ABSTRACT
Cdc45 is an essential protein that together with Mcm2-7 and GINS forms the eukaryotic replicative helicase CMG. Cdc45 seems to be rate limiting for the initial unwinding or firing of replication origins. In line with this view, Cdc45-overexpressing cells fired at least twice as many origins as control cells. However, these cells displayed an about 2-fold diminished fork elongation rate, a pronounced asymmetry of replication fork extension, and an early S phase arrest. This was accompanied by H2AX-phosphorylation and subsequent apoptosis. Unexpectedly, we did not observe increased ATR/Chk1 signaling but rather a mild ATM/Chk2 response. In addition, we detected accumulation of long stretches of single-stranded DNA, a hallmark of replication catastrophe. We conclude that increased origin firing by upregulated Cdc45 caused exhaustion of the single-strand binding protein RPA, which in consequence diminished the ATR/Chk1 response; the subsequently occurring fork breaks led to an ATM/Chk2 mediated phosphorylation of H2AX and eventually to apoptosis.
Introduction
In higher eukaryotes, initiation of DNA replication is achieved by the ordered assembly of a series of proteins at functionally defined origins of replication. In budding and fission yeasts, the cell division cycle protein 45, Cdc45, increases initial DNA unwinding at replication origins, i.e. origin firing efficiency.Citation1 In human cells, Cdc45 acts as proliferation-associated antigen.Citation2,3 Moreover, recruitment of Cdc45 at origins reflects the temporal replication program, i.e., Cdc45 is loaded at early-firing origins first, whereas it is assembled at late-firing ones late in S phase.Citation1,4,5
The CDC45 gene product is essential and the protein is part of the replicative helicase, the so-called CMG complex that consists of Cdc45, Mcm2-7, and GINS.Citation6 The DNA unwinding activity of the undecameric holoenzyme appears to be several hundred-fold stronger than that of the heterohexameric AAA+ ATPase Mcm2-7, providing strong evidence that CMG is the functional form of the eukaryotic replicative DNA helicase, whereas Mcm2-7 seems to reflect a dormant form.Citation7 Activated CMG is required for loading of the single-strand DNA binding protein RPA and DNA polymerase α onto the freshly exposed lagging strand,Citation8,9 as well as DNA polymerase ε onto the leading strand, where Dpb2, i.e. the second largest subunit of polymerase ε, forms a complex with the Psf1 subunit of GINS.Citation10,11 Then, DNA polymerase ε travels along with the activated CMG helicase on the leading strand.Citation12,13
The number of preformed replication origins per human cellCitation14-16 outnumbers the available Cdc45 molecules by far.Citation2,3 Therefore, Cdc45 may represent a limiting factor for the establishment and activation of the CMG helicase. In agreement with this view, Cdc45 has been suggested to activate nearby dormant origins of replication after an ongoing replication fork comes to an unscheduled halt.Citation5,15 On the other hand, Cdc45 is overexpressed in several tumor cells, where it helps to sustain rapid rounds of cell division.Citation2,17 Moreover, Myc-induced loading of Cdc45 into origins puts Cdc45 into a regulatory network of oncogene expression and carcinogenesis.Citation18 Indeed, there exist several studies that describe overexpressed Cdc45 in human cancer cells.Citation2,17,19,20 To get a deeper insight into the regulatory functions of Cdc45 for the initiation of DNA replication and for the maintenance of human tumors, we ectopically expressed this putative limiting factor and expected to see a faster S phase. Indeed, we observed increased firing of replication origins, but this caused severe replication stress, an early S-phase arrest, replication fork stalling, and eventually cell death by apoptosis.
Results
Increased levels of Cdc45 induced an early S phase arrest and apoptosis
Ectopic expression of Cdc45 was achieved in HeLa cells by creating a plasmid vector that encoded N-terminal human Cdc45 fused to a C-terminal enhanced green fluorescent protein (GFP). Cdc45 exclusively localized to the nucleus in essentially all GFP-positive cells inspected (), whereas transient transfection with a GFP-encoding control plasmid revealed an equal distribution of the control through the nucleus and cytoplasm.
Figure 1. Ectopic expression of Cdc45 caused apoptosis. (A) Transfection of HeLa cells with a vector containing Cdc45-GFP revealed an exclusive nuclear localization of the protein in transfected cells, whereas transfection with a vector carrying GFP alone led to staining of the entire cell. (B) The fraction of cells with a subgenomic DNA content increased dramatically between 36 and 48 h after transfection with Cdc45-GFP (lower panel), but not with GFP alone (upper panel), indicative for apoptotic DNA condensation and fragmentation. Both attached and detached cells were collected and stained with propidium iodide. The histograms represent the complete cell population from the FSC-SSC dot plot. (C) Accumulation of cleaved caspase 3 in Cdc45-GFP expressing cells 2 d after transfection. The dot plots represent only the GFP-positive (i.e., transfected) subpopulations.
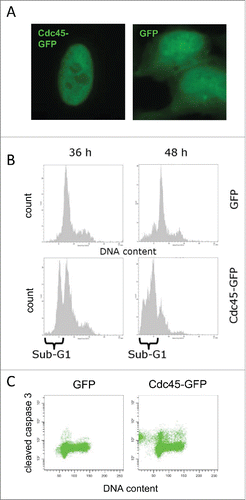
Cdc45-GFP expressing HeLa cells were analyzed by flow cytometry. Since one complete cell division cycle is needed for plasmid accumulation in the nucleus, we analyzed GFP-fluorescence starting at 36 h after transfection. GFP-fluorescent control cells showed a typical cell cycle distribution with only few cells with sub-G1 DNA content (, upper panel). In the Cdc45-GFP expressing cells, however, we observed an accumulation of cell with a G1 or early S-phase DNA content and a prominent sub-G1 peak indicating DNA fragmentation, a sign of apoptosis that became even more prominent 48 h after transfection (, lower panel). Flow cytometric detection of cleaved caspase confirmed that apoptosis had been induced by Cdc45 overexpression. Concordant with an early S-phase arrest, cells positive for a cleaved caspase 3-response displayed predominantly a G1 and early S-phase DNA content (, right panel).
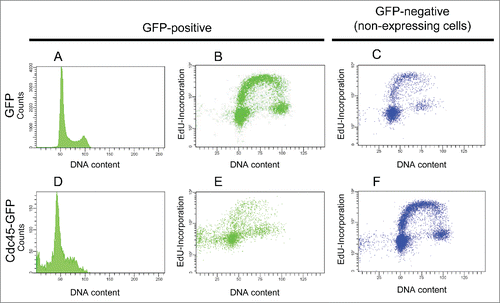
To characterize the events leading to cell death, we inspected DNA replication more directly. Cells were therefore pulse-labeled with the thymidine analog ethynyl deoxyuridine (EdU). Visualization of nascent DNA by fluorescent click conjugation of incorporated EdU revealed that GFP-transfected HeLa cells were normally passaged through S phase (). In contrast, a considerable proportion of cells with S phase DNA content incorporated nucleotides more slowly and apparently had difficulties to progress through S phase ().
