ABSTRACT
Copy number gain of the 8q24 region including the v-myc avian myelocytomatosis viral oncogene homolog (MYC) oncogene has been observed in many different cancers and is associated with poor outcomes. While the role of MYC in tumor formation has been clearly delineated, we have recently shown that co-operation between adjacent long non-coding RNA plasmacytoma variant transcription 1 (PVT1) and MYC is necessary for tumor promotion. Chromosome engineered mice containing an increased copy of Myc-Pvt1 (Gain Myc-Pvt1) accelerates mammary tumors in MMTV-Neu mice, while single copy increase of each is not sufficient. In addition, mammary epithelium from the Gain Myc-Pvt1 mouse show precancerous phenotypes, notably increased DNA replication, elevated -H2AX phosphorylation and increased ductal branching. In an attempt to capture the molecular signatures in pre-cancerous cells we utilized RNA sequencing to identify potential targets of supernumerary Myc-Pvt1 cooperation in mammary epithelial cells. In this extra view we show that an extra copy of both Myc and Pvt1 leads to increased levels of Rspo1, a crucial regulator of canonical β-catenin signaling required for female development. Human breast cancer tumors with high levels of MYC transcript have significantly more PVT1 transcript and RSPO1 transcript than tumors with low levels of MYC showing that the murine results are relevant to a subset of human tumors. Thus, this work identifies a key mechanism in precancerous and cancerous tissue by which a main player in female differentiation is transcriptionally activated by supernumerary MYC and PVT1, leading to increased premalignant features, and ultimately to tumor formation.
Introduction
The v-myc avian myelocytomatosis viral oncogene homolog (MYC) gene is one of the most studied oncogenes in the history of cancer research.Citation1 Though mutations in MYC are found with measurable frequency in human cancers,Citation2 MYC is commonly amplified in cancer as a part of the 8q24 amplicon.Citation3 We have recently shown that gain of the genomic region containing Myc, long non-coding RNA plasmacytoma variant transcription 1 (Pvt1), Ccdc26 and GasderminC (GsdmC) enhances tumor formation in mice compared to gain of an extra copy of Myc alone. Subsequently, we showed that Pvt1/PVT1co-operates with and potentiates Myc/MYC, in cancers with supernumerary MYC.Citation4 Chromosome engineering was used to generate sibling animals with a single extra copy of a genomic region and these animals allow the study of specific copy number gains in a clear and coordinated fashion along with control sibling mice.Citation5 Using this model we showed that a single copy of the 8q24.21 syntenic region containing both Myc and Pvt1 is sufficient to promote tumorigenesis in the MMTV-Neu mouse, while an extra copy of Myc or Pvt1 independently is not capable of this tumor promotion/acceleration.Citation4 Additionally, single copy number gain of the Myc, Pvt1, Ccdc26 and Gsdmc region (hereafter anointed as Gain(Myc-Pvt1)) gives rise to a precancerous phenotype in mouse mammary tissues, notably increased DNA replication, elevated γ-H2AX foci, and significantly increased ductal branching which also was not observed in mice harboring an increase copy of Myc only (Gain (Myc)) or Pvt1, Ccdc26, Gsdmc6 (hereafter anointed Gain (Pvt1)) alone (2). We hypothesized that a systematic analysis of the genes differentially expressed in the mammary epithelial cells from the Gain (Myc+Pvt1) relative to the Gain (Myc), Gain (Pvt1) and wild type (wt) siblings will reveal the molecular signatures associated with the precancerous lesions associated with the Gain (Myc+Pvt1) mammary glands. Here we report the analyses of RNA-SEQ data from primary mammary epithelial cells from these mice and compare the findings with transcriptome analysis of human breast cancer with increased MYC expression.
