRB-related p130 and p107 proteins in limb development. Genes Dev 1996; 10:1633-44; PMID:8682294; http://dx.doi.org/10.1101/gad.10.13.1633
- Heinrich AC, Pelanda R, Klingmuller U. A mouse model for visualization and conditional mutations in the erythroid lineage. Blood 2004; 104:659-66; PMID:15090451; http://dx.doi.org/10.1182/blood-2003-05-1442
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102:15545-50; PMID:16199517; http://dx.doi.org/10.1073/pnas.0506580102
- Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat.Genet 2006; 38:500-1
- An X, Schulz VP, Li J, Wu K, Liu J, Xue F, Hu J, Mohandas N, Gallagher PG. Global transcriptome analyses of human and murine terminal erythroid differentiation. Blood 2014; 123:3466-77; PMID:24637361; http://dx.doi.org/10.1182/blood-2014-01-548305
- Li J, Hale J, Bhagia P, Xue F, Chen L, Jaffray J, Yan H, Lane J, Gallagher PG, Mohandas N, et al. Isolation and transcriptome analyses of human erythroid progenitors: BFU-E and CFU-E. Blood 2014; 124:3636-45; PMID:25339359; http://dx.doi.org/10.1182/blood-2014-07-588806
- Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009; 37:1-13; PMID:19033363; http://dx.doi.org/10.1093/nar/gkn923
- Ruiz S, Santos M, Segrelles C, Leis H, Jorcano JL, Berns A, Paramio JM, Vooijs M. Unique and overlapping functions of pRb and p107 in the control of proliferation and differentiation in epidermis. Development 2004; 131:2737-48; PMID:15148303; http://dx.doi.org/10.1242/dev.01148
- MacLellan WR, Garcia A, Oh H, Frenkel P, Jordan MC, Roos KP, Schneider MD. Overlapping roles of pocket proteins in the myocardium are unmasked by germ line deletion of p130 plus heart-specific deletion of Rb. Mol.Cell Biol 2005; 25:2486-97
- Berman SD, West JC, Danielian PS, Caron AM, Stone JR, Lees JA. Mutation of p107 exacerbates the consequences of Rb loss in embryonic tissues and causes cardiac and blood vessel defects. Proc Natl Acad Sci USA 2009; 106:14932-6; PMID:19706423; http://dx.doi.org/10.1073/pnas.0902408106
- Robanus-Maandag E, Dekker M, van der Valk M, Carrozza ML, Jeanny JC, Dannenberg JH, Berns A, te Riele H. p107 is a suppressor of retinoblastoma development in pRb-deficient mice. Genes Dev 1998; 12:1599-609; PMID:9620848; http://dx.doi.org/10.1101/gad.12.11.1599
- Ajioka I, Martins RA, Bayazitov IT, Donovan S, Johnson DA, Frase S, Cicero SA, Boyd K, Zakharenko SS, Dyer MA. Differentiated horizontal interneurons clonally expand to form metastatic retinoblastoma in mice. Cell 2007; 131:378-90; PMID:17956737; http://dx.doi.org/10.1016/j.cell.2007.09.036
- Dannenberg JH, Schuijff L, Dekker M, van d V, te RH. Tissue-specific tumor suppressor activity of retinoblastoma gene homologs p107 and p130. Genes Dev 2004; 18:2952-62; PMID:15574596; http://dx.doi.org/10.1101/gad.322004
- Hsieh FF, Barnett LA, Green WF, Freedman K, Matushansky I, Skoultchi AI, Kelley LL. Cell cycle exit during terminal erythroid differentiation is associated with accumulation of p27(Kip1) and inactivation of cdk2 kinase. Blood 2000; 96:2746-54; PMID:11023508
- Sage J, Mulligan GJ, Attardi LD, Miller A, Chen S, Williams B, Theodorou E, Jacks T. Targeted disruption of the three Rb-related genes leads to loss of G(1) control and immortalization. Genes Dev 2000; 14:3037-50; PMID:11114892; http://dx.doi.org/10.1101/gad.843200
- Dannenberg JH, van RA, Schuijff L, te RH. Ablation of the retinoblastoma gene family deregulates G(1) control causing immortalization and increased cell turnover under growth-restricting conditions. Genes Dev 2000; 14:3051-64; PMID:11114893; http://dx.doi.org/10.1101/gad.847700
