ABSTRACT
Unscheduled DNA synthesis (UDS) is the final stage of the process of repair of DNA lesions induced by UVC. We detected UDS using a DNA precursor, 5-ethynyl-2′-deoxyuridine (EdU). Using wide-field, confocal and super-resolution fluorescence microscopy and normal human fibroblasts, derived from healthy subjects, we demonstrate that the sub-nuclear pattern of UDS detected via incorporation of EdU is different from that when BrdU is used as DNA precursor. EdU incorporation occurs evenly throughout chromatin, as opposed to just a few small and large repair foci detected by BrdU. We attribute this difference to the fact that BrdU antibody is of much larger size than EdU, and its accessibility to the incorporated precursor requires the presence of denatured sections of DNA. It appears that under the standard conditions of immunocytochemical detection of BrdU only fragments of DNA of various length are being denatured. We argue that, compared with BrdU, the UDS pattern visualized by EdU constitutes a more faithful representation of sub-nuclear distribution of the final stage of nucleotide excision repair induced by UVC. Using the optimized integrated EdU detection procedure we also measured the relative amount of the DNA precursor incorporated by cells during UDS following exposure to various doses of UVC. Also described is the high degree of heterogeneity in terms of the UVC-induced EdU incorporation per cell, presumably reflecting various DNA repair efficiencies or differences in the level of endogenous dT competing with EdU within a population of normal human fibroblasts.
Introduction
UVC (100–280 nm) induces DNA damage in the form of pyrimidine dimers and photoproducts. This type of damage is repaired by nucleotide excision repair (NER) pathway. In this report we focus on optimal detection and visualization of the sites of repair of UVC-induced damage and effectiveness of the repair process. In previous work done by other laboratories the sites of damage were visualized by immunofluorescence detection of pyrimidine dimers and photoproducts,Citation1,2 while the regions of active DNA repair were imaged by detecting recruitment and transient accumulation of NER repair factors.Citation1,3,4 The final stage of the repair process, unscheduled DNA synthesis, was detected by fluorescent labeling of newly incorporated DNA precursor analogs. Thus far most of the research aimed at detecting UDS employed tritiated thymidineCitation5,6 or 5-bromo-2′-deoxyuridine (BrdU).Citation7-9 Recently 5-ethynyl-2′-deoxyuridine (EdU)Citation10,11 was also used.Citation12,13 We have now revisited the issue of UDS detection with EdU, using widefield, confocal and super-resolution (sub-diffraction) fluorescence microscopy. Using normal human fibroblasts, derived from healthy subjects and grown under optimal conditions, we demonstrate that the sub-nuclear pattern of UDS detected via incorporation of EdU is different from the pattern revealed by BrdU. Specifically, EdU incorporation occurs all throughout the chromatin, as a uniformly distributed signal or a large number of very small foci, best detected by super-resolution imaging. In contract, just a few distinct repair foci are detected by confocal imaging of the incorporated BrdU.
We ascribe this difference to the fact that DNA denaturation (strand separation), which is required to ensure access of the BrdU antibody to the incorporated BrdU, is only partial. Unlike BrdU antibody, the click-reaction labeling of the incorporated EdU is not affected by the steric hindrance imposed by dsDNA. Moreover, denaturation of DNA that involves the harsh conditions of acid (or heat) treatment, and DNA strand separation, significantly alter nuclear structure, whereas the procedure of EdU labeling is expected to have lesser effect on chromatin organization. Given the above we argue that the UDS pattern visualized via EdU detection, when a sensitive image detector is used, constitutes a more accurate representation of sub-nuclear distribution of the final stage of NER in a process of repairing damage induced by UVC. Using the optimized EdU detection procedure we also measured the relative amounts of DNA precursors incorporated by cells during the process of UDS following exposure to various doses of UVC, presumably reflecting various repair efficiencies within a population of normal human fibroblasts recovering from UVC-induced damage.
Materials and methods
Cell culture and UV exposures
Secondary cultures of human fibroblasts (HSF) were cultured in Dulbecco's Modified Eagle's Medium (DMEM, cat. no D5523, Sigma-Aldrich) supplemented with 10% FBS (cat. no F7524, Sigma Aldrich), in a humidified atmosphere of 95% air and 5% CO2 (i.e. approx. 20% oxygen) at 37°C until the 15th cell passage. The cells were maintained in T-25 flasks and subcultured every 7 or 8 d using 0.25% trypsin solution. Two to 4 d before the experiment cells were seeded on round coverslips (0,17 mm thickness, 22 mm in diameter, Menzel-Glaser) and placed in Petri dishes (35 mm diameter). Prior to use the coverslips were degreased and sterilized by autoclaving.
UV was delivered by a Philips TUV PL-S 5 W/2P lamp, emitting at 254 nm, placed in a standard cell culture incubator (without CO2 control). The lamp delivered 10 W/m2s, measured 20 cm from the lamp. Cells were exposed to UVC for 3 – 300 s. During UVC exposure cells were maintained on coverslips, in a 35 mm diameter Petri dish, containing 1 ml of DMEM supplemented with 10% FBS, inside an incubator (37°C). Culture medium supplemented with EdU or BrdU was added to Petri dishes immediately following the UVC exposure.
Detection of UVC-induced UDS by EdU incorporation
In all experiments confluent fibroblast cultures at a plateau phase of growth were used. During exposure to UVC (254 nm) cells were maintained in 1 ml of DMEM supplemented with 10% FBS. Immediately after irradiation the cells were incubated with 20 µM EdU (C10338, Invitrogen) for 2 h under standard growth conditions. Cells were then washed with warm PBS, fixed (15 min) in 4% formaldehyde (EMS, Hatfield, PA), permeabilized (15 min) with 0,1% Triton X-100 (Sigma) and rinsed with 3% BSA (Sigma). EdU incorporated in DNA was detected by either of the 2 methods: (i) using fluorescent-azide coupling reaction (Click-iT, Invitrogen) for 30 min, according to the instructions provided by manufacturer, or (b) using the following procedure: rinsing with PBS containing 50 mM glycine and 50 mM NH4Cl for 5 min, washing with PBS, incubating with a freshly prepared staining mixture of a selected AlexaFluor azide, CuSO4, and sodium ascorbate. Azide-conjugated AlexaFluor 488 (C10337, Invitrogen) or 647 (A10277, Life Technologies) was used. For DNA staining DRAQ5 (5 μM, 10 min; DR50200, Biostatus) or YOYO-1 (0,1 nM, 30 min; Y3601, Life Technologies) was used. All procedures were carried out at room temperature, unless otherwise stated.