Transcriptome analysis reveals molecular signatures in precancerous mammary lesions with supernumerary Myc +Pvt1
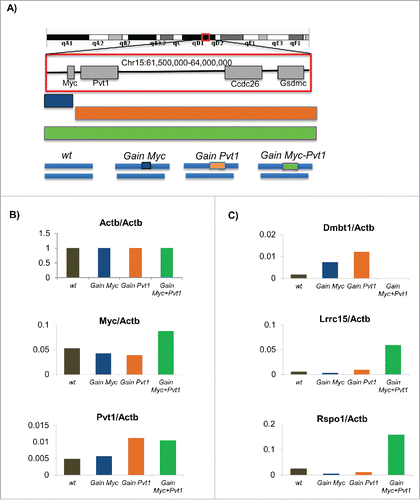
We rationalized that the identification of genes from engineered mammary epithelial cells which are differentially expressed between the pre-tumorigenic state (Gain(Myc-Pvt1)) and each of the non-tumorigenic states (Gain (Myc), Gain (Pvt1), wild type (wt)) would be useful in understanding the early molecular mechanisms of breast cancer formation. To validate that the RNA-SEQ libraries were of sufficient quality for meaningful analyses we compared the FPKM levels obtained for Myc and Pvt1 across the 4 sample groups relative to the FPKM value obtained for ActB. Plots of the relative transcript levels show that Myc and Pvt1 levels are increased at expected levels in Gain(Myc-Pvt1) animals relative to the other genotypes (). Because we could validate differential transcript levels within the region consistent with expectation, we concluded that we would be able to see differential expression in our data set. To look for differentially expressed genes we first carried out FPKM based analyses of the Gain(Myc-Pvt1) compared to the wt, Gain (Myc) and Gain (Pvt1) samples using CUFFLINKS. Dmbt1 and Lrcc15 transcript levels were observed by CUFFLINKS to be differentially expressed between Gain(Myc-Pvt1) and all other genotypes (). DMBT1 transcript level is decreased in breast cancerCitation6 and DMBT1 has been described as a breast cancer susceptibility locus in mouse tumors.Citation7 Mutations in DMBT1 have been associated with increased breast cancer risk in human population.Citation8 Dmbt1 is almost completely lost in the Gain(Myc-Pvt1) but remains expressed in wt, Gain (Myc) and Gain (Pvt1) mammary epithelial cells. We also find strong enrichment of Lrcc15 transcript in Gain(Myc-Pvt1) but not in wt, Gain (Myc) and Gain (Pvt1) (). Lrcc15 (LIB) is strongly expressed in breast cancer tumors.Citation9Additional genes were identified with greater than 5-fold change in each of the comparisons above and were called as significantly different in 2 of the 3 comparisons using Cuffdiff. Rspo1 was observed to be an average of 8-fold induced in GainMyc-Pvt1 precancerous mammary epithelial cells relative to the tissues not exhibiting the premalignant phenotype (). Rspo1 belongs to the R-spondin family of proteins which are secreted agonists of the canonical Wnt/β-catenin signaling pathway.Citation10 Rspo1 is involved in female specific differentiation.Citation11 Intriguingly, Rspo1 is required for normal epithelial morphogenesis during mammary gland development. Specifically, mammary tissue from Rspo1 null animals fail to exhibit ductal branching,Citation12 exactly the phenotype observed to be present in excess in the Gain(Myc-Pvt1) mammary tissue.
Figure 1. (A) Schematic of the engineered MMTV-Neu mouse model for the Gain (Myc), Gain (Pvt1) and Gain (Myc-Pvt1) animals from which mammary epithelium was obtained. Following RNA-SEQ analyses FPKM transcript abundance further normalized by the levels observed for Actb for (B) Actb, Myc and Pvt1 and (C) Dmbt1, Lrrc1, and Rspo1 are plotted using colors defined in the engineering schematic above.
