ABSTRACT
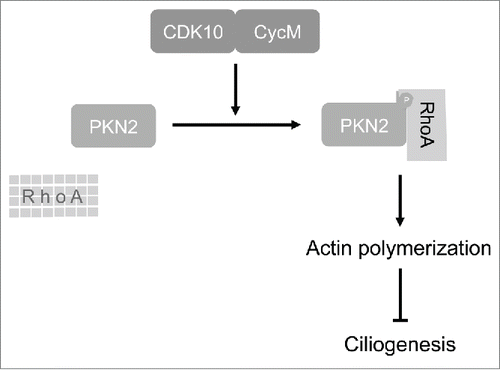
CDK10/CycM is a protein kinase deficient in STAR (toe Syndactyly, Telecanthus and Anogenital and Renal malformations) syndrome, which results from mutations in the X-linked FAM58A gene encoding Cyclin M. The biological functions of CDK10/CycM and etiology of STAR syndrome are poorly understood. Here, we report that deficiency of CDK10/Cyclin M promotes assembly and elongation of primary cilia. We establish that this reflects a key role for CDK10/Cyclin M in regulation of actin network organization, which is known to govern ciliogenesis. In an unbiased screen, we identified the RhoA-associated kinase PKN2 as a CDK10/CycM phosphorylation substrate. We establish that PKN2 is a bone fide regulator of ciliogenesis, acting in a similar manner to CDK10/CycM. We discovered that CDK10/Cyclin M binds and phosphorylates PKN2 on threonines 121 and 124, within PKN2′s core RhoA-binding domain. Furthermore, we demonstrate that deficiencies in CDK10/CycM or PKN2, or expression of a non-phosphorylatable version of PKN2, destabilize both the RhoA protein and the actin network architecture. Importantly, we established that ectopic expression of RhoA is sufficient to override the induction of ciliogenesis resulting from CDK10/CycM knockdown, indicating that RhoA regulation is critical for CDK10/CycM's negative effect on ciliogenesis. Finally, we show that kidney sections from a STAR patient display dilated renal tubules and abnormal, elongated cilia. Altogether, these results reveal CDK10/CycM as a key regulator of actin dynamics and a suppressor of ciliogenesis through phosphorylation of PKN2 and promotion of RhoA signaling. Moreover, they suggest that STAR syndrome is a ciliopathy.
Introduction
Primary ciliogenesis is the dynamic cellular process of building the primary cilium, a microtubule-based structure located at the cell surface.Citation1,2 After cell division, each daughter cell inherits a centrosome composed of a mother and a daughter centriole, surrounded by pericentriolar material.Citation3 When the cell enters quiescence, the mother centriole differentiates to form the so-called basal body through maturation of subdistal and distal appendages.Citation4 The centrosome migrates toward the plasma membrane where it anchors through its distal-end and nucleates microtubules for axoneme formation and elongation. During the course of migration, a vesicle can associate with the distal-end of the basal body to initiate axoneme formation prior to plasma membrane fusion.Citation1,5 Active vesicular trafficking at the basal body supplies essential components for primary cilium assembly and elongation.Citation6-8 A constant equilibrium between anterograde and retrograde intraflagellar transport regulates axonemal assembly and disassembly, to maintain primary cilium length.Citation9,10
Actin cytoskeleton dynamics plays a key role in ciliogenesis.Citation11,12 Extended cells can assemble contractile actin filaments, which repress ciliogenesis. In contrast, confined cells can organize a dorsal protrusive actin network, which promotes ciliogenesis.Citation13 Consistent with these observations, nucleating factors that promote actin polymerization repress primary cilium assembly and growth whereas actin severing enzymes promote ciliogenesis.Citation14-17 Actin depolymerization promotes ciliogenesis by stabilizing the pericentrosomal preciliary compartment, a vesiculotubular structure that contributes to ciliogenesis,Citation14,15,17 and by inhibiting transcription factors from the Hippo pathway, which induce the expression of cilium disassembly factors.Citation17
The small GTPase RhoA plays a key role in actin network regulation.Citation18 It is required to promote actin stress fiber assembly and maintenance, through a signaling pathway that includes activation of Rho kinase and phosphorylation of myosin light chain 2 (MLC2).Citation19 There is extensive evidence that RhoA plays key roles in the regulation of ciliogenesis through its effects on actin networks. In the context of multiciliogenesis, RhoA appears to act positively by enabling formation of an apical actin web-like structure and promoting basal body docking.Citation20,21 In contrast, in monociliated cells, activation of RhoA and its downstream signaling pathway has been shown to suppress primary ciliogenesis by promoting the formation of actin stress fiber networks.Citation13,22,23 The presence of abnormal ciliogenesis and/or abnormal primary cilium length during development has been linked to severe developmental syndromes, collectively dubbed ciliopathies.Citation11,24 Notably, hyperactivation of RhoA and consequent changes in actin polymerization have been observed in several prototypical ciliopathies, including Bardet-Biedl, Meckel and Joubert syndromes.Citation22,23,25
STAR syndrome is an X-linked dominant disorder caused by mutations in family with sequence similarity 58, member A (FAM58A).Citation26 We recently reported that Cyclin M (CycM), encoded by FAM58A, interacts with Cyclin-Dependent Kinase 10 (CDK10) to form an active protein kinase.Citation27 We demonstrated that STAR syndrome-associated CycM mutants fail to interact with CDK10, thus compromising CDK10 kinase activity. Young girls affected by STAR syndrome suffer from general growth retardation. Additionally, these individuals display a number of renal, retinal, anogenital and digital anomalies that are reminiscent of ciliary defects. This spurred us to investigate whether mammalian CDK10/CycM plays a role in the regulation of ciliogenesis. Here, we show that CDK10/CycM is a key repressor of ciliogenesis through the regulation of RhoA and modulation of actin dynamics. Moreover, we see evidence of ciliary defects in renal tissue from a STAR syndrome patient, suggesting that STAR syndrome is a bona fide ciliopathy.
Results
CDK10/CycM negatively regulates ciliogenesis and primary cilium length
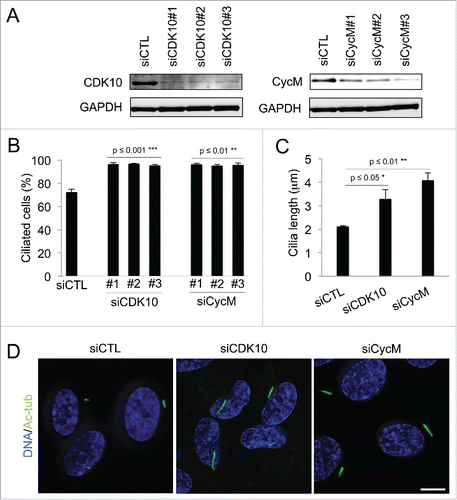
We investigated the role of CDK10/CycM in the regulation of ciliogenesis using the human telomerase reverse transcriptase retinal pigmented epithelial (hTERT RPE-1) cellular model. These immortalized cells maintain normal cell cycle checkpoints. In the absence of growth factors, they enter quiescence after mitosis and build a primary cilium.Citation28 We introduced 3 different siRNAs against CDK10 and CycM into hTERT RPE-1 cells and then withdrew serum 5 hours later to induce cell cycle exit. All of the siRNAs yielded efficient knockdown CDK10 or CycM (). We then determined the consequence on ciliogenesis by detection and quantification of primary cilia via acetylated-tubulin staining (Fig. S1A). There was no obvious difference in the number of cilia at 24 or 48 hours post-transfection (data not shown). However, by 72 hours post-transfection, we observed a significant increase in the number of ciliated cells in response to knockdown of either CDK10 (p ≤ 0.001) or CycM (p ≤ 0.01) such that almost all of the cells now bore cilia (). The time lag in the appearance of this phenotype suggests that CDK10/CycM knockdown is exerting its effect on cilia formation in serum starved cells. We also measured cilia length 72 hours after siRNA treatments (). This analysis revealed a significant increase of cilia length in response to CDK10 silencing (1.5 fold; p ≤ 0.05) and an even greater effect of CycM silencing (2 fold; p ≤ 0.01). Thus, CDK10 and CycM both act to repress cilia formation and elongation in serum starved cells.