Detection of UVC-induced UDS by BrdU incorporation
Following exposure to UVC cells were incubated with BrdU (10 µM; B23151, Invitrogen) for 2 h under standard growth conditions. Subsequently cells were washed with warm PBS, fixed (15 min) in 4% formaldehyde (EMS, Hatfield, PA), permeabilized (15 min) with 0,1% Triton X-100 (Sigma) and blocked with 3% BSA (Sigma) for 1 h. DNA was denatured using 4 M hydrochloric acid for 20 min at room temperature. Cells were incubated with primary (1 h) followed by a secondary antibody (1 h). Antibody dilutions were prepared in 0.1% BSA in PBS. The primary antibody was mouse anti-BrdU (1:100; B35128, Invitrogen). Secondary antibodies were goat anti-mouse Alexa Fluor 488 (1:1000; A11029, Invitrogen) or 647 (1:1000; A-21245, Life Technologies). DNA staining procedure was performed as described in section 2.2.
Detection of UVC-induced UDS by EdU incorporation
Imaging of sites of UDS
Confocal microscopy
A Leica SP5 SMD confocal microscope (Leica Microsystems) was used. The imaging conditions were: 63x HCX PL APO CS NA 1.4 oil immersion lens, excitation 488 (Ar+) and 633 nm (HeNe); emission detection bands 500–580 nm for Alexa Fluor 488 (Click-iT EdU or immunofluorescence - anti-BrdU) and 560–750 nm for DRAQ5; registration in sequential mode; 5 frames averaged. To compare the pattern of EdU incorporation into DNA during UDS images were collected using a photomultiplier (PMT) or an avalanche photodiode (APD) as light detectors. A wide-field fluorescence microscope (Leica DMI 3000B) equipped with an Andor iXon EMCCD camera was also used.
Super-resolution microscopy
The custom-made single molecule detection system (SMLM) used for one color imagingCitation14,15 was equipped with 491 nm laser excitation (Cobolt AB), focused through 63x, 1.4 NA objective lens (Leica Microsystems) and optionally collimated using a telescope system in order to expose a round area of the sample (diameter of ∼23 µm in imaging plane) to the high intensity excitation light necessary for induction of blinking of fluorophore molecules. Fluorescence was filtered using 500 – 550 nm emission bandpass (Semrock) and projected onto a CCD Camera (PCO Sensicam, Germany, effective pixel size 102 nm). Positions of the registered molecules were established using a center of the gravity fitting procedure, preceded by setting an appropriate threshold and background. Rendering of the super-resolution images was done by plotting the position of each single event as a Gaussian spot with standard deviation corresponding to the calculated precision of localization. Prior to rendering of the final image, a filter was applied to reject unsuitable events on the basis of brightness and spot dimension.
For single color localization microscopy imaging of EdU incorporated into UDS sites, a click reaction Alexa488 with the incorporated precursor was performed, followed by embedding the sample in ProlongGold® (Life Technologies). The quality of SMLM reconstructions was compared with images of microtubules labeled with secondary antibodies conjugated with Alexa488, used here as standards. ProlongGold® outperformed the switching bufferCitation16 as it yielded images of better quality and comparable single molecule localization density within a shorter time of acquisition. The average photon count in ProlongGold was higher than in the switching buffer (2482 photons versus only 1080 when detecting Alexa488) therefore, a higher structural resolution was achieved.Citation17 The imaging system and data collection parameters are described in detail inCitation18 and.Citation19
dSTORM imaging was performed using a Nikon N-STORM super-resolution microscope equipped with a 100x oil-immersion objective (N. A. 1.40) and an Andor iXon DU- 897E-CS0BV EMCCD camera (image pixel size, 160 nm) running at approximately 50 Hz (20 ms exposure time). The z position was maintained during the acquisition by a Nikon 'perfect focus system'. The set-up included an activation 405 nm laser (Coherent CUBE 405 – 100 mW), a 488 nm readout laser (Coherent Sapphire OPSL 488 nm – 50 mW), a 561 nm readout laser (Coherent Sapphire OPSL 561 nm-100 mW), and a 647 nm laser (MPBC's CW Visible Fiber Laser). Intensities at the sample were set in the range IUV < 1.0 W/cm2 for photoactivation and IReadout < 1.0 KW/cm2 for the readout. Dichroic mirrors and band-pass filters allowed selection of the emitted signals (ZET405ZT405/488/561/647, Chroma).
Single molecule localization was obtained by Gaussian fitting using the Nikon integrated software taking into account both drift and, for multi-color acquisitions, chromatic aberrations. Super-resolution images were rendered by superimposing the coordinates of each single molecule represented by 2D Gaussian curves of the intensity resulting from the associated fit.
Image processing and analysis of the amount of the incorporated EdU
ImageJ (Rasband, W.S., ImageJ, U. S. National Institutes of Health, Bethesda, Maryland, USA, http://imagej.nih.gov/ij/, 1997–2014) was used for basic image processing and analysis. Due to a low amount of the DNA precursors incorporated during UDS fluorescence intensities were low as well, therefore the brightness of images 1Aef, Bbcf, Cab, Da, Eab was increased using the contrast function. The typical signal-to-noise ratio of the images of BrdU and EdU incorporated during UDS is shown in fluorescence profiles in . To analyze the amount of the EdU incorporated in the process of UDS a specially written ImageJ macro, which sums the pixel values in the selected area of each cell nucleus, was used.
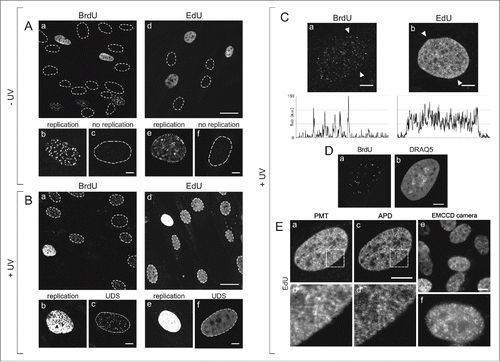
Figure 1. Patterns of DNA replication and UDS revealed by incorporation of BrdU or EdU, and various modes of fluorescence detection. (A) Images of BrdU (a) or EdU (d) incorporated into DNA of replicating cells in asynchronous cell cultures, under standard conditions, prior to exposure to UVC. Contours of nuclei of non-replicating cells, based on transmitted light images (not shown), are marked with dotted lines. Panels below show magnified images of replicating and non-replicating nuclei, with incorporated BrdU (b, c) or EdU (e, f), showing typical replication patterns (b, e) and the absence of fluorescence signals in non-replicating cells (c, f). No EdU or BrdU signals were detected in non-replicating cells at low or high instrumental gain (data not shown). Scale bar 20 μm (a,d) and 5 μm (b,c,e,f). (B) Images showing BrdU (a) or EdU (d) in cells, recorded 2 h after exposure to UVC. The gain of the fluorescence detector (PMT) was set higher than in images in panel A, therefore the signals of replicating cells are oversaturated (b,e). Non-replicating cells show UDS signals of the incorporated BrdU or EdU (c,f). Non-replicating cells that do not activate UDS are also occasionally detected. Scale bar 20 μm (a,d) and 5 μm (b,c,e,f). (C) High magnification images showing BrdU (a) or EdU (b) incorporated in a process of UDS (UVC dose 10 J/m2), and demonstrating an apparent difference between the patterns revealed by incorporation of BrdU and EdU. Fluorescence profiles plotted along the lines marked with arrowheads are shown below each image. Scale bar 5 μm. (D) A pattern of UDS revealed by BrdU (a), distribution of dsDNA stained with DRAQ5 (b), and UDS against the background of dsDNA (c). Scale bar 5 μm. E. Patterns of UDS revealed by the incorporated EdU, detected by a standard photomultiplier in a confocal microscope (a,b), an avalanche photodiode (c,d), and a sensitive EMCCD camera in a widefield fluorescence microscope (e,f). Scale bar 10 μm.