Figure 2. Cdc45-GFP mediated (S)phase arrest and reduced DNA synthesis. 48 h after transfection, GFP expressing cells displayed a normal cell cycle distribution (A) and incorporated nucleotides (B) like non-expressing cells (C). Cdc45-GFP expressing cells were accumulated in S phase and displayed a prominent sub-G1 peak (D). Moreover, these cells synthesized less DNA (E), whereas GFP-negative (non-transfected) cells of the same culture (C, F) or GFP transfected cells (B) displayed normal DNA synthesis.
Accumulation of γH2AX after Cdc45 transfection
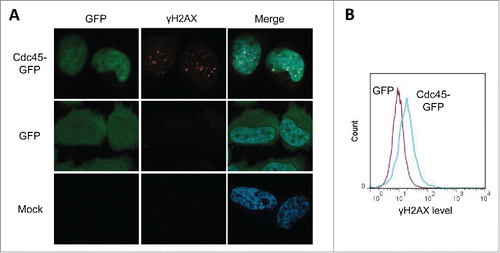
To further explore the problems with S phase progression, we probed Cdc45-GFP transfected cells for the presence of the DNA damage marker γH2AX. GFP-Cdc45 transfected cells displayed a prominent, granular nuclear γH2AX signal (, upper panels), whereas cells transfected with GFP alone (middle) or untransfected controls (lower panels) showed only residual DNA damage. Abundant Cdc45 thus clearly provoked DNA damage. Since background correction of the immunofluorescence images may exaggerate the γH2AX levels in Cdc45-transfected cells a more quantitative treatment by FACS was utilized. This revealed a twofold induction of the average γH2AX levels by transfection with Cdc45-GFP over transfection with GFP (). γH2AX induction was also confirmed in green monkey CV-1 cells expressing HA-tagged Cdc45 under a tetracyclin-inducible promoter (Supplemental Fig. 1).
Figure 3. Cdc45-GFP induced replication stress provoked focal accumulation of γH2AX in human HeLa cells. (A) Immunofluorescence imaging of γH2AX. Representative photographs of cells transfected with Cdc45-GFP (upper lane), GFP alone (middle lane), or pseudotransfected (lower lane). The same cells were subsequently immunostained for γH2AX, and subjected to immunofluorescence microscopy for GFP (green), γH2AX (Cy5 – red) and DNA (DAPI – blue). (B) Quantitation of the increase of γH2AX levels as measured by flow cytometry after transfection with vectors encoding GFP alone (magenta) or Cdc45-GFP (turquoise).
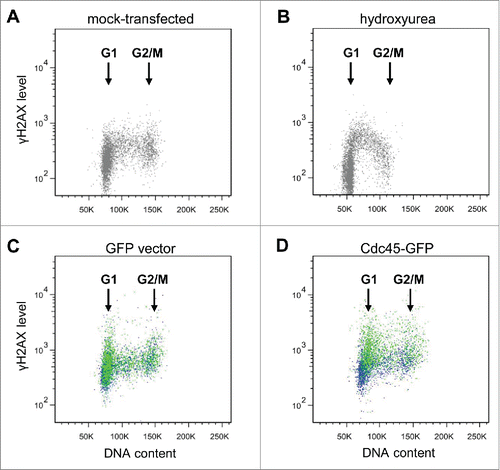
In an attempt to timely allocate this type of phosphorylation within the cell cycle, we transfected cells with either GFP or GFP-Cdc45 and analyzed the levels of γH2AX by flow cytometry (). 36 hours after transfection, in most GFP-only transfected cells no damage signal was visible over the entire cell cycle, as the γH2AX signal of GFP-positive cells (green dots) overlaid perfectly with that of non-transfected GFP-negative cells from the same culture (blue dots) () and was very similar to that of mock-transfected cells (). The Cdc45-GFP expressing cells, however, accumulated γH2AX signals just after entry into S-phase (green dots in , the blue dots represent GFP-negative cells from the same culture). This is consistent with the apparent difficulties of cells with elevated Cdc45 levels to traverse S phase to reach G2. In some contrast to this, addition of the ribonucleotide reductase inhibitor hydroxyurea led to an increase of γH2AX foci over the entire S-phase and a particular accumulation of foci in early/mid-S (). From these experiments we conclude that Cdc45 exhibits its deleterious effects mainly at the G1-S transition, maybe by affecting firing of replication origins.
Figure 4. Cdc45 transfection of asynchronous growing cells leads to an increased phosphorylation of H2AX predominantly at the transition from G1 to S. γH2AX levels were determined by flow cytometry (A) in untreated non-transfected cells, (B) after addition of 20 mM hydroxyurea for 3 h to non-transfected cells, (C) 36 h after transfection of HeLa cells with a vector encoding GFP alone, and (D) 36 h after transfection with a vector encoding Cdc45-GFP. In panels (C) and (D), an equal number of GFP-positive and -negative populations from the same transfection are shown in green and blue, respectively.
Cdc45 induced disproportionate firing of replication origins
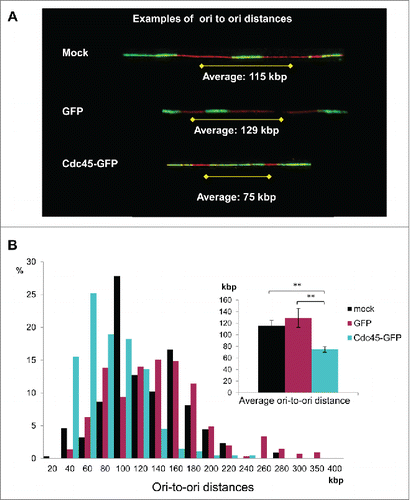
To determine whether Cdc45-GFP had any influence on the number of fired origins, DNA fiber assays were employed.Citation21 Asynchronous cells were consecutively pulse-labeled with the thymidine analogs CldU and IdU for various times, mostly 45 min each. After stretching of the fibers and immunological detection of the halogenated analogs, we identified the fraction of forks initiated during the CldU pulse and determined the distance between neighboring origins (). There appeared to be an increase in initiation events in Cdc45-GFP transfected cells ().Citation22,23 In control HeLa cells and in cells overexpressing GFP, we measured comparable inter-origin distances of 115 kb and 129 kb, respectively, which is in accordance with estimates from other groups.Citation21,24,25 In cells transfected with Cdc45-GFP this distance shrank to 75 kb (), indicating an increased firing of neighboring origins when Cdc45 was increased.