Figure 1. CDK10/Cyclin M represses primary cilium assembly and growth (A-D) hTERT-RPE1 cells were transfected with control (siCTL), CDK10 or CycM siRNAs, subjected to serum starvation, and analyzed 72 hour post transfection. (A) CDK10 and CycM knockdown was assessed by western blot. (B) The percent of siCTL, si CDK10 and siCycM cells with primary cilia was determined by visualization using acetylated-tubulin staining (Figure S1A) and quantification. 500 cells were counted for each condition, in 3 independent experiments. (C) Measurement of cilia length (225 cilia/condition, in 3 independent experiments) using the LAS AF software on images randomly selected across the coverslips. (D) Representative immunofluorescence images of primary cilia (acetylated-tubulin staining, shown in green), DNA (DAPI staining, shown in blue). Scale bar: 10 μm.
Ciliogenesis is a finely regulated process that occurs at the G0/G1 phase of the cell cycle.Citation29 Since CDK10/CycM has been previously linked to cell cycle regulation Citation30 it seems plausible that CDK10/CycM silencing might also contribute indirectly to ciliogenesis by promoting quiescence. Hence, we assessed the cell cycle status of siRNA-treated cells that were maintained in the presence of serum. As reported previously,Citation30 CDK10/CycM silencing yielded a slight increase in the G2/M population. However, we saw no detectable increase in the G0/G1 population in CDK10/CycM depleted cells 72 hours post transfection (Fig. S1B), arguing against an indirect effect on ciliogenesis by promoting cell cycle exit.
The disassembly of primary cilium acts to enable cell cycle progression at the G1/S transition.Citation28,31-33 Thus, to investigate whether CDK10/CycM silencing modulates cilium disassembly and cell cycle re-entry, we took cells that had been treated with control, CDK10, CycM siRNAs and serum-starved for 72 hours (as in ), and then reexposed them to serum for 14 hours. In this case, we found that CDK10 and CycM silencing promoted the maintenance of primary cilia (Fig. S1C) and impaired cell cycle resumption, as judged by a reduction in the percentage of post-G0/G1 cells (Fig. S1D). Taken together, these results establish key roles for CDK10 and CycM in ciliogenesis. First, they suppress primary cilium assembly and elongation in serum starved cells without promoting cell cycle exit. Second, in response to mitogenic signaling, they enable both cilia disassembly and cell cycle re-entry.
CDK10/CycM maintains actin network architecture and phosphorylates actin regulators
Maturation of the basal body and actin network reorganization both play central roles in governing ciliogenesis. Thus, we tested whether CDK10/CycM influenced either of these processes. With regard to the basal body, we detected intense immunofluorescence staining of CDK10 and CycM at the base of primary cilia (stained by acetylated tubulin) in control but not in siCDK10/CycM treated cells (Fig. S2A). Moreover, further co-staining with centrosomal-associated proteins suggested that CDK10/CycM was concentrated at the mother centriole (Fig. S2B). Given these observations, we examined the subcellular localization of 2 centrosomal proteins, Cep170 (a mature mother centriole marker) Citation34 and Cep290 (a centriolar satellite marker that is mutated in various ciliopathies) Citation35 in our siCDK10/CycM treated cells. We did not observe any alteration in the staining pattern of these proteins, compared to control cells (Fig. S2C). Thus, while we cannot rule out the possibility of subtle roles, CDK10/CycM deficiency does not grossly alter basal body integrity.
To assess the effect of CDK10/CycM knockdown on actin network regulation, we used 3D deconvolution microscopy. Our analyses revealed a striking disruption of the actin network throughout the cell in response to CDK10/CycM silencing that resulted in near complete loss of actin stress fibers (). To determine the magnitude of this disruption, we compared it to the consequences of treating the cells with cytochalasin D, which blocks actin polymerization by capping the fast growing, barbed ends of actin filaments. We found that separate treatment with cytochalasin D or CDK10/CycM silencing had a comparable impact on ciliogenesis (Fig. S3A, B). Indeed, combined cytochalasin D treatment and CDK10/CycM silencing had little or no additive effect on the percentage of ciliated cells (Fig. S3B). Thus, we conclude that CDK10/CycM silencing and cytochalasin D have a similar, profound impact on actin polymerization and thereby influence ciliogenesis.
Figure 2. CDK10/CycM maintains actin network architecture and phosphorylates actin dynamics regulators (A) Immunofluorescent visualization of primary cilia (acetylated-tubulin staining, shown in green) and F-actin (Rhodamine-Phalloidin staining, shown in red) without (siCTL) or with CDK10 or CycM silencing under the same conditions as . DNA was stained with DAPI (shown in blue). Scale bar, 25 μm. The XZ view shows XZ optical projections of the cilia structure and actin structure (taken from a horizontal middle cross section) in a baso-apical manner. (B) In vitro kinase assay on protein arrays without kinase (CTL) or with recombinant purified CDK10/CycM. Positive hits (differential signals between both arrays) are indicated with black arrows and listed on the right.
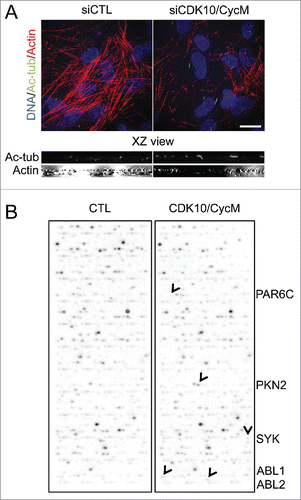
To better understand the mechanism by which CDK10/CycM inhibits ciliogenesis, we used an unbiased screen to identify phosphorylation substrates of this kinase. For this, we performed in vitro kinase assays using baculovirus-expressed, and purified, CDK10/CycM on a protein array containing 9,483 human recombinant proteins. Notably, this includes many core centrosomal proteins and ciliogenesis regulators (e.g. Cep72/290, BBS2/5/7/9/10/12, IFT88/52/81/57, KIF2A/3A/19, NDE1, Tctex, MARK4, TTBK2, Centrobin, AuroraA, HDAC6, PLK1, NEK2, MKS1) and also numerous actin network regulators. None of the centrosomal proteins were phosphorylated by CDK10/CycM. Strikingly, all 5 of the positive hits in this screen (; PKN2, PAR6C, ABL1, ABL2 and SYK) were known regulators of actin dynamics Citation36-39, reinforcing the rationale for focusing on this process.