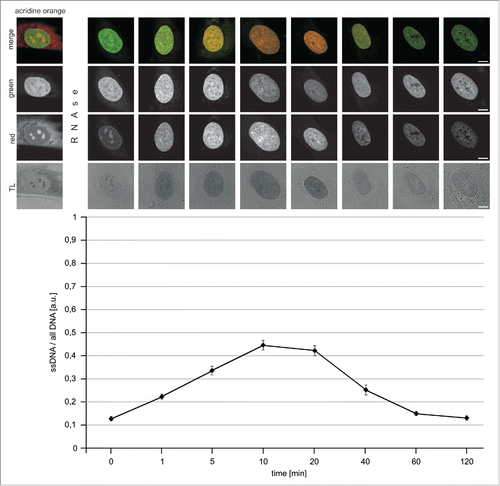
Assessing DNA denaturation
Single- and double-stranded DNA (dsDNA) was detected in situ, in nuclei of fixed cells, using acridine orange (AO), as described previously.Citation20-22 Briefly, AO intercalates between bases of double-stranded nucleic acids in a form of a monomer, and yields green fluorescence (∼530 nm). When molecules of this dye bind to single stranded nucleic acids, stacks are formed and yield red luminescence (>640 nm). Single- and double stranded nucleic acids can thus be differentiated by the color of the emitted luminescence when the concentration of AO is maintained within 5–10 μg/ml. AO was used here to assess the proportion of single-stranded DNA formed in RNase-treated cells (1 mg/ml RNase in PBS containing calcium and magnesium ions, for 1 h in 37°C) after exposure to acidic denaturing conditions (4 M hydrochloric acid; 1 – 120 minutes at room temperature).
Results and discussion
Subnuclear distribution of UDS sites detected by incorporation of BrdU or EdU
In order to investigate the sub-nuclear pattern of sites of UDS, and the efficiency of this process in cells repairing the damage inflicted by UVC, in an in vitro system, we have chosen asynchronous human skin fibroblasts isolated from healthy individuals, grown under optimal conditions. The fibroblasts were maintained in an in vitro culture, and exposed to various doses of UVC resulting in detectable levels of UDS. Immediately after an exposure to UVC, a DNA precursor analog, BrdU or EdU, was added to culture medium. Subsequently cells were incubated under standard conditions for 2 h, fixed, and the incorporated precursors were labeled and imaged as described in Materials & Methods.
S-phase cells were readily recognized in unexposed () and UV-exposed cultures () due to strong signals of the incorporated BrdU or EdU. These signals adopted the typical, readily recognizable patterns of early, mid or late S-phase replication Citation10,23 (). Many cells showed no precursor signal at all, as would be expected of non-replicating (G1 and G2/M) cells (). In a culture exposed to UVC, however, non-replicating cells revealed weak but detectable signals of the incorporated BrdU () or EdU (), which we attribute to UDS. At the instrumental settings that were adjusted for detection of UDS (i.e., a high gain required for detection of very weak signals), the intensity of S-phase replication signals (BrdU or EdU) exceeded the dynamic range of the PMT ( and ). Thus, distinguishing the scheduled replication from UDS on the basis of the pattern and intensity of fluorescence of the incorporated precursor analogs was straightforward.
In order to verify if the weak signals of EdU, that are shown in , represent UDS in non-replicating cells, and to establish whether they can reveal a pattern of UDS in the presence of the typical pattern of DNA replication in S-phase (i.e. weak signals among the strong signals), images of individual nuclei of cells in an untreated asynchronous population, and cells exposed to UVC, were carefully analyzed. Typical examples of high magnification images of the UDS patterns detected by incorporation of BrdU and EdU are shown in . The UDS pattern detected via incorporation of BrdU () was similar to those reported in numerous previous publications that also employed halogenated thymidine analogs.Citation2,8,9,24 These images consisted of a number of distinct foci of various brightness and size. In the published reports these foci were referred to as 'repair sites', and their number was in the order of a few dozen per nucleus.Citation8,9,24 In contrast, the mean distance between randomly spaced nucleotide excision sites in the DNA extracted from cells exposed to UV (3 J/m2) was reported to be 26–30 kbp,Citation25 corresponding to over 105 sites per nucleus of a human fibroblast.
The UDS pattern we detected by labeling the incorporated EdU was conspicuously different from BrdU. The fluorescently labeled EdU was detected by standard confocal imaging as a signal distributed almost uniformly throughout the nucleus (except for nucleoli) and showing no obvious, distinct foci (), or as several hundred closely spaced signals of various intensity all throughout chromatin, by high sensitivity or super-resolution imaging methods (see below). It is important to emphasize that EdU labeling yields no detectable non-specific signal, while immunofluorescence detection of BrdU always gives some weak non-specific staining, most likely arising from residual secondary antibody. A notion that the number of NER sites is to be counted in thousands (at moderate UV dose) rather than dozens or hundreds is consistent with the data based on imaging of the incorporated precursors in experiments that did not require DNA denaturation. Images of numerous NER sites distributed all throughout chromatin, visually similar to our images of the incorporated EdU, were obtained by detection of the incorporation of biotin-16-dUTP 1 hour after UV exposure.Citation26
As demonstrated by images shown in this report the incorporated EdU was not confined to a relatively low number of distinct repair foci, as was the case with BrdU (). This observation has important biological consequences in the context of the presence, structure and role of UV-induced 'repair foci'. UVC photons can reasonably be expected to interact with DNA in cell nuclei randomly, therefore there is no reason for the damage to be confined to a low number of distinct DNA regions. To a first approximation one should therefore expect the damage, and consequently the repair sites, to be distributed within DNA stochastically. Considering the available spatial resolution of standard optical microscopy, the regions of active UDS should be detected in fluorescence images all throughout chromatin in the nucleus, possibly with a higher density of UDS in areas of a higher density of DNA, like perinucleolar chromatin. In contrast, UDS detected via BrdU incorporation is seen in a low number of distinct foci. The low number of UDS sites characteristically detected by BrdU could hint at a nonuniform sensitivity of DNA to UVC, implying a significantly higher propensity to sustain damage in a relatively low number of specific regions of DNA. Alternatively, it could suggest that the process of DNA repair of UVC-induced damage occurs in distinct repair foci or regions, that might be formed as a result of chromatin rearrangements following damage induction. Thus, the very different patterns of UDS observed using 2 different DNA precursors have potentially important consequences for our understanding of DNA sensitivity to UV and the existence of hypersensitive DNA regions, and for an ability to detect the postulated chromatin rearrangements during repair.