- Rabinovich A, Jin VX, Rabinovich R, Xu X, Farnham PJ. E2F in vivo binding specificity: comparison of consensus versus nonconsensus binding sites. Genome Res 2008; 18:1763-77; PMID:18836037; http://dx.doi.org/10.1101/gr.080622.108
- Conboy CM, Spyrou C, Thorne NP, Wade EJ, Barbosa-Morais NL, Wilson MD, Bhattacharjee A, Young RA, Tavaré S, Lees JA, et al. Cell cycle genes are the evolutionarily conserved targets of the E2F4 transcription factor. PLoS One 2007; 2:e1061; PMID:17957245; http://dx.doi.org/10.1371/journal.pone.0001061
- Litovchick L, Sadasivam S, Florens L, Zhu X, Swanson SK, Velmurugan S, Chen R, Washburn MP, Liu XS, DeCaprio JA. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol Cell 2007; 26:539-51; PMID:17531812; http://dx.doi.org/10.1016/j.molcel.2007.04.015
- Pilon AM, Ajay SS, Kumar SA, Steiner LA, Cherukuri PF, Wincovitch S, Anderson SM; NISC Comparative Sequencing Center, Mullikin JC, Gallagher PG, Hardison RC, et al. Genome-wide ChIP-Seq reveals a dramatic shift in the binding of the transcription factor erythroid Kruppel-like factor during erythrocyte differentiation. Blood 2011; 118:e139-148; PMID:21900194; http://dx.doi.org/10.1182/blood-2011-05-355107
- Tallack MR, Whitington T, Yuen WS, Wainwright EN, Keys JR, Gardiner BB, Nourbakhsh E, Cloonan N, Grimmond SM, Bailey TL, et al. A global role for KLF1 in erythropoiesis revealed by ChIP-seq in primary erythroid cells. Genome Res 2010; 20:1052-63; PMID:20508144; http://dx.doi.org/10.1101/gr.106575.110
- Papadopoulos GL, Karkoulia E, Tsamardinos I, Porcher C, Ragoussis J, Bungert J, Strouboulis J. GATA-1 genome-wide occupancy associates with distinct epigenetic profiles in mouse fetal liver erythropoiesis. Nucleic Acids Res 2013; 41:4938-48; PMID:23519611; http://dx.doi.org/10.1093/nar/gkt167
- Defeo-Jones D, Huang PS, Jones RE, Haskell KM, Vuocolo GA, Hanobik MG, Huber HE, Oliff A. Cloning of cDNAs for cellular proteins that bind to the retinoblastoma gene product. Nature 1991; 352:251-4; PMID:1857421; http://dx.doi.org/10.1038/352251a0
- Iwase S, Lan F, Bayliss P, de la Torre-Ubieta L, Huarte M, Qi HH, Whetstine JR, Bonni A, Roberts TM, Shi Y. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell 2007; 128:1077-88; PMID:17320160; http://dx.doi.org/10.1016/j.cell.2007.02.017
- Christensen J, Agger K, Cloos PA, Pasini D, Rose S, Sennels L, Rappsilber J, Hansen KH, Salcini AE, Helin K. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell 2007; 128:1063-76; PMID:17320161; http://dx.doi.org/10.1016/j.cell.2007.02.003
- Benevolenskaya EV, Murray HL, Branton P, Young RA, Kaelin WG, Jr. Binding of pRB to the PHD protein RBP2 promotes cellular differentiation. Mol Cell 2005; 18:623-35; PMID:15949438; http://dx.doi.org/10.1016/j.molcel.2005.05.012
- Beshiri ML, Holmes KB, Richter WF, Hess S, Islam AB, Yan Q, Plante L, Litovchick L, Gévry N, Lopez-Bigas N, et al. Coordinated repression of cell cycle genes by KDM5A and E2F4 during differentiation. Proc Natl Acad Sci USA 2012; 109:18499-504; PMID:23093672; http://dx.doi.org/10.1073/pnas.1216724109
- Wong P, Hattangadi SM, Cheng AW, Frampton GM, Young RA, Lodish HF. Gene induction and repression during terminal erythropoiesis are mediated by distinct epigenetic changes. Blood 2011; 118:e128-138; PMID:21860024; http://dx.doi.org/10.1182/blood-2011-03-341404