Identification of PKN2 as a CDK10/CycM substrate and as a ciliogenesis regulator
Among the identified substrate candidates, Protein Kinase C-like 2 (PKN2, also known as Protein kinase C-related kinase 2, PRK2) exhibited the strongest phosphorylation signal. To further validate this finding, we performed in vitro phosphorylation assays using recombinant purified proteins. We observed a small amount of PKN2 autophosphorylation in the absence of our purified CDK10/CycM but inclusion of this kinase yielded a strong CDK10/CycM-dependent phosphorylation (), arguing that PKN2 is a direct CDK10/CycM substrate in vitro. To determine whether this interplay is relevant in vivo, we immunoprecipitated PKN2 from cell lysates and found that we could readily detect co-association of CDK10 and CycM by western blotting (). Thus, we propose that CDK10/CycM binds to PKN2 in vivo, and can directly phosphorylate this target.
Figure 3. PKN2 is a CDK10/CycM substrate and a ciliogenesis regulator (A) Quantification of 33P protein labeling in in vitro kinase assays conducted with CDK10/CycM alone, PKN2 alone or CDK10/CycM plus PKN2. CPM: counts per minute. (B) Western blot analysis of PKN2, CDK10 and CycM in anti-PKN2 or control rabbit IgG immunoprecipitates from serum-starved hTERT-RPE1 cells. Input is 1/25 of the total extract used for the immunoprecipitation. (C-F) hTERT-RPE1 cells were transfected with control (siCTL) or PKN2 (siPKN2) siRNAs, subjected to serum starvation, and analyzed 72 hour post transfection. (C) Western blot analysis of PKN2 protein levels. (D) Immunofluorescent visualization and XZ optical projections of primary cilia (acetylated-tubulin staining, shown in green) and F-actin (Rhodamine-Phalloidin staining, shown in red). Scale bar, 25 μm. (E, F) Percent ciliated cells and measurement of cilia length (quantified as described in ). (G,H) hTERT-RPE1 cells were serum starved for 24 hours, transfected with EGFP, EGFP-PKN2WT or EGFP-PKN2TATA and analyzed 16 hours post transfection. (G) Western blot analysis of PKN2 protein levels. (H) Percent ciliated cells (quantified as described in ) expression.
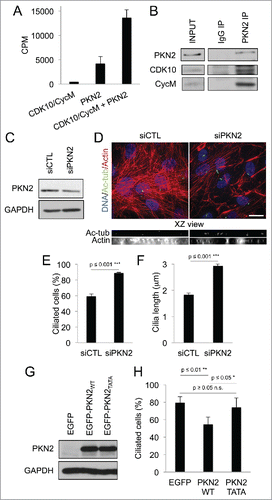
Several properties of PKN2 made it an extremely intriguing hit. First, PKN2 is known to be phosphorylated in vivo by an unidentified CDK.Citation40 Second, and directly relevant to the regulation of actin dynamics, PKN2 interacts with RhoA and it makes a positive contribution to various RhoA-regulated processes, including stress fiber formation.Citation39 Moreover, RhoA inhibition is known to promote ciliogenesis,Citation22 a result we have confirmed in our assay system (Fig. S3C, D). Notably, the nature of the interplay between PKN2 and RhoA is poorly understood, and PKN2 has not been examined for a role in ciliogenesis. Thus, we began by assessing whether PKN2 can modulate this process. Consistent with PKN2′s known role in actin regulation,Citation39 we found that a relatively modest knockdown of PKN2 () yielded a dramatic decrease of actin stress fibers (). Importantly, this was accompanied by a significant increase in both ciliogenesis (p ≤ 0.001, ) and cilia length (p ≤ 0.001, ). Moreover, these effects were comparable to those observed after CDK10/CycM silencing (). To complement this experiment, we also assessed the consequences of ectopic PKN2 expression in cells that had already been serum-starved for 24 hours (). In this case, upregulation of PKN2 was sufficient to significantly decrease the percentage of ciliated cells (p ≤ 0.01; ). Taken together, these results identify PKN2 as a direct phosphorylation substrate of CDK10/CycM. Moreover, they establish PKN2 as a previously unappreciated repressor of ciliogenesis.
CDK10/CycM maintains RhoA stability through PKN2 phosphorylation
Our data show that CDK10/CycM and PKN2 both promote maintenance of actin stress fibers and suppress ciliogenesis, and these act in the same direction as RhoA.Citation13,22,23 PKN2′s N-terminal domain is known to mediate its interaction with RhoA.Citation39 Thus, we hypothesized that CDK10/CycM might target this region, presumably in manner that would enhance RhoA and PKN2′s function in actin polymerization. Hence, we tested the ability of purified CDK10/CycM to phosphorylate a fragment of PKN2 (PKN21–174) that encompasses the core of its RhoA-binding domain and that lacks its kinase domain. We observed efficient phosphorylation of this fragment by CDK10/CycM in in vitro kinase assays, establishing that PKN21–174 is a direct CDK10/CycM target (Fig. S4A). We then analyzed the phosphorylated fragment by mass-spectrometry and identified T121 and T124 as the CDK10/CycM-targeted phosphorylation sites (Fig. S4B-D). Notably, a general phosphoproteomic endeavor verifies that these phosphorylated residues exist in vivo.Citation41 This, combined with our demonstration of CDK10/CycM-PKN2 coimmunoprecipitation (), provides strong evidence that CDK10/CycM phosphorylates these residues in the in vivo setting.
To investigate the biological impact of these 2 phosphorylation sites, we generated a non-phosphorylatable double mutant of PKN2 called PKN2TATA (T121A/T124A) and expressed this, or a PKN2WT control, in cells that had already been serum-starved for 24 hours. We consistently achieved similar expression levels of PKN2TATA versus PKN2WT (). We found that PKN2TATA, and not the wildtype PKN2 protein, triggered cell death that was first observed 24 hours post-transfection (data not shown) indicating that mutation of the phosphorylation sites disrupts PKN2 function. To assess the effect on ciliogenesis, we examined these cells at 16 hours post-transfection. As noted above, PKN2WT acts to suppress ciliogenesis, and we found that this ability is completely lost in PKN2TATA (). Thus, the CDK10/CycM phosphorylation sites are required for PKN2′s anti-ciliogenesis activity. At this timepoint, we did not see an increase in ciliogenesis in PKN2TATA expressing cells, relative to the vector controls. However, we note that it takes 72 hours for enhanced ciliogenesis to occur in our other transfections experiments (e.g., CDK10, CycM and PKN2 knockdown).