Figure 5. Cdc45 overexpression increased origin usage. (A) Representative DNA fiber spreads. Fiber assays were performed after pseudotransfection (mock) or transfection with a GFP- (GFP), or with a Cdc45-GFP-encoding vector (Cdc45), respectively. (B) The distribution of origin-to-origin distances as well as the average ori-to-ori distances (inlet) were evaluated from about 200 representative tracks by using a stretching factor of 2.59 kb/µm.Citation21
Cdc45-mediated over-firing decreased the elongation rate of replication and induced fork asymmetries
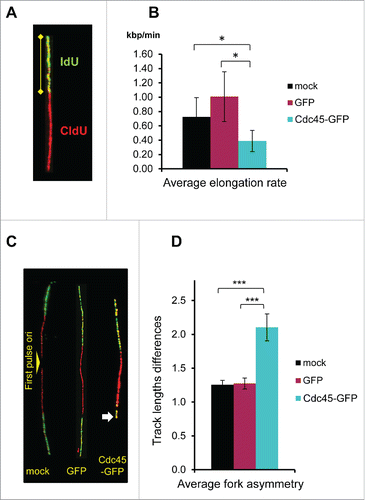
Unexpectedly, the lengths determination of IdU tracksCitation21 unveiled an about twofold decline of the progression rate of elongating forks in Cdc45-GFP transfected cells compared to its mock-transfected and GFP-alone controls (), despite a comparable fraction of elongating forks (). In addition, there was a dramatic asymmetry of elongation in Cdc45-GFP expressing cells, as shown by comparing the progression of the 2 forks from the same origin, again by using fiber assays (). Fork asymmetry indicates stalling of elongating replication forks.Citation26 Collectively, these results demonstrated that surplus Cdc45 increased the number of replication origins that became initiated at the same time, which then caused elongation problems and premature halts.
Figure 6. Diminished elongation rates and increased folk stalling in Cdc45-GFP expressing cells. (A) Depiction of a representative track displaying the method. (B) Summary of the measured elongation rates after mock transfection and transfection with a GFP- or a Cdc45-GFP-encoding vector. The elongation rates of mock-, GFP-, and Cdc45-GFP transfected cells were evaluated from about 600 representative tracks for each condition as the lengths of IdU tracks divided by the length of the IdU pulse time. (C) Depiction of representative tracks displaying the method. The shortened track in the Cdc45-GFP sample revealing fork stalling is indicated by an arrow. (D) Mock- and GFP-transfected cells displayed track-length differences of about 1.25 kb/min. After transfection with Cdc45-GFP the mean track-length difference increased to 2.1. Analysis was performed from the same set of images as used in .
Signaling in the course of Cdc45-induced S phase arrest
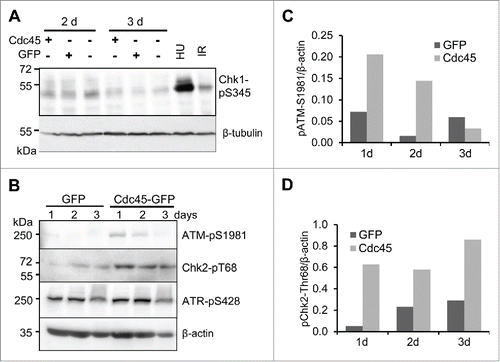
Since S phase replication control and checkpoint response is ascribed to the ATR/Chk1 protein kinase pathway, we firstly inspected this signaling cascade. While 5 h treatment with HU or irradiation with 2 Gy provoked phosphorylation of serine 345 (S345) of Chk1, no such modification was observed after Cdc45-GFP expression (). We rather observed some increase in Chk1 phosphorylation in all 3 samples 2 d after transfection, which probably reflected some replication stress. Noteworthy, this was not stronger in samples transfected with Cdc45-GFP compared to vector- or GFP-transfected cells (). Also immunofluorescence microscopy confirmed that Cdc45-GFP-transfected cells had Chk1 levels equal or lower than GFP- or mock-transfected cells (Supplemental ). Surprisingly, treatment of Cdc45-GFP-transfected cells with HU for one hour provided a normal Chk1 response, indicating that Cdc45-GFP does not compromise the response to nucleotide depletion (Supplemental ). There were also no apparent differences of phosphorylation of S428 of the upstream kinase ATR among the 3 different vectors (). However, we observed a slight but robust ATM response manifested by the phosphorylation of S1981 of ATM and T68 of Chk2 with Cdc45-GFP, but not with the controls (). Considering that only about one third of the cells were transfected to overexpress GFP or Cdc45-GFP, respectively, the magnitude of response observed in Cdc45-GFP likely represents an underestimate.
Figure 7. An excess of Cdc45 provoked an ATM- but not an ATR-mediated DNA damage response. (A) Abundant Cdc45 did not affect the phosphorylation state of Chk1 at S345. 100 000 GFP- and Cdc45-GFP-positive cells sorted by FACS were applied per well. (B) Autophosphorylation of ATM at S1981 and phosphorylation of its downstream effector kinase Chk2 at T68 were increased by Cdc45 overexpression, whereas ATR-phosphorylation at S428 was not altered. Equal amounts of cell extract were applied per well. Transfection efficiency was approximately 30% for both GFP and Cdc45-GFP. Phosphorylation levels of (C) ATM at S1981 and (D) Chk2 at T68 have been quantified relative to the β-actin protein level.
Accumulation of single-stranded DNA in Cdc45-GFP expressing cells
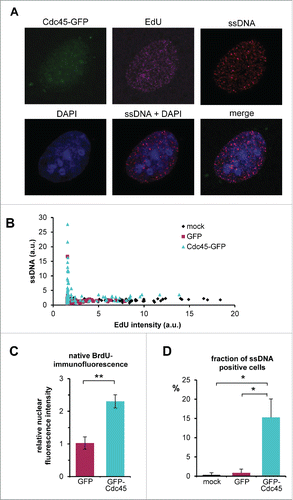
Considering the phenotype of our Cdc45-overexpression experiments we wondered why the observed difficulties in traversing S-phase, the γH2AX response and subsequent apoptosis apparently went along with ATM/Chk2 signaling but not with the expected ATR/Chk1 cascade. In a recent publication, the Lukas group demonstrated that depletion of the pool of the ssDNA binding protein RPA led to a “replication catastrophe,” which phenotypically was very similar to the effects observed here.Citation27 Since RPA is an essential component of ATR signaling,Citation28 a relative exhaustion of this factor by too many ongoing replication forks or by possible sequestration by soluble Cdc45 might have been responsible for the observed effects. We therefore asked whether Ccd45-GFP transfected cells displayed more ssDNA compared to mock or GFP transfected cells. To this end, cells were grown for 30 h in the presence of BrdU. Prior to transfection, BrdU was removed. To detect cells with ongoing DNA synthesis, EdU was added shortly before fixation. Exposure of ssDNA regions was visualized by anti-BrdU immunofluorescence without the canonically used DNA denaturing step. Cells that were transfected with GFP-Cdc45 displayed significantly more BrdU signal (), whereas transfection with GFP alone or mock transfected controls showed only residual amounts of ssDNA. Most important, most GFP-Cdc45 expressing cells with EdU-incorporation exhibited more ssDNA than the corresponding control (). Moreover, there was generally a strong negative correlation between EdU incorporation and ssBrdU exposure: The more cells actively synthesized DNA, the less ssDNA was present (). The quantitative evaluation of individual mock transfected or GFP-transfected controls revealed a largely undisturbed EdU-incorporation and only sporadic ssDNA exposure. In striking contrast, there was a pronounced ssDNA accumulation in Cdc45-GFP transfected cells (). This was particularly reflected by the average BrdU fluorescence intensity, which was increased more than 2-fold in Cdc45-GFP transfected cells, compared to GFP alone (), where about 15% of the Cdc45 transfected, but less than 2% of the GFP control cells were ssDNA-positive ().