- Pimentel H, Parra M, Gee S, Ghanem D, An X, Li J, Mohandas N, Pachter L, Conboy JG. A dynamic alternative splicing program regulates gene expression during terminal erythropoiesis. Nucleic Acids Res. 2014; 42:4031-4042; PMID:24442673; http://dx.doi.org/10.1093/nar/gkt1388
- Graveley BR, Maniatis T. Arginine/serine-rich domains of SR proteins can function as activators of pre-mRNA splicing. Mol Cell 1998; 1:765-71; PMID:9660960; http://dx.doi.org/10.1016/S1097-2765(00)80076-3
- Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, Sato Y, Sato-Otsubo A, Kon A, Nagasaki M, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 2011; 478:64-9; PMID:21909114; http://dx.doi.org/10.1038/nature10496
- Zhang SJ, Rampal R, Manshouri T, Patel J, Mensah N, Kayserian A, Hricik T, Heguy A, Hedvat C, Gönen M, et al. Genetic analysis of patients with leukemic transformation of myeloproliferative neoplasms shows recurrent SRSF2 mutations that are associated with adverse outcome. Blood 2012; 119:4480-5; PMID:22431577; http://dx.doi.org/10.1182/blood-2011-11-390252
- Lipinski MM, Macleod KF, Williams BO, Mullaney TL, Crowley D, Jacks T. Cell-autonomous and non-cell-autonomous functions of the Rb tumor suppressor in developing central nervous system. EMBO J 2001; 20:3402-13; PMID:11432828; http://dx.doi.org/10.1093/emboj/20.13.3402
- Zacksenhaus E, Jiang Z, Chung D, Marth JD, Phillips RA, Gallie BL. pRb controls proliferation, differentiation, and death of skeletal muscle cells and other lineages during embryogenesis. Genes Dev 1996; 10:3051-64; PMID:8957005; http://dx.doi.org/10.1101/gad.10.23.3051
- Ciavarra G, Zacksenhaus E. Rescue of myogenic defects in Rb-deficient cells by inhibition of autophagy or by hypoxia-induced glycolytic shift. J Cell Biol 2010; 191:291-301; http://dx.doi.org/10.1083/jcb.201005067
- Zhang J, Gray J, Wu L, Leone G, Rowan S, Cepko CL, Zhu X, Craft CM, Dyer MA. Rb regulates proliferation and rod photoreceptor development in the mouse retina. Nat.Genet 2004; 36:351-60
- Ajioka I, Dyer MA. A new model of tumor susceptibility following tumor suppressor gene inactivation. Cell Cycle 2008; 7:735-40; PMID:18239449; http://dx.doi.org/10.4161/cc.7.6.5612
- Liu Y, Zacksenhaus E. E2F1 mediates ectopic proliferation and stage-specific p53-dependent apoptosis but not aberrant differentiation in the ocular lens of Rb deficient fetuses. Oncogene 2000; 19:6065-73; PMID:11146559; http://dx.doi.org/10.1038/sj.onc.1203996
- Jiang Z, Zacksenhaus E, Gallie BL, Phillips RA. The retinoblastoma gene family is differentially expressed during embryogenesis. Oncogene 1997; 14:1789-97; PMID:9150384; http://dx.doi.org/10.1038/sj.onc.1201014
- Ciavarra G, Ho AT, Cobrinik D, Zacksenhaus E. Critical role of the Rb family in myoblast survival and fusion. PLoS One 2011; 6:e17682; PMID:21423694; http://dx.doi.org/10.1371/journal.pone.0017682
- Dimova DK, Stevaux O, Frolov MV, Dyson NJ. Cell cycle-dependent and cell cycle-independent control of transcription by the Drosophila E2F/RB pathway. Genes Dev 2003; 17:2308-20; PMID:12975318; http://dx.doi.org/10.1101/gad.1116703
- Frolov MV, Stevaux O, Moon NS, Dimova D, Kwon EJ, Morris EJ, Dyson NJ. G1 cyclin-dependent kinases are insufficient to reverse dE2F2-mediated repression. Genes Dev 2003; 17:723-8; PMID:12651890; http://dx.doi.org/10.1101/gad.1031803
- Korenjak M, Taylor-Harding B, Binne UK, Satterlee JS, Stevaux O, Aasland R, White-Cooper H, Dyson N, Brehm A. Native E2F/RBF complexes contain Myb-interacting proteins and repress transcription of developmentally controlled E2F target genes. Cell 2004; 119:181-93; PMID:15479636; http://dx.doi.org/10.1016/j.cell.2004.09.034