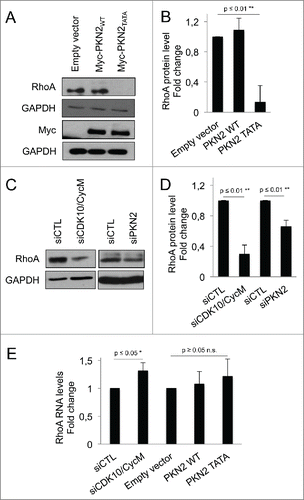
We next examined the effects on RhoA protein. Remarkably, at the 16 hour timepoint, we found that PKN2TATA caused loss of RhoA, while PKN2WT had no detectable effect on RhoA levels (). Thus, the absence of CDK10/CycM phosphorylation sites within ectopic PKN2 is sufficient to downregulate RhoA. Given this finding, we wondered whether depletion of endogenous CDK10/CycM or PKN2 could similarly affect RhoA. We used western blotting to assess RhoA levels in cells with partial CDK10/CycM or PKN2 knockdown, and observed a significant reduction of RhoA in both cases (). To determine whether this reflects any change at the RNA level, we performed quantitative PCR analysis in cells with partial CDK10/CycM knockdown, or ectopic expression of PKN2TATA or PKN2WT (). Contrary to the observed effect on RhoA protein levels, we found that RhoA RNA levels were either significantly upregulated (siCDK10/CycM) or not significantly altered (PKN2WT or PKN2TATA). Thus we conclude that depletion of CDK10/CycM, depletion of PKN2, or the presence of a non-phosphorylatable PKN2 mutant results in a destabilization of the RhoA protein, which perhaps activates a feedback loop that upregulates RhoA RNA levels.
Figure 4. CDK10/CycM maintains RhoA stability through PKN2 phosphorylation (A-D) Western blot analysis (A, C) and quantification using imageJ (B, D) of endogenous RhoA protein levels in serum-starved hTERT-RPE1 cells: (A, B) without (empty vector) or with ectopic expressed of Myc-PKN2WT or Myc-PKN2TATA; or (C, D) without (siCTL) or with CDK10/CycM or PKN2 silencing (as described in the prior figure legends). RhoA protein levels were normalized on GAPDH protein levels. (E) Quantification of RhoA RNA levels in different conditions described above, using quantitative RT-PCR. RhoA RNA levels were normalized on GAPDH RNA levels. n.s. = not significant.
CDK10/CycM represses ciliogenesis in a RhoA dependent manner
To further confirm that the perturbation of CDK10/CycM modulates RhoA signaling, we examined the effect of CDK10/CycM silencing on Phospho-Myosin Light Chain 2 (pMLC2), a downstream target of RhoA that is phosphorylated on serine 19 by the Rho-associated protein kinase, to promote actin stress fiber assembly.Citation19 Under normal conditions, pMLC2 staining roughly decorates stress fibers. In stark contrast to this expected pattern, CDK10/CycM knockdown resulted in either complete loss of stress fibers and complete loss of pMLC2 or a reduction in stress fibers and diffuse pMLC2 staining, presumably reflecting differences in the degree of CDK10/CycM depletion (). Thus, CDK10/CycM is required for appropriate RhoA signaling.
Figure 5. CDK10/CycM represses ciliogenesis through the modulation of RhoA signaling (A) Immunofluorescent visualization of Actin (Rhodamin-Phalloidin staining, shown in red), DNA (DAPI staining, shown in blue) and pMLC2 (shown in green) in serum-starved hTERT-RPE1 cells without (siCTL) or with CDK10/CycM silencing. The arrowhead indicates a cell with diffuse pMLC2 staining and reduced stress fibers. The arrow indicates a cell lacking pMLC2 staining and stress fibers. Scale bar, 25 μm. (B) Western blot analysis of exogenously-expressed Flag-RhoA protein levels in serum-starved hTERT-RPE1 cells, without (siCTL) or with CDK10/CycM silencing. (C) Percent ciliated cells (quantified as described in ) in the absence and presence of Flag-RhoA expression and CDK10/CycM silencing. n.s. = not significant.
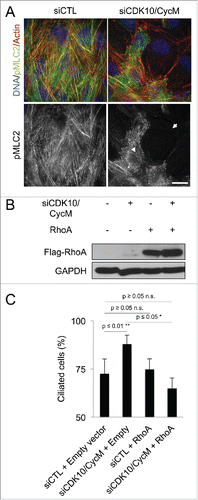
The requirement for CDK10/CycM to phosphorylate PKN2 and promote RhoA activity and stress fiber formation had the potential to explain CDK10/CycM's inhibitory effect on ciliogenesis. However, we could not rule out that another CDK10/CycM downstream pathway is actually responsible. To directly test whether CDK10/CycM regulates ciliogenesis via modulation of RhoA signaling, we overexpressed RhoA in cells treated with or without CDK10/CycM siRNAs to yield comparable levels in the 2 settings (). As we previously established, CDK10/CycM silencing significantly increased (p ≤ 0.01) the percentage of ciliated cells (). Importantly, expression of RhoA was sufficient to completely override this defect, while having no detectable effect on ciliogenesis in the absence of CDK10/CycM silencing (p ≥ 0.05) (). Taken together, these results demonstrate that CDK10/CycM promotes actin polymerization and suppresses ciliogenesis through phosphorylation of PKN2 and promotion of RhoA signaling.
A STAR patient presents dilated renal tubules with abnormal, elongated cilia
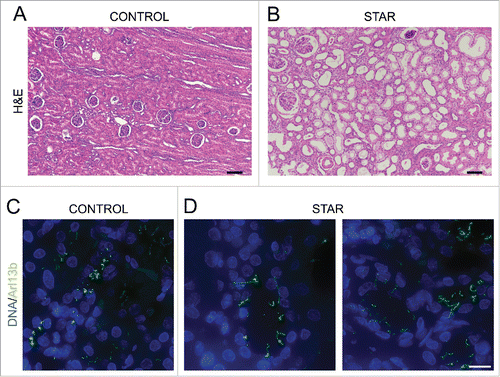
Our original rationale for testing whether CDK10/CycM modulates ciliogenesis was the fact that CycM is mutated in STAR syndrome, which displays a number of renal, retinal, anogenital and digital anomalies that are reminiscent of ciliary defects. Having established a bona fide role for CDK10/CycM in ciliogenesis, we wanted to investigate its relevance for the disease state. STAR syndrome is extremely rare, with only 9 patients having been reported to date, and thus it is almost impossible to access patient material. Fortunately, we were able to generate histological sections of a renal biopsy taken from a single STAR patient diagnosed with nephroblastoma.Citation26,42 We compared these sections with those from a control (non-STAR) nephroblastoma patient and, to ensure that the tumor did not interfere with our investigation, we focused specifically on the adjacent non-tumoral renal tissues. We observed dilated tubules in the STAR patient tissue, compared to the control (). Moreover, we stained these sections for Arl13b, a well-establish cilia marker, and found abnormal, elongated cilia specifically within the STAR patient tissue (). Thus, our analysis of this single, available sample supports our hypothesis that STAR syndrome is a ciliopathy.