In order to search for an explanation of the differences that we detected between the patterns of UDS revealed by BrdU or EdU incorporation, the precursor analogs incorporated in UVC-exposed fibroblasts were labeled as previously, subsequently the nuclei were counterstained with DRAQ5, and the cells were imaged by fluorescence confocal microscopy (). The images confirmed the notion that the subcellular distribution of UDS based on BrdU was indeed qualitatively different than the pattern of foci containing EdU (). At the same time the presence of the signal of DRAQ5 (), a DNA probe which labels dsDNA readily, but has only very low affinity for single stranded nucleic acids, confirmed that the nuclei of the cells that were subjected to the standard procedure required for BrdU detection, contained a considerable fraction of double stranded (not denatured) DNA.
In order to follow up on this observation and establish if an incomplete denaturation of DNA could be a factor interfering with detection of all BrdU incorporated in the process of UDS, we carefully analyzed the process of strand separation under conditions typically used in the detection of this precursor. Single- and double-stranded DNA can be differentiated by AO following hydrolysis of RNA.Citation20-22 Red luminescence is associated with ssDNA, while green fluorescence represents dsDNA. demonstrates that the amount of ssDNA increased for approximately 10 to 20 minutes during incubation of fixed cells under acidic conditions. The presence of strong green signals indicated, however, that dsDNA still existed at high local densities, confirming that DNA was not fully denatured. Longer exposures to hydrochloric acid resulted in a decrease of both, the green and the red signals of DNA-bound AO, indicating that DNA was progressively degraded (most likely ssDNA being more easily degraded than dsDNA). DNA degradation under the influence of low pH environment was also confirmed by the observation that signals of DNA-bound DRAQ5 were lower after longer acid exposures (data not shown). These observations are consistent with a notion that only a fraction of DNA in situ is denatured by a 10 to 20 min exposure to an acid, while a longer incubation in acidic conditions does not result in any further increase of the amount of ssDNA. They are also in agreement with a previous report, which emphasized the fact that DNA denaturation is incomplete under standard conditions typically used for BrdU incorporation.Citation27 We have also observed that, when the samples were returned to neutral pH, following a 10 min exposure to hydrochloric acid, a slow process of DNA renaturation was occurring (data not shown). These phenomena, that is an incomplete denaturation followed by degradation at low pH, or renaturation at neutral pH, are bound to adversely affect the ability to detect the incorporated BrdU.
Figure 2. Denaturation of DNA in situ, in fixed cells, by exposure to acidic conditions. Cells were treated by RNAse (1 h) and incubated with hydrochloric acid for 1 – 120 minutes, rinsed and stained with acridine orange in order to assess the amounts of single- and double-stranded nucleic acids (red and green AO emissions, respectively). Red and green signals, as well as overlays and transmitted light (TL) images are shown. Changes in the intensities of the green and red emissions show an initial increase of the proportion of ssDNA relative to all DNA (graph), and a subsequent decrease, which coincided with degradation of the polymer under acidic conditions.
Patterns of UDS revealed by different fluorescence detection methods
Confocal microscopy with PMT or APD detectors
As the number and subcellular distribution of UDS sites detected via EdU incorporation were apparently different than the sites detected using a generally accepted method based on BrdU incorporation, we focused on optimizing the EdU detection conditions to ensure faithful and optimal imaging of the DNA repair sites. It would be reasonable to expect that an almost uniform signal of EdU, which we observed by standard confocal microscopy, contained a significant contribution of electronic noise. This noise could overshadow the signals representing the repair sites. In order to ascertain that the signals of EdU were not confused with the noise, we used 2 other detection techniques that were expected to yield a better signal-to-noise ratio. Toward this end, in addition to using a standard PMT as a fluorescence detector (as in ), we also recorded images using APD in a confocal microscope (), and a sensitive EMCCD camera in a widefield fluorescence instrument ().
The fluorescence signals of EdU-Alexa detected by a PMT were found to be distributed throughout the nucleus, with areas showing a higher or lower signal intensity, however, as it is demonstrated by images shown in and , no distinct repair foci were detected. The pattern of signals detected in the same regions of the nucleus by APD was characterized by a higher signal-to-noise ratio, resulting in a better contrast and better effective spatial resolution. As the EdU images recorded using an APD were of a higher quality than the images collected by a PMT, they demonstrated numerous small features likely representing minute individual DNA repair (UDS) sites (). These images were similar to the confocal images recorded in the same area of the nucleus, however they were clearly qualitatively different than the typical images based on incorporation of BrdU. Further confirmation of the validity of EdU images as a representation of UDS was provided by wide-field images of the incorporated EdU recorded by a sensitive EMCCD camera (). Although the thickness of the imaged layer of the nucleus was larger than in the case of confocal imaging, this very sensitive detection approach also demonstrated numerous small regions of EdU fluorescence, similar to the patterns detected by APD, but dissimilar to the characteristic patterns observed in the case of BrdU. Thus, the optimized high sensitivity detection approaches support the view that imaging of the incorporated EdU provided a faithful representation of UDS following exposure to UVC, unbiased by regional chromatin differences in DNA denaturability as in the case of BrdU.
The image quality improvement, which was achieved by using highly sensitive fluorescence detectors, underscores the importance of optimization of the imaging technique for the benefit of an accurate biological interpretation of precursor incorporation data. While the standard images of BrdU incorporated during UDS could be interpreted as evidence in favor of a relatively low number of distinct repair foci, standard confocal images of EdU could be interpreted differently - as UDS active uniformly all throughout the nucleus. The optimally detected high sensitivity confocal images of EdU demonstrated that neither was true - in fact UDS was indeed taking place rather evenly throughout all chromatin, however precursor incorporation was not entirely homogenously distributed but could be resolved into a high number of minute individual sites.