- Cao L, Peng B, Yao L, Zhang X, Sun K, Yang X, Yu L. The ancient function of RB-E2F pathway: insights from its evolutionary history. Biol Direct 2010; 5:55; PMID:20849664; http://dx.doi.org/10.1186/1745-6150-5-55
- Humbert PO, Rogers C, Ganiatsas S, Landsberg RL, Trimarchi JM, Dandapani S, Brugnara C, Erdman S, Schrenzel M, Bronson RT, et al. E2F4 is essential for normal erythrocyte maturation and neonatal viability. Mol Cell 2000; 6:281-91; PMID:10983976; http://dx.doi.org/10.1016/S1097-2765(00)00029-0
- Rempel RE, Saenz-Robles MT, Storms R, Morham S, Ishida S, Engel A, Jakoi L, Melhem MF, Pipas JM, Smith C, et al. Loss of E2F4 activity leads to abnormal development of multiple cellular lineages. Mol Cell 2000; 6:293-306; PMID:10983977; http://dx.doi.org/10.1016/S1097-2765(00)00030-7
- Kinross KM, Clark AJ, Iazzolino RM, Humbert PO. E2f4 regulates fetal erythropoiesis through the promotion of cellular proliferation. Blood 2006; 108:886-95; PMID:16861343; http://dx.doi.org/10.1182/blood-2005-09-008656
- Li FX, Zhu JW, Hogan CJ, DeGregori J. Defective gene expression, S phase progression, and maturation during hematopoiesis in E2F1/E2F2 mutant mice. Mol Cell Biol 2003; 23:3607-22
- Murga M, Fernandez-Capetillo O, Field SJ, Moreno B, Borlado LR, Fujiwara Y, Balomenos D, Vicario A, Carrera AC, Orkin SH, et al. Mutation of E2F2 in mice causes enhanced T lymphocyte proliferation, leading to the development of autoimmunity. Immunity 2001; 15:959-70; PMID:11754817; http://dx.doi.org/10.1016/S1074-7613(01)00254-0
- Chong JL, Wenzel PL, Saenz-Robles MT, Nair V, Ferrey A, Hagan JP, Gomez YM, Sharma N, Chen HZ, Ouseph M, et al. E2f1-3 switch from activators in progenitor cells to repressors in differentiating cells. Nature 2009; 462:930-4; PMID:20016602; http://dx.doi.org/10.1038/nature08677
- Viatour P, Somervaille TC, Venkatasubrahmanyam S, Kogan S, McLaughlin ME, Weissman IL, Butte AJ, Passegué E, Sage J. Hematopoietic stem cell quiescence is maintained by compound contributions of the retinoblastoma gene family. Cell Stem Cell 2008; 3:416-28; PMID:18940733; http://dx.doi.org/10.1016/j.stem.2008.07.009
- Chicas A, Wang X, Zhang C, McCurrach M, Zhao Z, Mert O, Dickins RA, Narita M, Zhang M, Lowe SW. Dissecting the unique role of the retinoblastoma tumor suppressor during cellular senescence. Cancer Cell 2010; 17:376-87; PMID:20385362; http://dx.doi.org/10.1016/j.ccr.2010.01.023
- Hu T, Ghazaryan S, Sy C, Wiedmeyer C, Chang V, Wu L. Concomitant inactivation of Rb and E2f8 in hematopoietic stem cells synergizes to induce severe anemia. Blood 2012; 119:4532-42; PMID:22422820; http://dx.doi.org/10.1182/blood-2011-10-388231
- Ghazaryan S, Sy C, Hu T, An X, Mohandas N, Fu H, Aladjem MI, Chang VT, Opavsky R, Wu L. Inactivation of Rb and E2f8 synergizes to trigger stressed DNA replication during erythroid terminal differentiation. Mol.Cell Biol 2014; 34:2833-47
- Shibutani ST, de la Cruz AF, Tran V, Turbyfill WJ 3rd, Reis T, Edgar BA, Duronio RJ. Intrinsic negative cell cycle regulation provided by PIP box- and Cul4Cdt2-mediated destruction of E2f1 during S phase. Dev Cell 2008; 15:890-900; PMID:19081076; http://dx.doi.org/10.1016/j.devcel.2008.10.003
- Xu M, Sheppard KA, Peng CY, Yee AS, Piwnica-Worms H. Cyclin A/CDK2 binds directly to E2F-1 and inhibits the DNA-binding activity of E2F-1/DP-1 by phosphorylation. Mol Cell Biol 1994; 14:8420-31; PMID:7969176