Figure 8. Accumulation of ssDNA in Cdc45 overproducing cells. After one cell cycle in medium supplemented with BrdU, cells were transfected with vectors encoding GFP or Cdc45-GFP or empty vector. Newly synthesized DNA was labeled by a 15 min pulse of EdU immediately before fixation. ssDNA was visualized by immunofluorescence against BrdU under non-denaturating conditions (without the HCl step), and newly synthesized DNA was detected by fluorescent click conjugation of incorporated EdU. (A) Representative image of ssDNA foci in a Cdc45-GFP overexpressing cell. (B) Cdc45-induced increase of ssDNA foci as DNA synthesis decreases. Native BrdU fluorescence intensities of individual nuclei were plotted against the EdU-staining representing the DNA synthesis rate. (C) Average native nuclear BrdU fluorescence was increased in Cdc45-GFP transfected cells. Average fluorescence intensities were calculated relative to mock transfected cells (to allow comparison of multiple independent experiments). GFP-transfected cells displayed a mean fluorescence comparable to the control, whereas staining of the Cdc45-GFP transfected cells increased by a factor of two. (D) About 15 % of the Cdc45-GFP transfected cells showed elevated ssDNA levels, compared to 1-2 % of cells in the respective controls (D). At least 300 cells per treatment were analyzed. Error bars represent standard deviation between at least 3 independent experiments.
Discussion
Human Cdc45 is a helicase co-factor within the CMG complex, but its comprehensive functions during eukaryotic DNA replication are still not well understood. Upon initiation of replication, Cdc45 becomes loaded into origins, where it helps establishing the Mcm2-7 helicase function.Citation29,30 Apparently, the intracellular amount of Cdc45 is carefully controlled, since there are only 30,000 to 40,000 molcules per (growing) cell,Citation3,31,32 which is not enough to start all 250,000 preformed replication origins.Citation14-16 On the other hand, many tumor cells display a higher expression of Cdc45,Citation2,17 which may contribute a growth advantage due to a higher propensity of origin firing. Therefore, one important issue of this study was to analyze the effects of an increased number of intracellular Cdc45 molecules on cell growth.
Cdc45-GFP-transfected HeLa cells displayed a strict intranuclear localization. After transfection, cells entered the next S phase but had difficulties in reaching the following G2 state. Cells largely remained in S phase for up to 96 h. But already 24 to 36 h after transfection many cells underwent apoptosis, which became visible as a largely broadened sub-G1 peak during FACS analyses, and a dramatically increase of cleaved caspase 3 (). This was preceded by a significant increase in γH2AX formation indicative for double-strand breaks and/or disrupted replication forks.Citation33,34 Obviously, the firing of too many origins caused difficulties to initiate DNA synthesis of both strands (). But surprisingly, this did not provoke an ATR/Chk1 response, which is in contrast to replication stalling by e.g. HU-induced nucleotide depletion [() and ref. Citation35]. Commonly, ATR signaling inhibits both the initiation and elongation step of DNA replication, thereby limiting the adverse effects of fork stalling.Citation36-38 On the other hand, we did observe a significant increase in ATM/Chk2 signaling as well as accumulation of γH2AX, indicative for double-strand breaks.Citation33,34 Thus, firing of too many origins at a time provoked difficulties to complete DNA synthesis of both strands leading to subsequent fork disruption and breakage.Citation27
This scenario was directly supported by our observations of an accumulation of uncovered ssDNA in about 15% of all cells transfected with Cdc45-GFP, whereas only 1–2% of the controls displayed this type of DNA. Furthermore, the origin-to-origin distance shrank from 120 kb to about 75 kb indicating that about twice as many origins have been fired after ectopic expression of Cdc45. Comparably, a threefold increase in the number of replication foci has been observed after microinjection of Cdc45 into Chinese hamster ovary cells.Citation15 Both results confirm earlier data from Xenopus extracts and fission yeast, where Cdc45 was rate limiting for the initiation of DNA replication.Citation1,39 Intriguingly, after Cdc45 transfection the tracks indicating fork elongation were about 50% shorter than those in mock- or GFP-transfected control cells, suggesting that a twofold increase of origin usage resulted in an about twofold diminished elongation rate of individual forks.
This might have been due to an exhaustion of components of the DNA replication machinery or precursors for DNA replication, such as the deoxyribonucleotides (dNTPs).Citation40,41 Accordingly, cell treatment with the ribonucleotide reductase inhibitor HU is known to slow polymerase activity and cause γH2X formation but, different to the data reported here, activates the ATR-induced intra-S-phase checkpoint.Citation35,42 Moreover, dNTP depletion seems to be unlikely because the ribonucleotide reductase is a self-regulatory enzyme that counteracts this situation.Citation43
If not dNTP depletion, another factor may have become limiting. Because of the shortage of this component, the replication forks halted and this in turn provoked fork regression and fork collapse. Both events, however, should have been accompanied by severe ATR activation, as seen e.g., by DNA polymerase inhibition with aphidicolin (reviewed by ref. Citation44). A notable exception to this scenario is the relative depletion of RPA. RPA is required for stabilizing DNA after strand opening catalyzed by the CMG complex, and at the same time is implicated as a central component of the ATR activation cascade.Citation45 When too many origins are opened at a time, RPA may become short to cover all newly formed single-strands. This effect could be augmented if the surplus Cdc45 sequesters RPA independent of DNA (Szambowska et al. manuscript in preparation). Even though we cannot exclude that the excess ssDNA detected by native anti-BrdU staining may be in part covered by RPA, the results are consistent with a RPA exhaustion. Also the observed partial co-localization of ssDNA with sites of EdU incorporation (), which seems paradoxical, makes sense in the light of a RPA limitation. Mainly the freshly displaced lagging strands would suffer from insufficient amounts of RPACitation45. This in turn diminished both the loading,Citation46 and the catalytic rate of the DNA polymerase α-primase complex,Citation47,48 which would explain the deceleration of lagging strand DNA synthesis and the subsequent accumulation of ssDNA. At the same time, leading strand DNA synthesis should not be directly affected. RPA depletion would also explain the failure of ATR signaling. This, together with an increased amount of Cdc45 would allow the here observed initiation of novel origins elsewhere. In consequence, more and more uncovered single-strands were produced (as observed) that were prone for attacks by nucleases and hydroxyl radicals.