Increased RSPO1 expression in human breast cancer with high MYC and PVT1
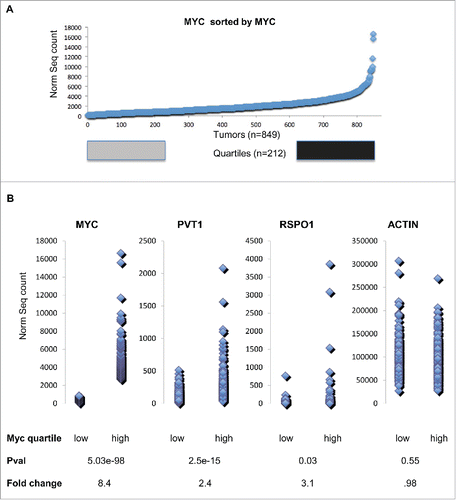
We next examined the expression levels of MYC, PVT1 and RSPO1 in The Cancer Genome Atlas breast cancer RNA-SEQ datasetCitation13 to find whether Myc-Pvt1 mediated increases in Rspo1 levels are important to a subset of human tumors. We hypothesized that sorting the data based on MYC expression would show that high MYC levels are associated with high levels of PVT1 and that a subset of these tumors would also show increases in RSPO1. The top quartile of the tumors sorted by MYC expression was compared with the bottom quartile of tumors by 2-group T-test (). As expected, highly significant increases in MYC transcript were observed between tumors with high levels of MYC and low levels of MYC (P-val < 10E-16, average fold change = 8.3) (). Confirmatory of a role for MYC and PVT1 co-operation in cancer, tumors with high levels of MYC showed highly significant increases in PVT1 transcript (P-val = 10E-11, average fold change 2.3). RSPO1 levels were also significantly increased in tumors with high levels of MYC compared to tumors with low levels of MYC (P-val = 0.03, average fold change 3.1). ACTB levels were not significantly different between the high MYC and low MYC group (P-val 0.55, average fold change 0.98).
Figure 2. (A) Myc transcript abundance for 849 TCGA Breast cancer tumors sorted by increasing Myc transcript abundance. Tumors within the bottom Quartile are shown in gray and tumors within the top quartile are shown in black. (B) Dot plot comparison of bottom quartile of transcript values compares to top quartile transcript for Myc, Pvt1, Rspo1 and ActB. A two group T-test shows that the Pvt1 and Rspo1 are both significantly higher and show a large average fold change in the top quartile relative to the bottom quartile based on Myc transcript level. ActB is not significantly different by T-test and also by fold change.
Role of RSPO1 in sex specification and cancer
By looking at low-level copy number changes within the context of normal organismal development we increase the likelihood of observing relevant events early in tumor formation. In an attempt to catch precancerous cells in the early stages of tumorigenesis we utilized RNA sequencing to identify potential targets of supernumerary MYC-PVT1 cooperation in mammary epithelial cells. Here we show that an extra copy of both Myc + Pvt1 leads to increased levels of Rspo1, a crucial regulator of canonical β-catenin signaling required for female development. We further show that human tumors with high levels of MYC also show high levels PVT1 and RSPO1.
Other members of the RSPO gene family have previously been implicated in cancer. Recurrent fusions in RSPO2 have been observed in human colon cancer.Citation14 High RSPO1 levels have been associated with poor outcome in glioma patients.Citation15 It has also been found to be a prominent susceptibility locus in ovarian cancer.Citation16 Further Rspo1 has been identified as common insertion site in murine forward genetic tumor screens and the T2/Onc insertions are oriented consistent with transcriptional activation of Rspo1.Citation17
That a gene involved in sex determination would also play a role in cancer formation has precedent. A key transcriptional regulator of the male expression pattern is also involved in tumorigenesis. Deletion of Dmrt1 leads to male to female sex reversalCitation18 and multiple variants in DMRT1 have been associated with germ cell tumors.Citation19 Likewise, RSPO1 mutations have been associated with female to male sex reversal and tumor formation.Citation20 In both mammary and testes tissue developmentally important mechanisms exist to allow cell division in mature tissues and these mechanisms appear to be misappropriated during the tumor formation processes. Therefore, sexual differentiation programs appear to have the potential to allow tumor promoting behavior, because they contain signal dependent proliferation programs for differentiated cell types.
