Transcription factors bind to specific DNA motifs in promoters to regulate gene expression. The cell-division cycle is supported by periodic transcription of selected genes which peak in expression when they are required. In fission yeast, 2 clusters of cell-cycle-regulated genes show maximal expression during mitosis and cell division, respectively, many of which are being regulated by a transcriptional cascade involving the forkhead transcription factor Sep1 which controls the zinc finger transcription factor Ace2.Citation1 The Sep1-Ace2 transcriptional pathway promotes the separation of daughter cells after mitosis by timely expression of genes required for the degradation of the cell-division septum. Sep1 is thought to act together with the MADS-box factor Mbx1 and another forkhead protein, Fkh2, which negatively controls transcription.Citation2 Recently, yet another transcription factor, Sak1, has been shown to be the major activator of mitotic genes.Citation3
In this issue, Suárez and colleges reveal additional complexities for the control of periodic transcription during cell division in fission yeast: Sep1, Fkh2 and Mbx1 not only regulate genes required for cell separation indirectly, via Ace2, but also directly by differentially binding to the promoters of some of these genes.Citation4 Most of the promoters of Ace2-regulated genes contain several copies of the conserved CCAGCC motif, previously described as the Ace2 binding motif. Moreover, these promoters contain different numbers and combinations of the Ace2 motif together with PCB and forkhead binding motifs. Ace2 binds to these promoters in a cell cycle-dependent manner when the corresponding genes show maximal expression. In addition, Sep1, Fkh2 and/or Mbx1 bind in different combinations and timings to some of these promoters to positively or negatively support transcription.
The authors analyze in detail 2 genes previously known to be controlled by Ace2, eng1 and agn1, which show different arrangements of regulatory motifs in their promoters. Ace2, Sep1 and Fkh2 bind during distinct times to their motifs at the eng1 promoter, reflecting their positive or negative roles in transcription. In contrast, functional studies show that Mbx1, besides Ace2, is required for the normal expression of agn1. Thus, eng1 and agn1 are regulated by different combinations of transcription factors, both directly and indirectly. Such regulatory circuits, where one factor regulates another factor and both factors together then regulate common target genes, are well known in network research as feed-forward loops (). Notably, another recent study shows that Sak1 also regulates Ace2, along with at least 2 Ace2 targets, eng1 and adg2.Citation3 This finding raises the possibility that Sak1 similarly participates in a feed-forward loop together with Ace2. Such feed-forward loops are widespread network motifs for cellular control, with the coherent feed-forward loop arrangements seen here leading to a delayed transcriptional response.Citation5 The feature of this regulatory circuit could therefore help to ensure that genes functioning in cell separation are activated only after those functioning in mitosis.
Figure 1. Simplified model for regulation of two Ace2 targets, eng1 and agn1, involved in cell separation. Both genes are regulated by coherent feed-forward loops,Citation5 involving Ace2 and different combinations of Sep1, Fkh2 and/or Mbx1. Continuous black lines depict previously known regulatory links,Citation1 while dashed gray lines show new aspects of control based on results by Suárez et al.Citation4
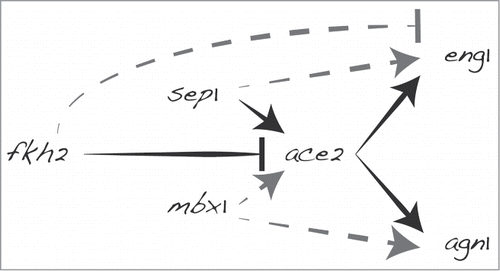
Regulatory processes are malleable and can evolve quite rapidly. It has been suggested that essential genes feature a lower number of regulatory promoter motifs, while genes with multiple, varied motifs tend to be under lower selective pressure.Citation6 This might explain differences in motif patterns of Ace2-regulated genes: agn1, which plays crucial roles in in cell shape maintenance and septum degradation close to the cell wall, is more tightly regulated than eng1 and accordingly shows fewer and less diverse promoter motifs. It is therefore possible that these regulatory differences mainly reflect random evolutionary changes under different selection pressures. It would be interesting to look at the arrangement of corresponding promoter motifs in natural isolates of fission yeast.Citation7 to analyze the extent of regulatory plasticity within this species.
In conclusion, the transcriptional control of Ace2-dependent genes in fission yeast is more complex and diverse than previously appreciated. Sep1, Mbx1 and Fkh2 (and probably Sak1) not only control gene transcription at the onset of mitosis, but also at the end of mitosis when cells divide. These factors indirectly control cell-separation genes by regulating Ace2, and they also directly control cell-separation genes in different combinations for different genes. These findings provide another example for the remarkable sophistication and complexity of transcriptional regulatory programs controlling the cell cycle, which without doubt still have many more surprises in store for us.
References
- Bähler J. Cell Cycle 2005; 4:39-41; PMID: 15611619; http://dx.doi.org/10.4161/cc.4.1.1336
- Papadopoulou K, et al. J Cell Sci 2008; 121:38-47; PMID:18057023; http://dx.doi.org/10.1242/jcs.019489
- Garg A, et al. Nucleic Acids Res 2015; 43:6874-88; PMID:25908789; http://dx.doi.org/10.1093/nar/gkv274
- Suárez MB, et al. Cell Cycle 2015; 14(19):3124-37; PMID:26237280; http://dx.doi.org/10.1080/15384101.2015.1078035
- Mangan A, Alon U. Proc Natl Acad Sci U S A 2003; 100:11980-5; PMID:14530388; http://dx.doi.org/10.1073/pnas.2133841100
- Bilu Y, Barkai N. Genome Biol 2005; 6:R103; PMID:16356266; http://dx.doi.org/10.1186/gb-2005-6-12-r103
- Jeffares DC, et al. Nat Genet 2015; 47:235-41; PMID:25665008; http://dx.doi.org/10.1038/ng.3215