High resolution single molecule localization microscopy of UDS
In order to obtain further, more detailed insight into the sub-nuclear distribution of UDS sites, we employed 2 super-resolution (sub-diffraction) microscopy methods. A single molecule localization microscope (SMLM, an in-house built dedicated instrument),Citation14,15 and a commercial dSTORM super-resolution microscope were used. SMLM images of regions with EdU showed a large number of minute foci or single molecules distributed all throughout the nucleus (). No large UDS foci or obvious clusters of EdU signals were detected. We interpret the SMLM images as further evidence supporting the notion that UDS occurred in a high number of very small regions distributed evenly throughout the chromatin. Subsequently, we extended sub-diffraction imaging to 2-color dSTORM experiments, where EdU representing UDS, as well as all DNA, were differentially labeled in order to relate the areas containing EdU to regions of various DNA density ().
Figure 3. Super-resolution imaging of UDS in cells exposed to UVC. (A) EdU detected by custom-built single molecule detection instrument, showing numerous sites of UDS; a standard widefield fluorescence image (a), a super-resolution image (b) and a magnified view of the area embraced by a square (c). Scale bar 5 μm. (B) UDS sites, revealed by EdU, shown against the background of nuclear DNA. Standard widefield images are shown in panels (a-c). Super-resolution images of UDS in the same cells (d-f), and enlarged selected areas (squares) (g-i), showing UDS sites and DNA density. Scale bar 2 μm (f) and 1 μm (i). (C) 3D dSTORM single molecule images of UDS sites (a, d) and enlarged images of selected UDS regions (square), shown at different angles to reveal their 3D structure (b,c,e,f). The depth of the position of individual foci within 3D space is color-coded (left). Scale bar 1 μm (a,d) and 0,5 μm (b,c,e,f,). (D) UDS sites, revealed by BrdU, shown against the background of nuclear DNA. Standard widefield images are shown in panels (a-c). Super-resolution images of UDS in the same cells (d-f), and enlarged selected areas (g-i) showing UDS sites and DNA density. As demonstrated in , the pattern of UDS seen via BrdU incorporation () is qualitatively different than the pattern seen through EdU incorporation (). Scale bar 2 μm (f) and 1 μm (i).
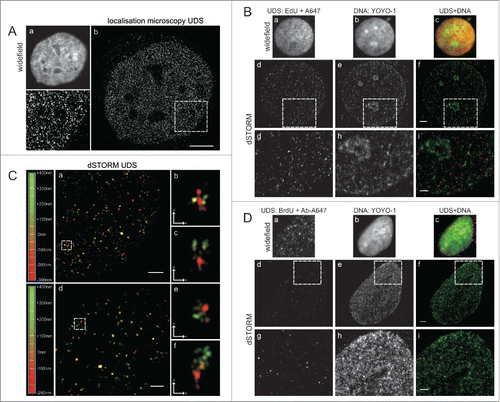
As standard confocal microscopy and SMLM, dSTORM images also showed a large number of small EdU regions distributed evenly in the equatorial slice of the nucleus. Two representative examples are shown in . No discrete large repair foci were detected. Occasionally objects resembling larger foci were seen in 2D projections. The depth-coded dSTORM images revealed, however, that these objects were groups of small foci located at small distances of each other (). The UDS foci found in dSTORM images showed no apparent relationship to the regions of high or low DNA density (). We also imaged BrdU with dSTORM (). In contrast to the findings provided by sensitive confocal and sub-diffraction imaging of EdU, the dSTORM images of BrdU again demonstrated a low number of foci of various sizes and brightness, including some large and bright objects. In this respect, sub-diffraction imaging confirmed the observations and conclusions based on confocal data, indicating that the distribution of BrdU foci is influenced by differences in sensitivity of DNA in situ to denaturation by hydrochloric acid. Because of this bias the pattern of BrdU incorporation is inconsistent with the actual sites of active UDS after UV exposure.
Since the patterns of UDS detected by confocal and super-resolution microscopy methods using BrdU or EdU as DNA precursor analogs are different, a question arises as to which of the 2 is a better representation of the spatial distribution of active repair regions. We note that the first successful in situ studies of UDS employed incorporation and detection of nucleoside analogs tritium labeled thymidineCitation5,6 or bromodeoxyuridine.Citation8,9,28 However, these approaches were characterized by several limitations. The main drawback of the method of detection of UDS based on incorporation of 3H-TdR arises from limited spatial resolution of 2-dimensional autoradiograms.Citation29 The highest resolution of silicon-based electron detectors in β-autoradiography is within the range between 0.1 to 1.0 μm.Citation30 In the case of detection of BrdU, as discussed earlier, the main problems arise from the need to denature DNA in order to facilitate access of an antibody to BrdU, and the fact that DNA denaturation is typically incomplete. Moreover, it has been shown that condensed chromatin in prophase, metaphase, and heterochromatin blocks are denatured more easily than euchromatin,Citation22 therefore acid denaturation may introduce bias in detection of BrdU (i.e., a preference for detecting BrdU in condensed chromatin). Achieving a complete DNA denaturation is not only difficult, but it is also undesired since leaving some dsDNA sections makes it possible to stain the cell nucleus with popular dsDNA-binding dyes like DAPI, Hoechst, or PI.Citation2,5,6,8 As a result, the standard procedure used in detection of the incorporated BrdU results in a limited rather than full access of anti-BrdU antibodies to the incorporated DNA precursors. In fact, it is apparent that the presence of stainable DNA is the evidence that denaturation is only partial and thus the BrdU incorporated into dsDNA section is undetectable. In contrast the click reaction occurs readily with the incorporated EdU molecules all throughout the cell nucleus. The label is covalently linked to the precursor after cell fixation, therefore its detachment and loss is unlikely. Even a low number of the incorporated fluorescently labeled EdU molecules can be detected by state-of-the art, high-sensitivity and super-resolution microscopy methods. Such sensitivity and resolution of microscopy images were not available in early studies of UDS. In conclusion, the fact that BrdU cannot reveal a pattern of UDS faithfully, even using the current sophisticated imaging methods, can be attributed to only partial denaturation of DNA causing limited accessibility of BrdU for immunolabeling. Needless to say the severe conditions of DNA denaturation by strong acid or high heat denature proteins and destroy nuclear architecture.