Figure 6. Renal tubules of a STAR patient are dilated and exhibit longer cilia (A, B) Histopathological analysis (H&E staining) of renal biopsies from a CONTROL (A) and a STAR patient (B). Scale bar, 100 μm. (C, D) Immunofluorescent visualization of primary cilia (Arl13b staining, shown in green) and DNA (DAPI staining, shown in blue) in renal tubules from a CONTROL (C) and a STAR patient (D). Scale bar, 15 μm.
Discussion
Although CDK10 was discovered more than 20 years ago, the elucidation of its functions has been largely hampered by the lack of an identified cyclin partner, which precluded analyses of its kinase activity. The recent discovery of Cyclin M as a CDK10 binding partner has enabled exploration of this novel protein kinase's function and downstream targets.Citation27 Here, we discover that CDK10/CycM is a negative regulator of ciliogenesis, by showing that knockdown of this kinase increases the percentage of ciliated cells and also cilia length. This effect was not attributable to a perturbation in cell cycle phasing, which is controlled by various CDKs.Citation43 Instead, our data show that CDK10/CycM regulates ciliogenesis, at least in part, by modulating actin dynamics via phosphorylation of PKN2 and consequent activation of the RhoA pathway. First, an unbiased search for CDK10/CycM phosphorylation substrates among more than 9,000 human recombinant proteins led to the identification of known actin dynamics regulators, of which the top hit is the RhoA interacting protein PKN2. Second, we establish PKN2 as a bone fide regulator of ciliogenesis, whereby depletion or ectopic expression of this kinase is sufficient to promote or suppress this process, respectively. Third, we show that CDK10/CycM associates with PKN2 in vivo, and can directly phosphorylate 2 residues, T121 and T124, within the N-terminal domain of PKN2 that is responsible for RhoA binding. Fourth, we discovered that partial knockdown of either CDK10/CycM or PKN2, or ectopic expression of a non-phosphorylatable version of PKN2 causes loss of the RhoA protein and disrupts its downstream signaling. Fifth, we find that ectopic expression of RhoA is sufficient to override the effect of CDK10/CycM silencing on ciliogenesis, establishing its key contribution in CDK10/CycM ciliogenesis role. Finally, we detect dilated kidney tubules, and ciliogenesis defects in tissue sections from a STAR syndrome patient.
Previous studies clearly established that PKN2 binds to RhoA, and makes a positive contribution to various RhoA-regulated processes.Citation39,40,44-47 Our study identifies CDK10/CycM as a key upstream regulator of this biology. Combining our and previous observations, we propose a model in which CDK10/CycM phosphorylates PKN2 within its RhoA binding domain, thereby promoting RhoA binding and stabilization, which results in actin polymerization and consequent repression of primary cilia formation at cell cycle exit (). Interestingly, deficiency of CDK10/CycM also delays both cilia disassembly and cell cycle re-entry in response to mitogenic signaling. In view of recent studies with several other ciliogenesis regulators,Citation33 it seems likely that the prolonged maintenance of primary cilia is acting to delay cell cycle entry, not vice versa.
Our data do not address the mechanism(s) by which loss of CDK10/CycM or PKN2 causes RhoA loss. However, we note that previous studies have shown that a cell division cycle 42-Partitioning defective 6-atypical Protein Kinase C (cdc42-Par6-aPKC) complex acts to recruit the SMAD ubiquitination regulatory factor 1 (Smurf1) E3-ubiquitin ligase to the membrane, where is it is able to drive RhoA degradation.Citation37 Thus, its tempting to speculate that CDK10/CycM and PKN2 act to protect RhoA from this Smurf1-mediated degradation. This could occur through 2 possible mechanisms. In the first mechanism, phosphorylation of PKN2 by CDK10/CycM could drive its interaction with RhoA and thereby protect it from Smurf1. This model is intriguing given the location of the CDK10/CycM phophosphorylation sites within PKN2, and the fact that cell-cell adhesion regulation depends upon the ability of PKN2 to bind RhoA, and vice versa.Citation44,45 If this model is correct, based on prior studies,Citation39 it must require active PKN2 kinase. In the second mechanism, CDK10/CycM-PKN2 could actively inhibit the Smurf1 complex. This latter model is also very intriguing because our protein kinase array identified Par6 homolog α (PAR6C) as a putative CDK10/CycM partner and/or substrate. Moreover, PKN2 has been reported to inhibit the activating phosphorylation of aPKC (also called PKCζ) by PDK1.Citation48 Thus, PKN2 has at least 2 known ways to modulate the activity of the cdc42-Par6-aPKC complex that is required to recruit Smurf1. Notably, these 2 potential mechanisms are not mutually exclusive, and it seems plausible that both could be involved. Obviously, additional experiments are required to explore the potential involvement of Smurf1.
While we have focused this study on the role of CDK10/CycM on actin dynamics, it is noteworthy that we also see a significant enrichment of CDK10/CycM at the basal body. Notably, CDK10/CycM did not phosphorylate any of the numerous core centrosomal proteins and ciliogenesis regulators present on the protein array. Moreover, based on immunofluorescence staining of the Cep290 and Cep170 protein, CDK10/CycM deficiency did not grossly disrupt the integrity of the basal body. We cannot rule out the possibility that CDK10/CycM plays a subtle role in basal body maturation that contributes to the regulation of ciliogenesis. However, this would need to play a more minor role, given our finding that RhoA overexpression is able to rescue the ciliogenesis defect caused by CDK10/CycM silencing. It is entirely possible that the positioning of CDK10/CycM at the basal body could enable coordination of actin remodeling and vesicle trafficking at this location. With regard to this notion, we note that RhoA is also highly enriched at the basal body.Citation25
Our study also provides key insight into STAR syndrome, which results from mutations in the FAM58A gene encoding CycM. These patients are extremely rare, but the existence of a biopsy from a STAR patient with nephroblastoma gave us the opportunity to screen for evidence of ciliogenesis defects. The STAR patient non-tumoral renal tissue shows abnormal, elongated cilia as compared to the non-STAR, non-tumoral control tissue. Thus, this FAM58A loss-of-function mutation mirrors our CDK10/CycM silencing experiments. Like all of the reported STAR patients,Citation26 this patient displayed kidney dysfunction soon after birth, well before being diagnosed with nephroblastoma in one kidney. Moreover, a recent paper reports the presence of left multicystic kidney within another STAR patient.Citation49 Renal dysfunction is one of the most common hallmarks of ciliopathies, due to abnormal cilia abundance and/or length. Other clinical features observed among known STAR patients, such as retinal dystrophy, facial, digital and anogenital abnormalities, are also frequently found in ciliopathies.Citation11 Based on these defects, and our cellular studies, we propose that STAR syndrome is a bona fide ciliopathy.
Materials and methods
Plasmids and recombinant purified proteins
The construction and production of these reagents is described in detail in Guen et al.Citation27 and/or the Supplementary material.
Cell culture, transfections and drug treatments
hTERT-RPE1 cells were cultured in DMEM/F12 Glutamax medium with 10% fetal bovine serum (Life Technologies) and transfected with plasmids or plasmids plus siRNAs, 24 hours after serum starvation using Lipofectamine 2000 (Life Technologies), or with siRNAs prior or not to serum starvation using Lipofectamine RNAi max (Life Technologies). Biochemical analysis, FACS analysis and immunofluorescence staining were performed 16 to 96 hours later.