The molecular mechanism of the MYC - PVT1 synergy leading to the activation of RSPO1 needs to be investigated in detail. We hypothesize that in addition to stabilization of MYC protein, PVT1 may retarget the MYC complex modifying the transcriptional response in the presence of Estrogen. The concept that MYC mediated responses such as proliferation could be retargeted in a cell dependent fashion is attractive considering the general role that MYC plays in tumorigenesis as well as the wide variety of molecular mechanisms attributed to MYC increase which run the gamut of apoptosis to proliferation to induced pluripotency. Tissue specific MYC retargeting opens the door to tissue specific MYC inhibition.
In summary our current work provides a specific molecular rational for the precancerous phenotypes observed with the Gain Myc-Pvt1 and expands the target list for pharmacological intervention downstream of MYC- PVT1 cooperation in both cancer and precancerous tissue. Further analyses of these biomarkers of precancerous state in “normal” human breast tissue may help to stratify populations at risk of developing breast cancer prior to the physical tumor diagnosis.
Disclosure of potential conflicts of interest
The authors declare that they have no conflict of interest.
1149660_Supplemental_Material.zip
Download Zip (1 MB)Additional information
Funding
References
- Soucek L, Evan G. Myc-Is this the oncogene from Hell? Cancer Cell 2002; 1:406-8; PMID:12124167; http://dx.doi.org/10.1016/S1535-6108(02)00077-6
- Wang X, Cunningham M, Zhang X, Tokarz S, Laraway B, Troxell M, Sears RC. Phosphorylation regulates c-Myc's oncogenic activity in the mammary gland. Cancer Res 2011; 71:925-36; PMID:21266350; http://dx.doi.org/10.1158/0008-5472.CAN-10-1032
- Huppi K, Pitt JJ, Wahlberg BM, Caplen NJ. The 8q24 gene desert: an oasis of non-coding transcriptional activity. Front Genet 2012; 3:69; PMID:22558003; http://dx.doi.org/10.3389/fgene.2012.00069
- Tseng YY, Moriarity BS, Gong W, Akiyama R, Tiwari A, Kawakami H, Ronning P, Reuland B, Guenther K, Beadnell TC, et al. PVT1 dependence in cancer with MYC copy-number increase. Nature 2014; 512:82-6; PMID:25043044
- Ramirez-Solis R, Liu P, Bradley A. Chromosome engineering in mice. Nature 1995; 378:720-4; PMID:7501018; http://dx.doi.org/10.1038/378720a0
- Braidotti P, Nuciforo PG, Mollenhauer J, Poustka A, Pellegrini C, Moro A, Bulfamante G, Coggi G, Bosari S, Pietra GG. DMBT1 expression is down-regulated in breast cancer. BMC Cancer 2004; 4:46; PMID:15301691; http://dx.doi.org/10.1186/1471-2407-4-46
- Blackburn AC, Hill LZ, Roberts AL, Wang J, Aud D, Jung J, Nikolcheva T, Allard J, Peltz G, Otis CN et al. Genetic mapping in mice identifies DMBT1 as a candidate modifier of mammary tumors and breast cancer risk. Am J Pathol 2007; 170:2030-41; PMID:17525270; http://dx.doi.org/10.2353/ajpath.2007.060512
- Tchatchou S, Riedel A, Lyer S, Schmutzhard J, Strobel-Freidekind O, Gronert-Sum S, Mietag C, D'Amato M, Schlehe B, Hemminki K, et al. Identification of a DMBT1 polymorphism associated with increased breast cancer risk and decreased promoter activity. Hum Mutat 2010; 31:60-6; PMID:19830809; http://dx.doi.org/10.1002/humu.21134