UDS dependence on the dose of UVC
Optimizing a sensitive and accurate method of detection of UDS opened the way for studies of an efficiency of the repair of UVC-induced lesions in individual cells. We were prompted to assess the efficiency of repair by the fact that, based on our images, the total amounts of EdU incorporated into DNA during NER varied significantly not only between different UVC doses, but also between individual cells. Since the extent of damage (the number of PPs and PDs) inflicted by a given dose of UV can reasonably be expected to be similar in all cellsCitation2,31 this large variation in the amounts of the incorporated precursor is likely to reflect different efficiencies or rates of UDS in individual cells. Thus, we measured the integrated intensities of fluorescence signals in each nucleus, and calculated the relative total amounts of the incorporated precursor. Two ways of plotting the data to describe the effectiveness of UDS were used - histograms of the amounts of the incorporated precursor within a population, and the proportions of cells exhibiting high, medium or low and no precursor incorporation in each group.
shows histograms of the amounts of the incorporated precursors within the whole cell population, for the 7 UVC doses used in our experiments. When fibroblasts were exposed to a low dose of UVC (5 J/m2), over 90% of non-replicating cells responded with detectable UDS. There was at least a 3-fold difference between the amounts of the precursor incorporated by various responding cells within this population. This wide range of the amounts of the incorporated EdU is likely to represent different rates or efficiency of the repair processes in individual cells. It is important to note, however, that we cannot exclude a possibility that 2 other factors might contribute to the intercellular differences between the amounts of the incorporated EdU within a group treated with one dose of UV. These two factors are (i) different levels of endogenous thymidine and the resulting competition between the endogenous precursor and EdU, and (ii) higher numbers of UVC-induced lesions in late S and G2 cells, as compared to G0/G1 cells, due to a higher amount of DNA (assuming a random nature of DNA damage induction). We were unable to estimate the influence, if any, of different levels of intracellular thymidine on the amount of the incorporated EdU. Different amounts of DNA in G1, S and G2 cells can account for no more than a 2-fold difference in the amount if the incorporated EdU within an asynchronous cell population.
Figure 4. Efficiency of UDS in fibroblasts following exposure to UVC. (A) Incorporation of EdU during UDS. Histograms representing distribution of the amounts of EdU incorporated during repair in cells exposed to various doses of UVC. Gray bars represent cells that do not exhibit detectable EdU signals. Black bars represent cells that incorporate various amounts of EdU during repair. The number of cells measured in each sample was 58 to 160. (B) Three images representing the nuclei of cells with different UDS efficiencies. A cell that did not incorporate any EdU (top image; the contour of this nucleus is shown), and cells that incorporated medium (middle), and high amounts of EdU (bottom image) during repair of UVC-induced damage. Scale bar 5 μm. (C) Proportions of cells that do not respond to UVC damage, or respond by incorporating medium or high amounts of EdU, as a function of the UVC dose delivered. The percentage of 'non-responders' decreases for doses between 5 and 10 J/m2, and increases again with higher doses of UVC (20 – 3,000 J/m2), while the proportion of cells showing a standard response decreases. The proportion of cells with very efficient UDS is independent of the UVC dose.
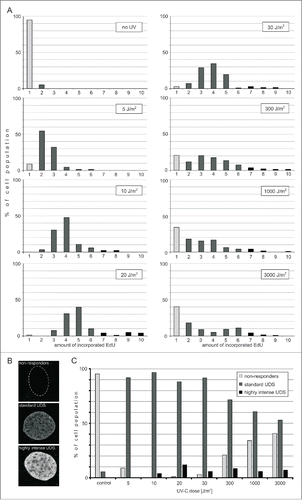
We note, that when the increasing UVC doses were used (5, 10 or 20 J/m2), the mean and maximum amounts of the incorporated EdU also increased, presumably reflecting the growing number of DNA lesions. Interestingly, when we used even higher doses of UVC (30 up to 3,000 J/m2) the extent of EdU incorporation did not significantly increase.
Considering that the doses of UVC delivered to these cells varied by 2 orders of magnitude, the fact that the amounts of the incorporated EdU at doses 30 J/m2 and higher were similar, suggests that saturation of the repair capacity occurred. This saturation may be a consequence of a limited amount of some critical component of the NER pathway. A shortage of thymidine is unlikely since, if it were to occur, EdU, which is in plentiful supply, would have been incorporated at an even higher rate than in the presence of thymidine. Thus, a limiting step of the repair process at high damage levels may be the availability of one or several NER repair factors.
The percentage of cells showing no EdU incorporation ('non-responders') was decreasing for doses of 5–20 J/m2, but begun to increase with the larger UVC doses (above 30 J/m2). Non-responders constituted a large proportion (40%) of a population exposed to a very high dose of UVC (3,000 J/m2). The observation that, within the range of large UVC doses, the higher was the dose the higher was the number of cells that did not respond by detectable EdU incorporation, suggests that heavy DNA damage resulted in the lower chance to complete the steps of repair processes preceding incorporation of EdU. The data available currently do not allow to propose a clear mechanism of this phenomenon. We note, however, that it is possible that some cells classified here as 'nonresponders' activate the repair processes at many sites but fail to reach UDS and complete the repair. It is also possible that at the maximal UVC dose cells recognize the high extent of DNA damage as lethal and activate the apoptotic instead of repair pathways.
In the second method of plotting the data described above we defined the efficiency of UDS as zero to low, medium or high when the amount of the incorporated EdU fell within the range of 0–10%, 11–60% and 61–100% of the maximum value of the incorporated precursor, respectively. The proportions of cells exhibiting various levels of efficiency of UDS are shown in . Apparently the size of these groups is strongly dependent on the dose of UVC delivered to cells. The proportion of cells showing none or only a low level response decreased for doses up to 10 J/m2, but increased significantly for doses higher than 20 J/m2. The cells characterized by standard efficiency of UDS constituted over 90% after a low dose of UVC (5 J/m2), but only 40% in response to the highest dose used (3,000 J/m2). The cells capable of very efficient repair constituted 3–12% of the population and apparently their proportion was independent of the UVC dose (for doses 10 – 3,000 J/m2). This observation hints at the presence of high levels of repair factors in these few highly UVC-resistant cells.
Time course of EdU incorporation after UVC irradiation
Unscheduled DNA synthesis is the last stage of repair of the UVC-induced lesions, thus we turned our attention to the time, which elapsed between UVC exposure and incorporation of DNA precursors. We studied the kinetics of the UDS process by measuring the extent of EdU incorporated into DNA of the cells that were exposed to the dose of 30 J/m2. Incorporation of EdU representing repair of endogenous damage was only barely detectable in some cells in control cultures (). Incorporation of the DNA precursor analog in irradiated cells was detectable already 15 minutes after the UVC exposure, but a significant increase of the amount of the incorporated EdU occurred one hour after the insult ().
Figure 5. The timecourse of UDS in cells exposed to UVC. (A) Images of EdU in nuclei (central cross-sections) of human fibroblasts at various time points following a UVC insult. Scale bar 5 μm; (B) The relative amounts of EdU incorporated within 0.25 h to 6 h after UVC exposure. Control refers to unirradiated cells. (C) The surface areas of central cross-sections of nuclei of cells, measured at different times after exposure to UVC (10 J/m2). The number of cells measure in each sample was 60–83.