Renal biopsies
Renal biopsies were fixed in formalin and paraffin-embedded. 6 μm sections were stained with haematoxylin and eosin or dewaxed for immunoflurecence.
Immunofluorescence and image analysis
Cells were fixed in 4% paraformaldehyde for 10 min, permeabilized with 0.3% Triton X100 for 15 min and blocked with 1% BSA for 1 h before staining with primary antibodies against: acetylated tubulin (Sigma T7451, 1:1000), Arl13b (Antibodies Inc. 73-287, 1:200), CDK10 (Covalab pab0847p, 1:100), CycM (Covalab pab0882-P, 1:100), γ-tubulin (Abcam ab27074, 1:500), Cep164 (Santa-Cruz Biotechnology sc-240226, 1:500), Cep170 (Life Technologies 41–3200, 1:500), Sas6 (Santa-Cruz Biotechnology sc-81431, 1:300) or Phospho-Myosin Light Chain 2 (Cell Signaling 3675, 1:100). Secondary antibodies were: anti-mouse Alexa Fluor 488 (Life Technologies A11001, 1:1000) or anti-rabbit Alexa Fluor 594 (Life Technologies A21207, 1:1000. Rhodamine-Phalloidin (Life Technologies R415, 1:10) was used to probe F-actin. Mounted coverslips were examined using Leica DMI6000B, Zeiss CellObserver, Deltavision Olympus X71 microscopes. Images were acquired using 20X, 63X and 100X objectives and CoolSNAP HQ and HQ2 cameras. Z-stacks were deconvolved (Metamorph, Softworx) and processed with ImageJ.
Protein kinase assays
ProtoArray Human Protein Microarrays v5.0 (Life Technologies PAH0525065) were incubated with recombinant purified CDK10/StrepII-CycM in kinase buffer A (25 mM Tris-HCl, 10 mM MgCl2, 1 mM EGTA, 1 mM DTT, 3.7 μM heparin, pH 7.5). The array was scanned using a Typhoon 9200 (Amersham Biosciences) and analyzed by ProtoArray Prospector software (Life Technologies). Positive hits were manually confirmed.
To assess PNK2 phosphorylation, GST-CDK10/StrepII-CycM was incubated for 30 min at 30°C in kinase buffer A containing 22.7 μM BSA, 15 mM DTT and either 1 μM GST-PKN2, 15 μM ATP and 2 μCi ATP[γ-33P] or 1 μM GST-PKN21–174, 100 μM ATP and 5 μCi ATP[γ-32P]. [γ-33P]-GST-PKN2 was quantified on Whatman P81 phosphocellulose papers. Papers were washed 5X with 1% phosphoric acid, and put in 1 ml ACS scintillation fluid (Amersham) to perform scintillation counting (Tri-Carb, PerkinElmer). [γ-32P]- GST-PKN21–174 was determined by running 10% Bis-Tris SDS-PAGE gels. Substrate was stained by Coomassie (R-250, Biorad) and the incorporated radioactivity was revealed by autoradiography.
FACS, Immunoprecipitation and western blot experiments
These experiments were conducted using standard procedures, as detailed in the Supplementary material. Briefly, DNA content was determined by FACS, on 30, 000 propidium iodide-stained cells for each condition. Immunoprecipitations were conducted on 500 µg of precleared cell lysates using 5 μg of antibodies against Myc (Abcam ab9106) or PKN2 (Abcam ab138514). Western blots were performed using primary antibodies against CDK10 (Covalab pab0847p, 1:500), CycM (Covalab pab0882-P, 1:500), GAPDH (Covalab 6357, 1:10000), PKN2 (Abcam ab138514, 1:1000), Myc (Abcam ab9106, 1:1000), GFP (Roche 11814460001, 1:1000), RhoA (Cell Signaling 2117, 1:1000), Flag (Sigma-Aldrich F3165, 1:500) and secondary antibodies HRP-coupled anti-mouse (Biorad 170–6516, 1:3000) or anti-rabbit (Biorad 172–1019, 1:5000).
Quantitative Real Time-PCR
Total RNA was isolated using the RNeasy Plus kit (Qiagen), and cDNAs were generated with random primers and SuperScript III Reverse Transcriptase (Life Technologies). Real-time quantitative PCR reactions were performed with a StepOnePlus Real-Time PCR system (Life Technologies) using the SYBR Green PCR Master Mix (Life Technologies) and the following primers. For: RhoA 5′-ATGTGCCCACAGTGTTTGAGAAC-3′, 5′-CGTTGGGACAGAAATGCTTGAC-3′; for GAPDH 5′-CTGGGCTACACTGAGCACC-3′ and 5′-AAGTGGTCGTTGAGGGCAATG-3′ (used for normalization).
Mass spectrometry
Procedures are detailed in the Supplementary material.
Statistical analyses
Excel (Microsoft) was used to analyze data, draw graphs and perform statistical analysis. Significance was determined using data from 3 independent experiments and Student T-tests were used to determine p-values.
Abbreviations
| CDK10 | = | Cyclin-Dependent Kinase 10 |
| CycM | = | Cyclin M |
| STAR | = | Toe syndactyly, telecanthus and anogenital and renal malformations |
| PKN2 | = | Protein kinase C-like 2 |
| pMLC2 | = | phospho-myosin light chain 2 |
| FAM58A | = | Family with sequence similarity 58, member A |
| hTERT-RPE1 | = | human telomerase reverse transcriptase retinal pigmented epithelial 1 |
| cdc42 | = | cell division control protein 42 |
| Par6 | = | Partitioning defective 6 |
| PKC | = | Protein Kinase C |
| Smurf1 | = | SMAD specific E3 ubiquitin protein ligase 1 |
| Cep | = | Centrosomal protein |
| Sas6 | = | Spindle assembly abnormal protein 6 |
| GAPDH | = | Glyceraldehyde-3-phosphate dehydrogenase |
| EGFP | = | Enhanced green fluorescent protein |
| Arl13b | = | ADP-ribosylation factor-like protein 13B. |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
2015CC6842R-file002.docx
Download MS Word (58.7 MB)Acknowledgments
We are grateful for technical advice from Xavier Fant and Sandrine Ruchaud, Nathalie Desban and Tiziana Parisi. We are grateful to Anna Greka for her expert opinion on renal tissue observations. We thank Saïd El Alaoui (Covalab) for CDK10 and CycM antibodies; Daniel Fisher for the gift of protein arrays; Laurent Meijer for his hospitality in ManRos Therapeutics for FACS analysis. Cell imaging was conducted using imaging core facilities of the Station Biologique de Roscoff and the Koch Institute Swanson Biotechnology Center.
Funding
This project was funded by the Région Bretagne, the Ligue Nationale contre le Cancer Grand Ouest, the Département Finistère, the Association pour la Recherche contre le Cancer, and the Fondation Jérôme Lejeune. J.A.L. is a Ludwig Scholar at MIT. V.J.G. was a recipient of a doctoral fellowship from the French Ministry of Research.