- Satoh K, Hata M, Yokota H. High lib mRNA expression in breast carcinomas. DNA Res 2004; 11:199-203; PMID:15368894; http://dx.doi.org/10.1093/dnares/11.3.199
- de Lau WB, Snel B, Clevers HC. The R-spondin protein family. Genome Biol 2012; 13:242; PMID:22439850; http://dx.doi.org/10.1186/gb-2012-13-3-242
- Chassot AA, Ranc F, Gregoire EP, Roepers-Gajadien HL, Taketo MM, Camerino G, de Rooij DG, Schedl A, Chaboissier MC. Activation of beta-catenin signaling by Rspo1 controls differentiation of the mammalian ovary. Hum Mol Genet 2008; 17:1264-77; PMID:18250098; http://dx.doi.org/10.1093/hmg/ddn016
- Chadi S, Buscara L, Pechoux C, Costa J, Laubier J, Chaboissier MC, Pailhoux E, Vilotte JL, Chanat E, Le Provost F. R-spondin1 is required for normal epithelial morphogenesis during mammary gland development. Biochem Biophys Res Commun 2009; 390:1040-3; PMID:19857464; http://dx.doi.org/10.1016/j.bbrc.2009.10.104
- Comprehensive molecular portraits of human breast tumours. Nature 2012; 490:61-70; PMID:23000897; http://dx.doi.org/10.1038/nature11412
- Seshagiri S, Stawiski EW, Durinck S, Modrusan Z, Storm EE, Conboy CB, Chaudhuri S, Guan Y, Janakiraman V, Jaiswal BS, et al. Recurrent R-spondin fusions in colon cancer. Nature 2012; 488:660-4; PMID:22895193; http://dx.doi.org/10.1038/nature11282
- Gu X, Wang X, Xiao H, Ma G, Cui L, Li Y, Zhou H, Liang W, Zhao B, Li K. Silencing of R-Spondin1 increases radiosensitivity of glioma cells. Oncotarget 2015; 6:9756-65; PMID:25865226; http://dx.doi.org/10.18632/oncotarget.3395
- Kuchenbaecker KB, Ramus SJ, Tyrer J, Lee A, Shen HC, Beesley J, Lawrenson K, McGuffog L, Healey S, Lee JM, et al. Identification of six new susceptibility loci for invasive epithelial ovarian cancer. Nat Genet 2015; 47:164-71; PMID:25581431; http://dx.doi.org/10.1038/ng.3185
- Takeda H, Wei Z, Koso H, Rust AG, Yew CC, Mann MB, Ward JM, Adams DJ, Copeland NG, Jenkins NA. Transposon mutagenesis identifies genes and evolutionary forces driving gastrointestinal tract tumor progression. Nat Genet 2015; 47:142-50; PMID:25559195; http://dx.doi.org/10.1038/ng.3175
- Matson CK, Murphy MW, Sarver AL, Griswold MD, Bardwell VJ, Zarkower D. DMRT1 prevents female reprogramming in the postnatal mammalian testis. Nature 2011; 476:101-4; PMID:21775990; http://dx.doi.org/10.1038/nature10239
- Kanetsky PA, Mitra N, Vardhanabhuti S, Vaughn DJ, Li M, Ciosek SL, Letrero R, D'Andrea K, Vaddi M, Doody DR, et al. A second independent locus within DMRT1 is associated with testicular germ cell tumor susceptibility. Hum Mol Genet 2011; 20:3109-17; PMID:21551455; http://dx.doi.org/10.1093/hmg/ddr207
- Parma P, Radi O, Vidal V, Chaboissier MC, Dellambra E, Valentini S, Guerra L, Schedl A, Camerino G. R-spondin1 is essential in sex determination, skin differentiation and malignancy. Nat Genet 2006; 38:1304-9; PMID:17041600; http://dx.doi.org/10.1038/ng1907