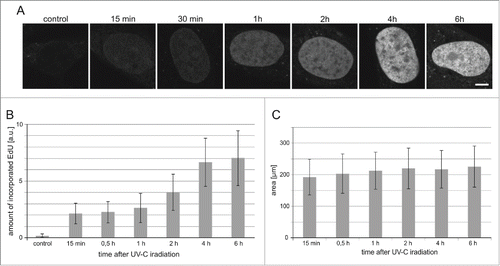
There was no increase of the signal of the incorporated EdU between 4 and 6 hours indicating that the repair process, which gathered pace one hour after UV exposure, was completed within approximately 4 hours. This time course of the repair is in agreement with observations described in other reports.Citation32-34 We have previously noticed significant increase of the nuclear volume following DNA damage (dsDNA breaks) induced by topotecan.Citation35 This increase may be caused by global chromatin decondensation observed during DNA repair.Citation35-37 Here, in the case of UV-induced damage, no significant increase of nuclear volumes was observed within 6 hours of UV insult (), suggesting that large scale chromatin decondensation may not be required for NER at that time point after the UV exposure. It was observed previously, however, that chromatin decondensation is a rapid and transient event peaking at 30 min after cell exposure to UV (50 J/m2) followed by a decline after 60 min.Citation37
Conclusions
Detection of UDS by means of EdU incorporation (as opposed to BrdU) argues in favor of induction of randomly distributed UVC damage and repair, and an absence of a low number of distinct repair foci. The UDS detection based on EdU reveals that the capacity to repair UVC-induced damage, measured using EdU incorporation, varies significantly between individual cells, with some cells demonstrating no active UDS. At UVC doses 30 J/m2 and above progressively more cells show no EdU incorporation. A small fraction (approx. 5%) of the cell population shows a very high propensity to activate and complete UDS after DNA damage; their proportion is not influenced by the dose of UVC within the range of 10 J/m2 to as high as 3,000 J/m2.
Based on the data presented above we conclude that when a sensitive detection is used, the incorporated DNA precursor analog EdU provides a more relevant representation of UDS than BrdU. Shortcomings of the BrdU method likely arise from incomplete denaturation of DNA and limited access of an anti-BrdU antibody, as well as global changes of nuclear and chromatin structure induced by the procedure required for BrdU detection.
Abbreviations
| APD | = | avalanche photodiode |
| BrdU | = | 5-bromo-2-deoxyuridine |
| EdU | = | 5-ethynyl-2′-deoxyuridine |
| EMCCD | = | electron multiplying charged coupled device |
| NER | = | nucleotide excision repair |
| PMT | = | photomultiplier |
| STORM | = | Stochastic Optical Reconstruction Microscopy |
| 3H-TdR - | = | tritium labeled thymidine |
| UVC | = | ultraviolet C |
| UDS | = | unscheduled DNA synthesis |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We acknowledge the Nikon Imaging Center at the Istituto Italiano di Tecnologia for N-STORM imaging and Precoptic, Warsaw, for analysis tools. We are grateful to Prof. Paul J. Smith for helpful discussions.
Funding
This research was supported by a grant from Polish National Science Center (2013/11/B/NZ3/00189) (JD) and The Robert A. Welke Cancer Research Foundation (ZD). Confocal instrumentation was purchased through the funds awarded by BMZ no. POIG.02.01.00-12-064/08. Faculty of Biochemistry, Biophysics and Biotechnology is a partner of the Leading National Research Center (KNOW) supported by the Ministry of Science and Higher Education in Warsaw. AP-M is a recipient of a SET fellowship from JU.
References
- Moser J, Volker M, Kool H, Alekseev S, Vrieling H, Yasui A, van Zeeland AA, Mullenders LHF. The UV-damaged DNA binding protein mediates efficient targeting of the nucleotide excision repair complex to UV-induced photo lesions. DNA Repair (Amst) 2005; 4:571-82; PMID:15811629.
- Nakagawa A, Kobayashi N. Three-dimensional visualization of ultraviolet-induced DNA damage and its repair in human cell nuclei. J Invest Dermatol 1998; 110:143-8; PMID:9457909.
- Moné MJ, Bernas T, Dinant C, Goedvree FA, Manders EMM, Volker M, Houtsmuller AB, Hoeijmakers JHJ, Vermeulen W, van Driel R. In vivo dynamics of chromatin-associated complex formation in mammalian nucleotide excision repair. Proc Natl Acad Sci U S A 2004; 101:15933-7.
- Solimando L, Luijsterburg MS, Vecchio L, Vermeulen W, van Driel R, Fakan S. Spatial organization of nucleotide excision repair proteins after UV-induced DNA damage in the human cell nucleus. J Cell Sci 2009; 122:83-91; PMID:19066286.
- Darzynkiewicz Z. Radiation-induced DNA synthesis in nuclei of hen erythrocytes reactivated in heterokaryons. Exp Cell Res 1971; 69(2):477-81; PMID:4360911.
- Darzynkiewicz Z, Chelmicka-Szorc E, Arnason BG. UV-induced DNA synthesis in Xeroderma pigmentosum nuclei in heterokaryons. Exp Cell Res 1972; 74:602-6; PMID:4343024.
- Li W, Choy DF, Post JM, Sullivan ME. A dual-labeling method to quantify unscheduled DNA synthesis in primary cells. J Pharmacol Toxicol Methods 2008; 57:220-6; PMID:18396065.
- Kao GD, McKenna WG, Yen TJ. Detection of repair activity during the DNA damage-induced G2 delay in human cancer cells. Oncogene 2001; 20:3486-96; PMID:11429695.
- Rubbi CP, Milner J. Analysis of nucleotide excision repair by detection of single-stranded DNA transients. Carcinogenesis 2001; 22:1789-96; PMID:11698340.
- Berniak K, Rybak P, Bernas T, Zarębski M, Darzynkiewicz Z, Dobrucki JW, Zarebski M. Relationship between DNA Damage Response, initiated by camptothecin or oxidative stress, and DNA replication, analyzed by quantitative image analysis. Cytom Part A 2013; 83:913-24.
- Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci U S A 2008; 105:2415-20; PMID:18272492.
- Limsirichaikul S, Niimi A, Fawcett H, Lehmann A, Yamashita S, Ogi T. A rapid non-radioactive technique for measurement of repair synthesis in primary human fibroblasts by incorporation of ethynyl deoxyuridine (EdU). Nucleic Acids Res 2009; 37:e31; PMID:19179371.
- Nakazawa Y, Yamashita S, Lehmann AR, Ogi T. A semi-automated non-radioactive system for measuring recovery of RNA synthesis and unscheduled DNA synthesis using ethynyluracil derivatives. DNA Repair (Amst) 2010; 9:506-16; PMID:20171149.