References
- Sorokin SP. Reconstructions of centriole formation and ciliogenesis in mammalian lungs. J Cell Sci 1968; 3:207-30; PMID:5661997
- Bloodgood RA. From central to rudimentary to primary: the history of an underappreciated organelle whose time has come. The primary cilium. Method Cell Biol 2009; 94:3-52; PMID:20362083
- Nigg EA, Stearns T. The centrosome cycle: Centriole biogenesis, duplication and inherent asymmetries. Nat Cell Biol 2011; 13:1154-60; PMID:21968988; http://dx.doi.org/10.1038/ncb2345
- Kobayashi T, Dynlacht BD. Regulating the transition from centriole to basal body. J Cell Biol 2011; 193:435-44; PMID:21536747; http://dx.doi.org/10.1083/jcb.201101005
- Benmerah A. The ciliary pocket. Curr Opin Cell Biol 2013; 25:78-84; PMID:23153502; http://dx.doi.org/10.1016/j.ceb.2012.10.011
- Nachury MV, Loktev AV, Zhang Q, Westlake CJ, Peranen J, Merdes A, Slusarski DC, Scheller RH, Bazan JF, Sheffield VC, et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell 2007; 129:1201-13; PMID:17574030; http://dx.doi.org/10.1016/j.cell.2007.03.053
- Knodler A, Feng S, Zhang J, Zhang X, Das A, Peranen J, Guo W. Coordination of Rab8 and Rab11 in primary ciliogenesis. Proc Natl Acad Sci U S A 2010; 107:6346-51; PMID:20308558; http://dx.doi.org/10.1073/pnas.1002401107
- Westlake CJ, Baye LM, Nachury MV, Wright KJ, Ervin KE, Phu L, Chalouni C, Beck JS, Kirkpatrick DS, Slusarski DC, et al. Primary cilia membrane assembly is initiated by Rab11 and transport protein particle II (TRAPPII) complex-dependent trafficking of Rabin8 to the centrosome. Proc Natl Acad Sci U S A 2011; 108:2759-64; PMID:21273506; http://dx.doi.org/10.1073/pnas.1018823108
- Marshall WF, Rosenbaum JL. Intraflagellar transport balances continuous turnover of outer doublet microtubules: implications for flagellar length control. J Cell Biol 2001; 155:405-14; PMID:11684707; http://dx.doi.org/10.1083/jcb.200106141
- Ishikawa H, Marshall WF. Ciliogenesis: building the cell's antenna. Nat Rev Mol Cell Biol 2011; 12:222-34; PMID:21427764; http://dx.doi.org/10.1038/nrm3085
- Avasthi P, Marshall WF. Stages of ciliogenesis and regulation of ciliary length. Different Res Biol Diver 2012; 83:S30-42; PMID:22178116; http://dx.doi.org/10.1016/j.diff.2011.11.015
- Yan X, Zhu X. Branched F-actin as a negative regulator of cilia formation. Exp Cell Res 2013; 319:147-51; PMID:22975729; http://dx.doi.org/10.1016/j.yexcr.2012.08.009
- Pitaval A, Tseng Q, Bornens M, Thery M. Cell shape and contractility regulate ciliogenesis in cell cycle-arrested cells. J Cell Biol 2010; 191:303-12; PMID:20956379; http://dx.doi.org/10.1083/jcb.201004003
- Kim J, Lee JE, Heynen-Genel S, Suyama E, Ono K, Lee K, Ideker T, Aza-Blanc P, Gleeson JG. Functional genomic screen for modulators of ciliogenesis and cilium length. Nature 2010; 464:1048-51; PMID:20393563; http://dx.doi.org/10.1038/nature08895
- Cao J, Shen Y, Zhu L, Xu Y, Zhou Y, Wu Z, Li Y, Yan X, Zhu X. miR-129-3p controls cilia assembly by regulating CP110 and actin dynamics. Nat Cell Biol 2012; 14:697-706; PMID:22684256; http://dx.doi.org/10.1038/ncb2512
- Bershteyn M, Atwood SX, Woo WM, Li M, Oro AE. MIM and cortactin antagonism regulates ciliogenesis and hedgehog signaling. Dev Cell 2010; 19:270-83; PMID:20708589; http://dx.doi.org/10.1016/j.devcel.2010.07.009
- Kim J, Jo H, Hong H, Kim MH, Kim JM, Lee JK, Heo WD, Kim J. Actin remodelling factors control ciliogenesis by regulating YAP/TAZ activity and vesicle trafficking. Natu commun 2015; 6:6781; PMID:25849865; http://dx.doi.org/10.1038/ncomms7781
- Hall A. Rho GTPases and the actin cytoskeleton. Science 1998; 279:509-14; PMID:9438836; http://dx.doi.org/10.1126/science.279.5350.509
- Totsukawa G, Yamakita Y, Yamashiro S, Hartshorne DJ, Sasaki Y, Matsumura F. Distinct roles of ROCK (Rho-kinase) and MLCK in spatial regulation of MLC phosphorylation for assembly of stress fibers and focal adhesions in 3T3 fibroblasts. J Cell Biol 2000; 150:797-806; PMID:10953004; http://dx.doi.org/10.1083/jcb.150.4.797
- Pan J, You Y, Huang T, Brody SL. RhoA-mediated apical actin enrichment is required for ciliogenesis and promoted by Foxj1. J Cell Sci 2007; 120:1868-76; PMID:17488776; http://dx.doi.org/10.1242/jcs.005306
- Park TJ, Mitchell BJ, Abitua PB, Kintner C, Wallingford JB. Dishevelled controls apical docking and planar polarization of basal bodies in ciliated epithelial cells. Nat Gen 2008; 40:871-9; PMID:18552847; http://dx.doi.org/10.1038/ng.104
- Hernandez-Hernandez V, Pravincumar P, Diaz-Font A, May-Simera H, Jenkins D, Knight M, Beales PL. Bardet-Biedl syndrome proteins control the cilia length through regulation of actin polymerization. Hum Mol Genet 2013; 22:3858-68; PMID:23716571; http://dx.doi.org/10.1093/hmg/ddt241
- Dawe HR, Adams M, Wheway G, Szymanska K, Logan CV, Noegel AA, Gull K, Johnson CA. Nesprin-2 interacts with meckelin and mediates ciliogenesis via remodelling of the actin cytoskeleton. J Cell Sci 2009; 122:2716-26; PMID:19596800; http://dx.doi.org/10.1242/jcs.043794
- Fliegauf M, Benzing T, Omran H. When cilia go bad: cilia defects and ciliopathies. Nat Revi Mol Cell Biol 2007; 8:880-93; PMID:17955020; http://dx.doi.org/10.1038/nrm2278
- Valente EM, Logan CV, Mougou-Zerelli S, Lee JH, Silhavy JL, Brancati F, Iannicelli M, Travaglini L, Romani S, Illi B, et al. Mutations in TMEM216 perturb ciliogenesis and cause Joubert, Meckel and related syndromes. Nat Genet 2010; 42:619-25; PMID:20512146; http://dx.doi.org/10.1038/ng.594