What then is the phenotype of RPA depletion without Cdc45 overexpression? In two studies RPA depleted cells passed rather slowly through S- and accumulated in G2-phase, where they halted and induced an ATM-, but not an ATR-dependent checkpoint response with subsequent apoptosis.Citation49,50 In the third study, an apparently stronger RPA depletion provoked a cell cycle arrest in early S-phase. Moreover, these cells accumulated stalled replication forks and double-strand breaks as indicated by γH2AX foci, and finally underwent apoptosis.Citation51 These features amazingly parallel the phenotypes of Cdc45-GFP expression. Most strikingly, the Lukas group demonstrated recently that exhaustion of the RPA pool by different means led to a “replication catastrophe” with a very similar phenotype as described here.Citation27
Recently, the Gautier group reported on Myc-induced replication stress in the Xenopus oocyte replication system that was attributed to a Myc-facilitated loading of Cdc45 into replication origins.Citation18 Addition of purified Myc-protein to Xenopus extracts approximately doubled the amount of fired replication origins and caused severe fork asymmetries indicating halted replication forks. These authors also observed γH2AX foci and an ATM response indicating stress-induced fork collapses, again confirming our data. Particularly the recent findings put Cdc45 into a regulatory network of oncogene expression and carcinogenesis. Indeed, there exist several studies that describe overexpressed Cdc45 in human cancer cells.Citation2,17,19,20 How can this be accomplished when too much Cdc45 causes cell death? Since tumor cells are usually selected to divide rapidly, it is of advantage to start as many replication origins as possible. This study shows that Cdc45 can become rate limiting for initiation of replication. Therefore, an overexpression of this protein in tumor cells comes as no surprise. This however works only up to a 2-to-3-fold excess,Citation15 as long as no other factor becomes limiting. Interestingly, an elevated expression of RPA has been widely observed in many malignant tumors (see e.g.Citation52-54). Therefore, it will be of interest to find out whether Cdc45-overproducing tumors also overproduce RPA. Is it then possible to further overexpress Cdc45 in cancer cells to drive them into apoptosis? Since Cdc45 expression is largely under the control of the E2F promoter,Citation55 this might be technically difficult. However, the Cdc45 protein contains several PEST and KEN boxes that facilitate cell cycle-specific degradation.Citation31 Indeed, the half-life of Cdc45 has been determined to be about 10 h, just enough for one passage through S phase.Citation2 Therefore, inhibiting ubiquitylation and/or degradation of Cdc45 might be a promising strategy for combatting cancer.
Materials and methods
Cloning and vectors
For the preparation of vector pCdc45-eGFP, the complete ORF of human Cdc45 was amplified using primers Cdc45-1F_Xho (5′-GAC CTC GAG ATG TTC GTG TCC GAT TTT CC) and Cdc45R_N1Bam (5′-GAC GGA TCC CGG GAC AGG AGG GAA ATA AG). The PCR product was then cloned into the XhoI and BamHI sites of vector peGFP-N1 (Life Technologies, Darmstadt, Germany). The vector constitutively expresses Cdc45 with a C-terminal eGFP fluorescent protein tag (Cdc45-GFP) in mammalian cells. For the preparation of vector pcDNA4/TO-Cdc45HA, the complete ORF of human Cdc45 was amplified using primers Cdc45_Bam_for (5′-GAC GGA TGG GCC ACC ATG TTC GTG TCC GAT TTC C) and Cdc45HA_Xho_rev (5′-GAC CTC GAG CTA AGC GTA GTG TGG GAC GTC GTA TGG GTA GGA CAG). The PCR product was then cloned into the BamHI and XhoI sites of vector pcDNA4/TO (Life Technologies). The vector expresses Cdc45 with a C-terminal haemagglutinin (HA) tag under the control of a tetracycline inducible vector. All used expression vectors were verified by Sanger sequencing before use.
Cell culture
HeLa (ATCC-CCL-2) were cultivated in Dulbecco's Modified Eagle's Medium supplemented with 10 % fetal bovine serum (PAN Biotech GmbH, Aidenbach, Germany) at 37°C, with 5% CO2 and 95% relative humidity. Six-well plates were seeded with 2×105 cells per well and incubated overnight. For transient transfection, cells were transfected using FuGene HD according to the instructions of the manufacturer (Roche Applied Science, Mannheim, Germany) and incubated for the times indicated. Where indicated cells were incubated with 10 µM EdU 15 min before harvesting.
Flow cytometry
Cells were trypsinized, washed with phosphate buffered saline (PBS), fixed with 4% formaldehyde for 10 min at room temperature (RT) and then stored at −20°C in 70% ethanol. After washing with PBS, the cells were permeabilized with 0.2% Triton X-100 (Sigma-Aldrich, Steinheim, Germany) in PBS for 2 min at RT and blocked with 5 % (v/v) goat serum (Vector Laboratories, Burlingame) in PBS for 1 h followed by incubation with the primary antibody in PBS supplemented with 0.1% triton X-100 and 1% BSA (TPB) for 2 h at RT (see for a list of antibodies used). The cells were washed 3 times with PBS containing 1% BSA and incubated for 1 h at RT with appropriate secondary antibodies in TPB followed by washes as above. EdU detection was performed with the Click-iT® EdU Alexa Fluor® 647 Flow Cytometry Assay Kit according to the instructions of the manufacturer (Life Technologies, Darmstadt, Germany). DNA was visualized with 4′,6-diamidino-2-phenylindole (DAPI, Sigma-Aldrich) or Cell Cycle 405 Blue (Life Technologies) dyes. Flow cytometry was performed with a FACSCanto flow cytometer equipped with violet, blue and red lasers, using the FACSDiva software (BD Biosciences, Heidelberg, Germany) and FlowJo (FlowJo, LLC, Ashland/Oregon, USA).
Table 1. Antibodies used in this study.
Immunofluorescence microscopy
Cells grown on glass cover slips were washed with PBS, fixed with 4% formaldehyde and stored in PBS. For immunostaining, the cells were permeabilized with 0.2% Triton X-100 (Sigma-Aldrich) in PBS for 2 min at RT and blocked with 5 % (v/v) goat serum (Vector Laboratories, Burlingame) in PBS for 1 h followed by incubation with the primary antibody in block buffer for 2 h at RT. The cells were washed 3 times with PBS and incubated for 1 h at RT with appropriate secondary antibodies in block buffer, followed by PBS washes. EdU detection was performed by click chemistry as described. Cells were then mounted in VectorShield containing DAPI (Vector Laboratories, Burlingame, USA). Immunofluorescence studies were performed with an Axio Imager Z1 Apotome and the Axiovision software (Carl Zeiss Jena, Germany).
Labeling of DNA replication sites and DNA fiber assays
Twenty-four h after transfection with GFP or Cdc45-GFP encoding vectors, HeLa CCL2 cells were grown in DMEM supplemented with 25 µM iodo-deoxyuridine (IdU, Sigma-Aldrich) for 30 or 45 min. After the first pulse labeling, the medium was removed and fresh DMEM containing 250 µM chloro-deoxyuridine (CldU, Sigma-Aldrich) was added for the second pulse label. Preparation of DNA spreads was done as describedCitation21 with minor modifications. Briefly, after labeling with halogenated nucleotide analogs and resuspension in PBS the cells were washed with ice-cold PBS and diluted to 5×105 cells/ml. Approximately 1000 cells (2 µl) were lysed on glass slides by addition of 7 µl lysis buffer (200 mM Tris-HCl pH 7.4, 50 mM EDTA, 0.5 % SDS). DNA spreads were generated by slightly tilting the microscope slide and fixed using methanol/acetic acid (3:1) after air drying.