- Lemmer P, Gunkel M, Baddeley D, Kaufmann R, Urich a., Weiland Y, Reymann J, Müller P, Hausmann M, Cremer C. SPDM: light microscopy with single-molecule resolution at the nanoscale. Appl Phys B 2008; 93:1-12.
- Reymann J, Baddeley D, Gunkel M, Lemmer P, Stadter W, Jegou T, Rippe K, Cremer C, Birk U. High-precision structural analysis of subnuclear complexes in fixed and live cells via spatially modulated illumination (SMI) microscopy. Chromosome Res 2008; 16:367-82; PMID:18461478.
- Heilemann M, van de Linde S, Schüttpelz M, Kasper R, Seefeldt B, Mukherjee A, Tinnefeld P, Sauer M. Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew Chemie 2008; 47:6172-6.
- Thompson RE, Larson DR, Webb WW. Precise nanometer localization analysis for individual fluorescent probes. Biophys J 2002; 82:2775-83; PMID:11964263.
- Szczurek AT, Prakash K, Lee H-K, Zurek-Biesiada DJ, Best G, Hagmann M, Dobrucki JW, Cremer C, Birk U. Single molecule localization microscopy of the distribution of chromatin using Hoechst and DAPI fluorescent probes. Nucleus 2014; 5:331-40; PMID:25482122.
- Żurek-Biesiada D, Szczurek AT, Prakash K, Mohana GK, Lee H-K, Roignant J-Y, Birk U, Dobrucki JW, Cremer C. Localization microscopy of DNA in situ using Vybrant® DyeCycle™ Violet fluorescent probe: A new approach to study nuclear nanostructure at single molecule resolution. Exp Cell Res 2015; ( in press); PMID:26341267; http://dx.doi.org/10.1016/j.yexcr.2015.08.020
- Darzynkiewicz Z. Acid-induced denaturation of DNA in situ as a probe of chromatin structure. Methods Cell Biol 1994; 41:527-41; PMID:7532268.
- Darzynkiewicz Z, Kapuscinski J. Acridine orange: a versatile probe of nucleic acids and other cell constituents. In: Flow Cytometry and Sorting, 2nd ed. (Melamed, M. R., Lindmo, T., and Mendelsohn, M. L., eds.). New York: Wiley-Liss; 1990; 291-314.
- Dobrucki J, Darzynkiewicz Z. Chromatin condensation and sensitivity of DNA in situ to denaturation during cell cycle and apoptosis - a confocal microscopy study. Micron 2001; 32:645-52; PMID:11334733.
- Leonhardt H, Rahn HP, Weinzierl P, Sporbert A, Cremer T, Zink D, Cardoso MC. Dynamics of DNA replication factories in living cells. J Cell Biol 2000; 149:271-80; PMID:10769021.
- Svetlova M, Solovjeva L, Tomilin N. Application of new methods for detection of DNA damage and repair. Int Rev Cell Mol Biol 2009; 277:217-51; PMID:19766971.
- Cohn SM, Lieberman MW. The distribution of DNA excision-repair sites in human diploid fibroblasts following ultraviolet irradiation. J Biol Chem 1984; 259:12463-9; PMID:6490626.
- Jackson DA, Balajee AS, Mullenders L, Cook PR. Sites in human nuclei where DNA damaged by ultraviolet light is repaired: visualization and localization relative to the nucleoskeleton. J Cell Sci 1994; 107(Pt 7):1745-52; PMID:7983144.
- Moran R, Darzynkiewicz Z, Staiano-Coico L, Melamed MR. Detection of 5-bromodeoxyuridine (BrdUrd) incorporation by monoclonal antibodies: role of the DNA denaturation step. J Histochem Cytochem 1985; 33:821-7; PMID:3860561.
- Liu SC, Parsons CS, Hanawalt PC. DNA repair response in human epidermal keratinocytes from donors of different age. J Invest Dermatol 1982; 79:330-5; PMID:7130746.
- Cavanagh BL, Walker T, Norazit A, Meedeniya ACB. Thymidine analogues for tracking DNA synthesis. Molecules 2011; 16:7980-93; PMID:21921870.
- Cabello J, Wells K. The spatial resolution of silicon-based electron detectors in beta-autoradiography. Phys Med Biol 2010; 55:1677-99; PMID:20197603.
- Moredock S, Nairn RS, Johnston DA, Byrom M, Heaton G, Lowery M, Mitchell DL. Mechanisms underlying DNA damage resistance in a Xiphophorus melanoma cell line. Carcinogenesis 2003; 24:1967-75; PMID:12949045.
- Nishiwaki Y, Kobayashi N, Imoto K, Iwamoto T, Yamamoto A, Katsumi S, Shirai T, Sugiura S, Nakamura Y, Sarasin A, et al. Trichothiodystrophy fibroblasts are deficient in the repair of ultraviolet-induced cyclobutane pyrimidine dimers and (6–4)photoproducts. J Invest Dermatol 2004; 122:526-32; PMID:15009740; http://dx.doi.org/10.1046/j.0022-202X.2004.22226.x.
- Volker M, Moné MJ, Karmakar P, van Hoffen A, Schul W, Vermeulen W, Hoeijmakers JH, van Driel R, van Zeeland AA, Mullenders LH. Sequential assembly of the nucleotide excision repair factors in vivo. Mol Cell 2001; 8:213-24; PMID:11511374; http://dx.doi.org/10.1016/S1097-2765(01)00281-7.
- Verbruggen P, Heinemann T, Manders E, von Bornstaedt G, van Driel R, Höfer T. Robustness of DNA repair through collective rate control. PLoS Comput Biol 2014; 10:e1003438; PMID:24499930; http://dx.doi.org/10.1371/journal.pcbi.1003438.
- Rybak P, Waligórska A, Bujnowicz Ł, Hoang A, Dobrucki J. Activation of new replication foci under conditions of replication stress. Cell Cycle 2015; 14:2634-47; PMID:26212617; http://dx.doi.org/10.1080/15384101.2015.1064566.
- Kruhlak MJ, Celeste A, Dellaire G, Fernandez-Capetillo O, Müller WG, McNally JG, Bazett-Jones DP, Nussenzweig A. Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J Cell Biol 2006; 172:823-34; PMID:16520385; http://dx.doi.org/10.1083/jcb.200510015.
- Halicka HD, Zhao H, Podhorecka M, Traganos F, Darzynkiewicz Z. Cytometric detection of chromatin relaxation, an early reporter of DNA damage response. Cell Cycle 2009; 8:2233-7; PMID:19502789; http://dx.doi.org/10.4161/cc.8.14.8984.