- Unger S, Bohm D, Kaiser FJ, Kaulfuss S, Borozdin W, Buiting K, Burfeind P, Bohm J, Barrionuevo F, Craig A, et al. Mutations in the cyclin family member FAM58A cause an X-linked dominant disorder characterized by syndactyly, telecanthus and anogenital and renal malformations. Nat Genet 2008; 40:287-9; PMID:18297069; http://dx.doi.org/10.1038/ng.86
- Guen VJ, Gamble C, Flajolet M, Unger S, Thollet A, Ferandin Y, Superti-Furga A, Cohen PA, Meijer L, Colas P. CDK10/cyclin M is a protein kinase that controls ETS2 degradation and is deficient in STAR syndrome. Proc Natl Acad Sci U S A 2013; 110:19525-30; http://dx.doi.org/10.1073/pnas.1306814110
- Pugacheva EN, Jablonski SA, Hartman TR, Henske EP, Golemis EA. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell 2007; 129:1351-63; PMID:17604723; http://dx.doi.org/10.1016/j.cell.2007.04.035
- Santos N, Reiter JF. Building it up and taking it down: the regulation of vertebrate ciliogenesis. Dev Dyn 2008; 237:1972-81; PMID:18435467; http://dx.doi.org/10.1002/dvdy.21540
- Li S, MacLachlan TK, De Luca A, Claudio PP, Condorelli G, Giordano A. The cdc-2-related kinase, PISSLRE, is essential for cell growth and acts in G2 phase of the cell cycle. Cancer Res 1995; 55:3992-5; PMID:7664269
- Kim S, Zaghloul NA, Bubenshchikova E, Oh EC, Rankin S, Katsanis N, Obara T, Tsiokas L. Nde1-mediated inhibition of ciliogenesis affects cell cycle re-entry. Nat Cell Biol 2011; 13:351-60; PMID:21394081; http://dx.doi.org/10.1038/ncb2183
- Li A, Saito M, Chuang JZ, Tseng YY, Dedesma C, Tomizawa K, Kaitsuka T, Sung CH. Ciliary transition zone activation of phosphorylated Tctex-1 controls ciliary resorption, S-phase entry and fate of neural progenitors. Nat Cell Biol 2011; 13:402-11; PMID:21394082; http://dx.doi.org/10.1038/ncb2218
- Kim S, Tsiokas L. Cilia and cell cycle re-entry: more than a coincidence. Cell Cycle 2011; 10:2683-90; PMID:21814045; http://dx.doi.org/10.4161/cc.10.16.17009
- Guarguaglini G, Duncan PI, Stierhof YD, Holmstrom T, Duensing S, Nigg EA. The forkhead-associated domain protein Cep170 interacts with Polo-like kinase 1 and serves as a marker for mature centrioles. Mol Biol Cell 2005; 16:1095-107; PMID:15616186; http://dx.doi.org/10.1091/mbc.E04-10-0939
- Coppieters F, Lefever S, Leroy BP, De Baere E. CEP290, a gene with many faces: mutation overview and presentation of CEP290base. Hum Mutat 2010; 31:1097-108; PMID:20690115; http://dx.doi.org/10.1002/humu.21337
- Jaumouille V, Farkash Y, Jaqaman K, Das R, Lowell CA, Grinstein S. Actin cytoskeleton reorganization by syk regulates fcgamma receptor responsiveness by increasing its lateral mobility and clustering. Dev Cell 2014; 29:534-46; PMID:24914558; http://dx.doi.org/10.1016/j.devcel.2014.04.031
- Wang HR, Zhang Y, Ozdamar B, Ogunjimi AA, Alexandrova E, Thomsen GH, Wrana JL. Regulation of cell polarity and protrusion formation by targeting RhoA for degradation. Science 2003; 302:1775-9; PMID:14657501; http://dx.doi.org/10.1126/science.1090772
- Woodring PJ, Hunter T, Wang JY. Regulation of F-actin-dependent processes by the Abl family of tyrosine kinases. J Cell Sci 2003; 116:2613-26; PMID:12775773; http://dx.doi.org/10.1242/jcs.00622
- Vincent S, Settleman J. The PRK2 kinase is a potential effector target of both Rho and Rac GTPases and regulates actin cytoskeletal organization. Mol Cell Biol 1997; 17:2247-56; PMID:9121475; http://dx.doi.org/10.1128/MCB.17.4.2247
- Schmidt A, Durgan J, Magalhaes A, Hall A. Rho GTPases regulate PRK2/PKN2 to control entry into mitosis and exit from cytokinesis. The EMBO J 2007; 26:1624-36; PMID:17332740; http://dx.doi.org/10.1038/sj.emboj.7601637
- Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci U S A 2008; 105:10762-7; PMID:18669648; http://dx.doi.org/10.1073/pnas.0805139105
- Rakenius A, Zappel H, Borozdin W, Craig A, Gärtner J, Kohlhase J. STAR syndrome and nephroblastoma–FAM58A linking organogenesis and tumorigenesis? Eur J Pediatr 2010; 169:389; http://doi 10.1007/s00431-009-1136-4
- Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer 2009; 9:153-66; PMID:19238148; http://dx.doi.org/10.1038/nrc2602
- Calautti E, Grossi M, Mammucari C, Aoyama Y, Pirro M, Ono Y, Li J, Dotto GP. Fyn tyrosine kinase is a downstream mediator of Rho/PRK2 function in keratinocyte cell-cell adhesion. J Cell Biol 2002; 156:137-48; PMID:11777936; http://dx.doi.org/10.1083/jcb.200105140
- Wallace SW, Magalhaes A, Hall A. The Rho target PRK2 regulates apical junction formation in human bronchial epithelial cells. Mol Cell Biol 2011; 31:81-91; PMID:20974804; http://dx.doi.org/10.1128/MCB.01001-10
- Quilliam LA, Lambert QT, Mickelson-Young LA, Westwick JK, Sparks AB, Kay BK, Jenkins NA, Gilbert DJ, Copeland NG, Der CJ. Isolation of a NCK-associated kinase, PRK2, an SH3-binding protein and potential effector of Rho protein signaling. J Biol Chem 1996; 271:28772-6; PMID:8910519; http://dx.doi.org/10.1074/jbc.271.46.28772
- Morissette MR, Sah VP, Glembotski CC, Brown JH. The Rho effector, PKN, regulates ANF gene transcription in cardiomyocytes through a serum response element. Am J Physiol Heart Circulat Physiol 2000; 278:H1769-74; PMID:10843871
- Hodgkinson CP, Sale GJ. Regulation of both PDK1 and the phosphorylation of PKC-zeta and -delta by a C-terminal PRK2 fragment. Biochemistry 2002; 41:561-9; PMID:11781095; http://dx.doi.org/10.1021/bi010719z
- Zarate YA, Farrell JM, Alfaro MP, Elhassan NO. STAR syndrome is part of the differential diagnosis of females with anorectal malformations. Am J Med Gen Part A 2015; 167:1940-3; http://dx.doi.org/10.1002/ajmg.a.37078