- Inuzuka M, Hayakawa M, Ingi T. Serinc, an activity-regulated protein family, incorporates serine into membrane lipid synthesis. J.Biol Chem 2005; 280:35776-83; http://dx.doi.org/10.1074/jbc.M505712200
- Hurford RK, Jr., Cobrinik D, Lee MH, Dyson N. pRB and p107/p130 are required for the regulated expression of different sets of E2F responsive genes. Genes Dev 1997; 11:1447-63; PMID:9192872; http://dx.doi.org/10.1101/gad.11.11.1447
- Shou Y, Ma Z, Lu T, Sorrentino BP. Unique risk factors for insertional mutagenesis in a mouse model of XSCID gene therapy. Proc Natl Acad Sci USA 2006; 103:11730-5; PMID:16864781; http://dx.doi.org/10.1073/pnas.0603635103
- Frolov MV, Dyson NJ. Molecular mechanisms of E2F-dependent activation and pRB-mediated repression. J.Cell Sci 2004; 117:2173-81; http://dx.doi.org/10.1242/jcs.01227
- Korenjak M, Anderssen E, Ramaswamy S, Whetstine JR, Dyson NJ. RBF binding to both canonical E2F targets and noncanonical targets depends on functional dE2F/dDP complexes. Mol Cell Biol 2012; 32:4375-87
- Chen C, Lodish HF. Global analysis of induced transcription factors and cofactors identifies Tfdp2 as an essential coregulator during terminal erythropoiesis. Exp Hematol 2014; 42:464-76; PMID:24607859; http://dx.doi.org/10.1016/j.exphem.2014.03.001
- Pilon AM, Arcasoy MO, Dressman HK, Vayda SE, Maksimova YD, Sangerman JI, Gallagher PG, Bodine DM. Failure of terminal erythroid differentiation in EKLF-deficient mice is associated with cell cycle perturbation and reduced expression of E2F2. Mol.Cell Biol 2008; 28:7394-401
- Tallack MR, Keys JR, Humbert PO, Perkins AC. EKLF/KLF1 controls cell cycle entry via direct regulation of E2f2. J Biol Chem 2009; 284:20966-74; http://dx.doi.org/10.1074/jbc.M109.006346
- Kadri Z, Shimizu R, Ohneda O, Maouche-Chretien L, Gisselbrecht S, Yamamoto M, Romeo PH, Leboulch P, Chretien S. Direct binding of pRb/E2F-2 to GATA-1 regulates maturation and terminal cell division during erythropoiesis. PLoS Biol 2009; 7:e1000123; PMID:19513100; http://dx.doi.org/10.1371/journal.pbio.1000123
- Bracken AP, Ciro M, Cocito A, Helin K. E2F target genes: unraveling the biology. Trends Biochem Sci 2004; 29:409-17
- Xu J, Shao Z, Li D, Xie H, Kim W, Huang J, Taylor JE, Pinello L, Glass K, Jaffe JD, et al. Developmental control of polycomb subunit composition by GATA factors mediates a switch to non-canonical functions. Mol Cell 2015; 57:304-16; PMID:25578878; http://dx.doi.org/10.1016/j.molcel.2014.12.009
- Lange S, Perera S, Teh P, Chen J. Obscurin and KCTD6 regulate cullin-dependent small ankyrin-1 (sAnk1.5) protein turnover. Mol Biol Cell 2012; 23:2490-504; PMID:22573887; http://dx.doi.org/10.1091/mbc.E12-01-0052
- Blanchet E, Annicotte JS, Lagarrigue S, Aguilar V, Clapé C, Chavey C, Fritz V, Casas F, Apparailly F, Auwerx J, et al. E2F transcription factor-1 regulates oxidative metabolism. Nat Cell Biol 2011; 13:1146-52
- Xu Y, Swartz KL, Siu KT, Bhattacharyya M, Minella AC. Fbw7-dependent cyclin E regulation ensures terminal maturation of bone marrow erythroid cells by restraining oxidative metabolism. Oncogene 2014; 33:3161-71; PMID:23873023; http://dx.doi.org/10.1038/onc.2013.289
- Reynolds MR, Lane AN, Robertson B, Kemp S, Liu Y, Hill BG, Dean DC, Clem BF. Control of glutamine metabolism by the tumor suppressor Rb. Oncogene 2014; 33:556-66; PMID:23353822; http://dx.doi.org/10.1038/onc.2012.635