DNA spreads were denatured by incubation with 2.5 M HCl for 75 min at RT. The microscope slides were washed with PBS and blocked with TPB for 1 h. After incubation with a monoclonal rat anti-BrdU antibody (Clone BU1/75, 1:1000) in TPB for 1 h RT, which binds CldU but not IdU, the slides were washed twice with PBS and once with TPB and subsequently fixed using 4 % paraformaldehyde in PBS (10 min, RT), washed again twice with PBS and 3 times with TPB and incubated with goat anti-rat antibody conjugated with Alexa Fluor® 555 (Life Technologies, 1:500 in TPB) for 2 h at RT. DNA spreads were washed again as described above and incubated overnight at 4° C with a monoclonal mouse anti-BrdU antibody (Clone 44, 1:1500) which binds IdU, but not CldU. After washing, an Alexa Fluor® 488 conjugated goat anti-mouse antibody (Life Technologies, 1:500 in TPB) was added for 2 h at RT. Then the slides were extensively washed again and mounted. Pictures of fluorescent immunolabeled DNA spreads were taken using an Axio Imager.Z1 at 400-fold magnification.
Detection of ssDNA
Visualization of ssDNA was performed as described.Citation56 Genomic DNA of HeLa or U2OS cells (the latter gave lesser backgrounds) was halogenated by growing in DMEM supplemented with 10 µM BrdU for 30 h. BrdU was removed by addition of fresh DMEM before transfection. After 24 h, the replicating cells were pulse-labeled with 10 µM EdU for 15 min, then fixed with methanol for 30 min at −20°C, and shortly rinsed with cold acetone. Inc. BrdU exposed in the single-stranded template strand was detected with a monoclonal rat-anti-BrdU antibody (Clone BU1/75) in TPB for 1 h without a DNA denaturing step, and anti-rat-IgG antibody-Alexa Fluor® 555 conjugate as described above. Newly synthesized DNA was stained using the Click-iT® EdU imaging kit with Alexa Fluor® 647 azide (Life Technologies) according to the manufactures instructions. DNA was counterstained with DAPI. Images were taken using the Zeiss Axio Imager.Z1 at 630-fold magnification.
Data analysis
DNA fibers were analyzed using ImageJ 1.46r (http://imagej.nih.gov/ij). The pixel to µm ratio was determined and DNA-track lengths were calculated using a DNA stretching factor of 2.59 kbp/µm as described.Citation21 Nuclear fluorescence intensities for ssDNA were quantified with ImageJ 1.46r using DAPI staining to determine region of interest. At least 3 independent experiments were performed; the given error bars represent standard deviation of the mean values. Significances were determined using 2-tailed Student's t-test.
Cell extract preparation and Western analysis
For Western analysis, cells were lysed directly into Laemmli buffer followed by a brief sonication. Proteins were separated by SDS- polyacrylamide gel electrophoresis and transferred onto an Immobilon P PVDF membrane (Merck Millipore, Billerica, USA). Membranes were blocked for at least one hour with 5% skim milk in Tris-buffered saline with Tween 20 (TBST, 10mM Tris–HCl, pH 7.5, 150mM NaCl and 0.5% Tween 20) and washed thrice with TBST, followed by incubation with the indicated primary antibody () for at least 2 h. After washing 3 times with TBST, the membranes were incubated with horseradish peroxidase–conjugated secondary antibody () for 1 h at RT. Immunoblots were developed with SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific, Rockford, USA) as described by the manufacturer, using a gel documentation device (G-box, Syngene, Cambridge, UK) or film exposure (Hyperfilm ECL, GE Healthcare, Freiburg, Germany).
Abbreviations
| AAA+ | = | ATPase associated with various activities |
| ATM | = | ataxia telangiectasia mutated |
| ATR | = | ATM and Rad3 related |
| BrdU | = | Bromodeoxyuridine |
| Cdc45 | = | cell division cycle protein 45 |
| Chk1 | = | Checkpoint kinase 1 |
| Chk2 | = | Checkpoint kinase 2 |
| CldU | = | chloro-deoxyuridine |
| CMG | = | Cdc45-Mcm2-7-GINS |
| DAPI | = | 4′,6-diamidino-2-phenylindole |
| EdU | = | ethynyl deoxyuridine |
| γH2AX | = | phosphorylated histone H2AX |
| GFP | = | green fluorescent protein |
| HA | = | haemagglutinin |
| HU | = | hydroxyurea |
| IdU | = | iodo-deoxyuridine |
| Mcm | = | minichromosome maintenance |
| PBS | = | phosphate buffered saline |
| RPA | = | replication protein A |
| RT | = | room temperature |
| ssDNA | = | single-stranded DNA |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Author contributions
CK and DK performed all the experiments presented in the manuscript, ACP and KB contributed to the fiber analysis, and AF contributed to immunofluorescence experiments. HP and FG are responsible for this study and contributed to the design and interpretation of the experiments. All authors contributed to the writing of the manuscript.
Supplementary Files
Download Zip (6.5 MB)Acknowledgments
The authors are grateful to A. Gleiche for technical assistance.
Funding
This study was supported by UFA Grant 3610S30016 by the German Federal Office for Radiation Protection as part of the “Kompetenzverbund für Strahlenforschung.” The Fritz Lipmann Institute (FLI) is member of the Science Association ‘Gottfried Wilhelm Leibniz’ (WGL) and is financially supported by the Federal Government of Germany and the State of Thuringia.
References
- Wu PY, Nurse P. Establishing the program of origin firing during S phase in fission Yeast. Cell 2009; 136:852-64; PMID:19269364; http://dx.doi.org/10.1016/j.cell.2009.01.017
- Pollok S, Bauerschmidt C, Sänger J, Nasheuer H-P, Grosse F. Human Cdc45 is a proliferation-associated antigen. FEBS J 2007; 274:3669-84; PMID:17608804; http://dx.doi.org/10.1111/j.1742-4658.2007.05900.x
- Broderick R, Ramadurai S, Tóth K, Togashi DM, Ryder AG, Langowski J, Nasheuer HP. Cell cycle-dependent mobility of Cdc45 determined in vivo by fluorescence correlation spectroscopy. PLoS One 2012; 7:e35537; PMID:22536402; http://dx.doi.org/10.1371/journal.pone.0035537
- Aparicio OM, Stout AM, Bell SP. Differential assembly of Cdc45p and DNA polymerases at early and late origins of DNA replication. Proc Natl Acad Sci USA 1999; 96:9130-5; http://dx.doi.org/10.1073/pnas.96.16.9130
- Aparicio OM. Location, location, location: it's all in the timing for replication origins. Genes Dev 2013; 27:117-28; PMID:23348837; http://dx.doi.org/10.1101/gad.209999.112
- Costa A, Ilves I, Tamberg N, Petojevic T, Nogales E, Botchan MR, Berger JM. The structural basis for MCM2-7 helicase activation by GINS and Cdc45. Nat Struct Mol Biol 2011; 18:471-7; PMID:21378962; http://dx.doi.org/10.1038/nsmb.2004
- Boos D, Frigola J, Diffley JF. Activation of the replicative DNA helicase: breaking up is hard to do. Curr Opin Cell Biol 2012; 24:423-30; http://dx.doi.org/10.1016/j.ceb.2012.01.011
- Heller RC, Kang S, Lam WM, Chen S, Chan CS, Bell SP. Eukaryotic origin-dependent DNA replication in vitro reveals sequential action of DDK and S-CDK kinases. Cell 2011; 146:80-91; http://dx.doi.org/10.1016/j.cell.2011.06.012
- Zou L, Stillman B. Assembly of a complex containing Cdc45p, replication protein A, and Mcm2p at replication origins controlled by S-phase cyclin-dependent kinases and Cdc7p-Dbf4p kinase. Mol Cell Biol 2000; 20:3086-96; PMID:10757793; http://dx.doi.org/10.1128/MCB.20.9.3086-3096.2000
- Langston LD, Zhang D, Yurieva O, Georgescu RE, Finkelstein J, Yao NY, Indiani C, O'Donnell ME. CMG helicase and DNA polymerase epsilon form a functional 15-subunit holoenzyme for eukaryotic leading-strand DNA replication. Proc Natl Acad Sci U S A 2014; 111:15390-5; PMID:25313033; http://dx.doi.org/10.1073/pnas.1418334111
- Sengupta S, van Deursen F, de Piccoli G, Labib K. Dpb2 integrates the leading-strand DNA polymerase into the eukaryotic replisome. Curr Biol 2013; 23:543-52; PMID:23499531; http://dx.doi.org/10.1016/j.cub.2013.02.011.
