The unidirectional and deterministic development of a zygote to trillions of cells that constitute a human produces the impression that it is an irreversible process, perhaps driven by reduction in free energy (ΔG), as postulated in the Waddington Model, the prevailing doctrine in developmental biology. This model however, has great difficulties in explaining induced pluripotency,Citation1 where any somatic cells (with presumed low ΔG) can be reprogrammed epigenetically to pluripotent stem cells (with presumed high ΔG); nor the reestablishment of totipotency by the fusion of an egg and a sperm – two terminally differentiated cells. A first principle analysis suggests that epigenetic reprogramming is an isoenergetic process if we assume that epigenetic information of the original cell and the converted cell are similar. With no significant change in enthalpy and at constant temperature, ΔG reflects changes in information content of the cell. As long as there is no significant change in the total epigenetic information content, there is no mythical cell type at the apex of energy landscape; it is largely flat. The reason that one cell type does not readily change to another is the kinetic barriers erected by the cell to maintain the local energy minimum.
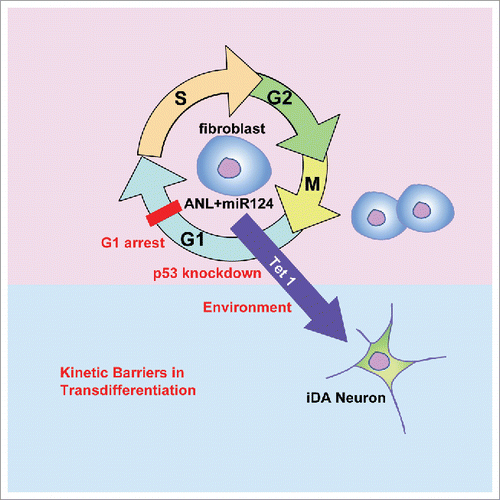
Our recent studies on the epigenetic conversion of human fibroblasts to induced dopaminergic neuronsCitation2 and induced serotonergic neuronsCitation3 have shown that these kinetic barriers include p53, cell cycle and extracellular environment (). When p53 is attenuated, the conversion becomes significantly more efficient.Citation2,3 Overexpression of p53 significantly decreases reprogramming efficiency.Citation2 Previous studies have shown that the p53 pathway serves as a barrier in the reprogramming of somatic cells to induced pluripotent stem cells (iPSCs).Citation1,4 The effects of p53 attenuation on epigenetic reprogramming and cell cycle progression go hand-in-hand in the derivation of iPSCs, a length process that requires many rounds of cell divisions.Citation5 In the conversion of a mitotic cell (fibroblast) to a postmitotic neuron, p53 attenuation exerts two opposing actions – it facilitates the transdifferentiation of receptive cells to postmitotic neurons and promotes the division of those fibroblasts that are unresponsive to conversion.Citation2
Figure 1. Overcoming kinetic barriers such as p53, cell cycle and extracellular environment enables highly efficient transdifferentiation of human fibroblasts to induced dopaminergic (iDA) neurons by Ascl1, Nurr1, Lmx1a and miR124. This epigenetic conversion is dependent on the DNA hydroxylase Tet1.
The responsiveness of the fibroblasts has to do with their cell cycle status. Fibroblasts that are labeled with EdU after the start of reprogramming never become neurons.Citation2 Almost none of the induced neurons are labeled with EdU, suggesting that the receptive cells never enters S phase.Citation2 Indeed, when we arrest cell cycle at G1/S checkpoint by a variety of independent mechanisms, the conversion becomes significantly more efficient.Citation2
Epigenetic reprogramming requires changes in epigenetic modifiers. We have found that p53 knockdown, G1 arrest and induction of reprogramming factors synergistically induce the expression of Tet1,Citation2 a DNA hydroxylase that significantly regulates the epigenome by oxidizing 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) and other products.Citation6 Tet1 appears to be a converging point in the actions of p53 knockdown, G1 arrest and induction of reprogramming factors, as it directly modifies the epigenome to a neural lineage, which has the highest 5hmC content.Citation6 Overexpression of Tet1 significantly increases the yield of induced neurons, while its knockdown causes massive cell death only during transdifferentiation.Citation2 These results are consistent with previous studies on the critical functions of Tet proteins in other epigenetic reprogramming events, such as induced pluripotency and demethylation of paternal genome during fertilization.Citation6
With p53 knockdown and G1 arrest, the transdifferentiation of human fibroblasts to induced neurons is highly efficient and fast. In 10 days, 93% of the cells in the dish are neurons; even the remaining 7% no longer express fibroblast markers.Citation2 Exit of cell cycle occurs within the first 24 hours of reprogramming, mainly driven by Ascl1, a neurogenic pioneering transcription factor that shuts down cell cycle by inducing the cyclin-dependent kinase inhibitor p27Kip1.Citation7 The rapid pace of conversion means that cell culture conditions need to be changed in synchrony with transdifferentiation in order for the highly efficient process to manifest itself. Interestingly, the media additives do not change the expression of Tet1 and are dispensable for the first 2 d without significant detriment,Citation2 suggesting that they do not enhance transdifferentiation per se, but are needed for the neurons to survive and reveal the highly efficient intrinsic program being conducted during the first two days.
The artificiality of transdifferentiation allows us to explore novel ways to read our genome. The natural way of reading the genome is what we call development. Life is a DNA-based software system embodied in the cell. Transdifferentiation is an efficient method to decipher this software system. The speed and efficiency of this method, once we remove kinetic barriers imposed by p53, cell cycle and extracellular environment, enable us to read the software at will. It is particularly useful for us to decipher the human version, where invasive techniques of developmental biology routinely used to query the mouse version cannot be carried out. The practical applications are many, as the method allows us to generate valuable but inaccessible cells such as dopaminergic neuronsCitation2 and serotonergic neuronsCitation3 from skin fibroblasts of any patients. It will greatly facilitate mechanistic study and drug discovery research on diseases by making the human model system readily available to many more researchers.
References
- Takahashi K, Yamanaka S. A developmental framework for induced pluripotency. Development 2015; 142:3274-85; PMID:26443632; http://dx.doi.org/10.1242/dev.114249
- Jiang H, et al. Cell cycle and p53 gate the direct conversion of human fibroblasts to dopaminergic neurons. Nat Commun 2015; 6:1-14; PMID:26639555; http://dx.doi.org/10.1038/ncomms10100
- Xu Z, et al. Direct conversion of human fibroblasts to induced serotonergic neurons. Mol Psychiatry 2016; 21:62-70; PMID:26216300; http://dx.doi.org/10.1038/mp.2015.101
- Zhao Y, et al. Two supporting factors greatly improve the efficiency of human iPSC generation. Cell Stem Cell 2008; 3:475-9; PMID:18983962; http://dx.doi.org/10.1016/j.stem.2008.10.002
- Hanna J, et al. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature 2009; 462:595-601; PMID:19898493; http://dx.doi.org/10.1038/nature08592
- Wu H, Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell 2014; 156:45-68; PMID:24439369; http://dx.doi.org/10.1016/j.cell.2013.12.019
- Farah MH, et al. Generation of neurons by transient expression of neural bHLH proteins in mammalian cells. Development 2000; 127:693-702; PMID:10648228