- Galal WC, Kang YH, Hurwitz J. Establishing the human rolling circle reaction. Cell Cycle 2012; 11:2771-2; PMID:22833048; http://dx.doi.org/10.4161/cc.21258
- Bauerschmidt C, Pollok S, Kremmer E, Nasheuer H-P, Grosse F. Interactions of human Cdc45 with the Mcm2-7 complex, the GINS complex, and DNA polymerases delta and epsilon during S phase. Genes Cells 2007; 12:745-58; PMID:17573775
- Besnard E, Babled A, Lapasset L, Milhavet O, Parrinello H, Dantec C, Marin JM, Lemaitre JM. Unraveling cell type-specific and reprogrammable human replication origin signatures associated with G-quadruplex consensus motifs. Nat Struct Mol Biol 2012; 19:837-44; http://dx.doi.org/10.1038/nsmb.2339
- Wong PG, Winter SL, Zaika E, Cao TV, Oguz U, Koomen JM, Hamlin JL, Alexandrow MG. Cdc45 limits replicon usage from a low density of preRCs in mammalian cells. PLoS One 2011; 6:e17533; PMID:21390258; http://dx.doi.org/10.1371/journal.pone.0017533
- Cayrou C, Coulombe P, Puy A, Rialle S, Kaplan N, Segal E, Méchali M. New insights into replication origin characteristics in metazoans. Cell Cycle 2012; 11:658-67; PMID:22373526; http://dx.doi.org/10.4161/cc.11.4.19097
- Tomita Y, Imai K, Senju S, Irie A, Inoue M, Hayashida Y, Shiraishi K, Mori T, Daigo Y, Tsunoda T, et al. A novel tumor-associated antigen, cell division cycle 45-like can induce cytotoxic T-lymphocytes reactive to tumor cells. Cancer Sci 2011; 102:697-705; PMID:21231984; http://dx.doi.org/10.1111/j.1349-7006.2011.01865.x
- Srinivasan SV, Dominguez-Sola D, Wang LC, Hyrien O, Gautier J. Cdc45 Is a Critical Effector of Myc-Dependent DNA Replication Stress. Cell Rep 2013; 3:629-39; http://dx.doi.org/10.1016/j.celrep.2013.04.002
- Di Paola D, Zannis-Hadjopoulos M. Comparative analysis of pre-replication complex proteins in transformed and normal cells. J Cell Biochem 2012; 113:1333-47; PMID:22134836; http://dx.doi.org/10.1002/jcb.24006
- Li JN, et al. mRNA expression of the DNA replication-initiation proteins in epithelial dysplasia and squamous cell carcinoma of the tongue. BMC Cancer 2008; 8:395; PMID:18194582; http://dx.doi.org/10.1186/1471-2407-8-395
- Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol 1998; 140:1285-95; PMID:9508763; http://dx.doi.org/10.1083/jcb.140.6.1285
- Conti C, Seiler JA, Pommier Y. The mammalian DNA replication elongation checkpoint: implication of Chk1 and relationship with origin firing as determined by single DNA molecule and single cell analyses. Cell Cycle 2007; 6:2760-7; PMID:17986860; http://dx.doi.org/10.4161/cc.6.22.4932.
- Petermann E, Woodcock M, Helleday T. Chk1 promotes replication fork progression by controlling replication initiation. Proc Natl Acad Sci U S A 2010; 107:16090-5; http://dx.doi.org/10.1073/pnas.1005031107.
- Conti C, Saccà B, Herrick J, Lalou C, Pommier Y, Bensimon A. Replication fork velocities at adjacent replication origins are coordinately modified during DNA replication in human cells. Mol Biol Cell 2007; 18:3059-67; http://dx.doi.org/10.1091/mbc.E06-08-0689.
- Estefania MM, Ganier O, Hernández P, Schvartzman JB, Mechali M, Krimer DB. DNA replication fading as proliferating cells advance in their commitment to terminal differentiation. Sci Rep 2012; 2:279; PMID:22359734; http://dx.doi.org/10.1038/srep00279.
- Marheineke K, Hyrien O, Krude T. Visualization of bidirectional initiation of chromosomal DNA replication in a human cell free system. Nucleic Acids Res 2005; 33:6931-41; PMID:16332696; http://dx.doi.org/10.1093/nar/gki994.
- Toledo LI, Altmeyer M, Rask MB, Lukas C, Larsen DH, Povlsen LK, Bekker-Jensen S, Mailand N, Bartek J, Lukas J. ATR Prohibits Replication Catastrophe by Preventing Global Exhaustion of RPA. Cell 2013; 155:1088-103; PMID:24267891; http://dx.doi.org/10.1016/j.cell.2013.10.043.
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 2003; 300:1542-8; PMID:12791985; http://dx.doi.org/10.1126/science.1083430.
- Masuda T, Mimura S, Takisawa H. CDK- and Cdc45-dependent priming of the MCM complex on chromatin during S-phase in Xenopus egg extracts: possible activation of MCM helicase by association with Cdc45. Genes Cells 2003; 8:145-61; PMID:12581157; http://dx.doi.org/10.1046/j.1365-2443.2003.00621.x.
- Gambus A, Khoudoli GA, Jones RC, Blow JJ. MCM2-7 form double hexamers at licensed origins in Xenopus egg extract. J Biol Chem 2011; 286:11855-64; PMID:21282109; http://dx.doi.org/10.1074/jbc.M110.199521.
- Pollok S, Grosse F. Cdc45 degradation during differentiation and apoptosis. Biochem Biophys Res Commun 2007; 362:910-5; PMID:17767920; http://dx.doi.org/10.1016/j.bbrc.2007.08.069.
- Takaya J, Kusunoki S, Ishimi Y. Protein interaction and cellular localization of human CDC45. J Biochem 2013; 153:381-8; PMID:23364835; http://dx.doi.org/10.1093/jb/mvt004.
- Kinner A, Wu W, Staudt C, Iliakis G. Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res 2008; 36:5678-94; PMID:18772227; http://dx.doi.org/10.1093/nar/gkn550.
- Lukas J, Lukas C, Bartek J. More than just a focus: The chromatin response to DNA damage and its role in genome integrity maintenance. Nat Cell Biol 2011; 13:1161-9; http://dx.doi.org/10.1038/ncb2344.
- Wei F, Xie Y, He L, Tao L, Tang D. ERK1 and ERK2 kinases activate hydroxyurea-induced S-phase checkpoint in MCF7 cells by mediating ATR activation. Cell Signal 2011; 23:259-68; PMID:20840867; http://dx.doi.org/10.1016/j.cellsig.2010.09.010.
- Merrick CJ, Jackson D, Diffley JF. Visualization of altered replication dynamics after DNA damage in human cells. J Biol Chem 2004; 279:20067-75; http://dx.doi.org/10.1074/jbc.M400022200.
- Seiler JA, Conti C, Syed A, Aladjem MI, Pommier Y. The intra-S-phase checkpoint affects both DNA replication initiation and elongation: single-cell and -DNA fiber analyses. Mol Cell Biol 2007; 27:5806-18; http://dx.doi.org/10.1128/MCB.02278-06.
- Chastain PD, 2nd, Heffernan TP, Nevis KR, Lin L, Kaufmann WK, Kaufman DG, Cordeiro-Stone M. Checkpoint regulation of replication dynamics in UV-irradiated human cells. Cell Cycle 2006; 5:2160-7; PMID:16969085; http://dx.doi.org/10.4161/cc.5.18.3236.
- Edwards MC, Tutter AV, Cvetic C, Gilbert CH, Prokhorova TA, Walter JC. MCM2-7 complexes bind chromatin in a distributed pattern surrounding the origin recognition complex in Xenopus egg extracts. J Biol Chem 2002; 277:33049-57; PMID:12087101; http://dx.doi.org/10.1074/jbc.M204438200.
- Tanaka S, Nakato R, Katou Y, Shirahige K, Araki H. Origin association of Sld3, Sld7, and Cdc45 proteins is a key step for determination of origin-firing timing. Curr Biol 2011; 21:2055-63; http://dx.doi.org/10.1016/j.cub.2011.11.038.
- Bester AC, Roniger M, Oren YS, Im MM, Sarni D, Chaoat M, Bensimon A, Zamir G, Shewach DS, Kerem B. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell 2011; 145:435-46; http://dx.doi.org/10.1016/j.cell.2011.03.044.
- Sabatinos SA, Green MD, Forsburg SL. Continued DNA synthesis in replication checkpoint mutants leads to fork collapse. Mol Cell Biol 2012; 32:4986-97; http://dx.doi.org/10.1128/MCB.01060-12.
- Reichard P. Ribonucleotide reductases: substrate specificity by allostery. Biochem Biophys Res Commun 2010; 396:19-23; PMID:20494104; http://dx.doi.org/10.1016/j.bbrc.2010.02.108.
- Melo J, Toczyski D. A unified view of the DNA-damage checkpoint. Curr Opin Cell Biol 2002; 14:237-45; PMID:11891124; http://dx.doi.org/10.1016/S0955-0674(02)00312-5.
- Wu X, Shell SM, Zou Y. Interaction and colocalization of Rad9/Rad1/Hus1 checkpoint complex with replication protein A in human cells. Oncogene 2005; 24:4728-35; PMID:15897895; http://dx.doi.org/10.1038/sj.onc.1208674.
- Mimura S, Masuda T, Matsui T, Takisawa H. Central role for cdc45 in establishing an initiation complex of DNA replication in Xenopus egg extracts. Genes Cells 2000; 5:439-52; PMID:10886370; http://dx.doi.org/10.1046/j.1365-2443.2000.00340.x.
- Tsurimoto T, Stillman B. Multiple replication factors augment DNA synthesis by the two eukaryotic DNA polymerases, a and d. EMBO J 1989; 8:3883-9; PMID:2573521.
- Braun KA, Lao Y, He Z, Ingles CJ, Wold MS. Role of protein-protein interactions in the function of replication protein A (RPA): RPA modulates the activity of DNA polymerase a by multiple mechanisms. Biochemistry 1997; 36:8443-54; PMID:9214288; http://dx.doi.org/10.1021/bi970473r.
- Dodson GE, Shi Y, Tibbetts RS. DNA replication defects, spontaneous DNA damage, and ATM-dependent checkpoint activation in replication protein A-deficient cells. J Biol Chem 2004; 279:34010-4; http://dx.doi.org/10.1074/jbc.C400242200.
- Araya R, Hirai I, Meyerkord CL, Wang HG. Loss of RPA1 induces Chk2 phosphorylation through a caffeine-sensitive pathway. FEBS Lett 2005; 579:157-61; http://dx.doi.org/10.1016/j.febslet.2004.11.066.
- Haring SJ, Mason AC, Binz SK, Wold MS. Cellular functions of human RPA1. Multiple roles of domains in replication, repair, and checkpoints. J Biol Chem 2008; 283:19095-111; http://dx.doi.org/10.1074/jbc.M800881200.
- Levidou G, Gakiopoulou H, Kavantzas N, Saetta AA, Karlou M, Pavlopoulos P, Thymara I, Diamantopoulou K, Patsouris E, Korkolopoulou P. Prognostic significance of replication protein A (RPA) expression levels in bladder urothelial carcinoma. BJU Int 2011; 108:E59-65; PMID:21062395; http://dx.doi.org/10.1111/j.1464-410X.2010.09828.x.
- Givalos N, Gakiopoulou H, Skliri M, Bousboukea K, Konstantinidou AE, Korkolopoulou P, Lelouda M, Kouraklis G, Patsouris E, Karatzas G. Replication protein A is an independent prognostic indicator with potential therapeutic implications in colon cancer. Mod Pathol 2007; 20:159-66; PMID:17361204; http://dx.doi.org/10.1038/modpathol.3800719.
- Tomkiel JE, Alansari H, Tang N, Virgin JB, Yang X, VandeVord P, Karvonen RL, Granda JL, Kraut MJ, Ensley JF, et al. Autoimmunity to the M(r) 32,000 subunit of replication protein A in breast cancer. Clin Cancer Res 2002; 8:752-8; PMID:11895905.
- Arata Y, Fujita M, Ohtani K, Kijima S, Kato JY. Cdk2-dependent and -independent pathways in E2F-mediated S phase induction. J Biol Chem 2000; 275:6337-45; PMID:10692433; http://dx.doi.org/10.1074/jbc.275.9.6337.
- Raderschall E, Golub EI, Haaf T. Nuclear foci of mammalian recombination proteins are located at single-stranded DNA regions formed after DNA damage. Proc Natl Acad Sci U S A 1999; 96:1921-6; PMID:10051570; http://dx.doi.org/10.1073/pnas.96.5.1921.
