ABSTRACT
We previously reported real-time monitoring of cell cycle dynamics of cancer cells throughout a live tumor intravitally using a fluorescence ubiquitination cell cycle indicator (FUCCI). Approximately 90% of cancer cells in the center and 80% of total cells of an established tumor are in G0/G1 phase. Longitudinal real-time FUCCI imaging demonstrated that cytotoxic agents killed only proliferating cancer cells at the surface and, in contrast, and had little effect on the quiescent cancer cells. Resistant quiescent cancer cells restarted cycling after the cessation of chemotherapy. Thus cytotoxic chemotherapy which targets cells in S/G2/M, is mostly ineffective on solid tumors, but causes toxic side effects on tissues with high fractions of cycling cells, such as hair follicles, bone marrow and the intestinal lining. We have termed this phenomenon tumor intrinsic chemoresistance (TIC). We previously demonstrated that tumor-targeting Salmonella typhimurium A1-R (S. typhimurium A1-R) decoyed quiescent cancer cells in tumors to cycle from G0/G1 to S/G2/M demonstrated by FUCCI imaging. We have also previously shown that when cancer cells were treated with recombinant methioninase (rMETase), the cancer cells were selectively trapped in S/G2, shown by cell sorting as well as by FUCCI. In the present study, we show that sequential treatment of FUCCI-expressing stomach cancer MKN45 in vivo with S. typhimurium A1-R to decoy quiescent cancer cells to cycle, with subsequent rMETase to selectively trap the decoyed cancer cells in S/G2 phase, followed by cisplatinum (CDDP) or paclitaxel (PTX) chemotherapy to kill the decoyed and trapped cancer cells completely prevented or regressed tumor growth. These results demonstrate the effectiveness of the praradigm of “decoy, trap and shoot” chemotherapy.
Introduction
We previously reported intravitally monitoring of real-time cell cycle dynamics of cancer cells throughout a live tumor using a fluorescence ubiquitination cell cycle indicator (FUCCI) .Citation1,2 In a mature tumor, approximately 90% of cancer cells in the center and 80% of total cells of an established tumor are in G0/G1 phase. Longitudinal real-time FUCCI imaging demonstrated that cytotoxic agents killed only proliferating cancer cells at the surface and, in contrast, had little effect on quiescent cancer cells, the vast majority of an established tumor. Resistant quiescent cancer cells restarted cycling after the cessation of chemotherapy. We have termed this phenomenon tumor intrinsic chemoresistance (TIC).Citation1
We previously developed the tumor-targeting bacterial strain Salmonella typhimurium A1-R (S. typhimurium A1-R).Citation3 S. typhimurium A1-R is auxotrophic for Leu—Arg, which prevents it from mounting a continuous infection in normal tissues. S. typhimurium A1-R was able to inhibit primary and metastatic tumor growth as monotherapy in mouse models of major cancers,Citation4 including prostate,Citation5,6 breast,Citation7-9 lung,Citation10,11 pancreatic,Citation12-16 ovarian,Citation17,18 stomach,Citation19 and cervical cancer,Citation20 as well as sarcoma cell linesCitation21-24 and glioma,Citation25,26 as well as on pancreatic cancerCitation15 and sarcomaCitation24 patient-derived orthotopic xenograft (PDOX) models, all of which are highly aggressive tumor models.
Time-lapse FUCCI imaging demonstrated that tumor-targeting S. typhimurium A1-R decoyed quiescent cancer cells in tumors growing in nude mice to cycle from G0/G1 to S/G2/M, thereby acquiring chemosensitivity.Citation19
We previously demonstrated a selective growth arrest of cancer cells by depletion of their source of methionine in vitro. This growth arrest resulted in a reduction in the percentage of mitotic cells. Fluorescence-activated cell sorting demonstrated that the cells were arrested in the S and G2 phases of the cell cycle.Citation27 Methionine depletion of co-cultures of cancer and normal cells enabled the selective elimination of the cancer cells by chemotherapy drugs.Citation28
Subsequently we induced the tumor-specific cell cycle block in S/G2 in vivo by depriving Yoshida sarcoma-bearing nude mice of dietary methionine. Methionine depletion caused the tumor to eventually regress.Citation29
Cancer cells treated with recombinant methioninase (rMETase), were also selectively trapped in S/G2 as visualized with FUCCI imaging. rMETase-induced S/G2-phase blockage and sensitized the cancer cells to doxorubicin (DOX), cisplatinum (CDDP), or 5-fluorouracil (5-FU).Citation30 Cancer cells may be generally methinoine dependent compared to normal cells.Citation31-33
In the present study, we show that sequential treatment of FUCCI-expressing MKN45 human stomach cancer in vivo with S. typhimurium A1-R to decoy quiescent cancer cells to cycle; rMETase to selectively trap the decoyed cancer cells in S/G2 phase; and CDDP or paclitaxel (PTX), completely prevented or regressed tumor growth, demonstrating the effectiveness of the paradigm of “decoy, trap and shoot” chemotherapy.
Results and discussion
S. typhimurium A1-R decoys quiescent cancer cells to cycle visualized by FUCCI imaging
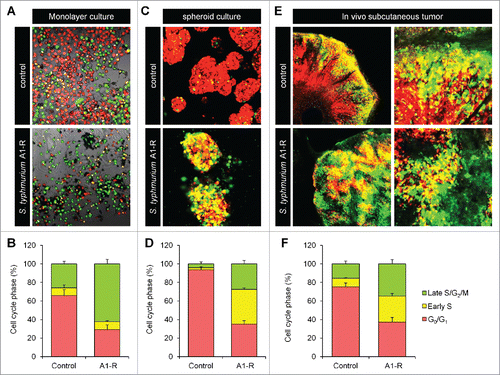
S. typhimurium A1-R treatment significantly decoyed HeLa-FUCCI cells in monolayer culture to cycle from G0/G1 to S/G2 phase (S. typhimurium A1-R treatment vs control: 62.3% vs 25.9% in S/G2, respectively, p < 0.01) (). In tumor spheres, S. typhimurium A1-R treatment significantly decoyed MKN45-FUCCI cells to cycle to S/G2 phase (S. typhimurium A1-R treatment vs control: 62.5 % vs 6.3% in S/G2, respectively, p < 0.01) (). S. typhimurium A1-R significantly decoyed MKN45-FUCCI cells in tumors in vivo to cycle to late-S/G2 phase (S. typhimurium A1-R treatment vs control; 62.6 % vs 24.6% in S/G2, respectively, p < 0.01) ().
Figure 1. S. typhimurium A1-(R) decoyed quiescent cancer cells to cycle. S. typhimurium A1-R targeted quiescent cancer cells and decoyed cell cycle transit from G0/G1 to S/G2/M phases. (A) Representative images of control HeLa-FUCCI cancer cells and HeLa-FUCCI cancer cells in monolayer culture treated with S. typhimurium A1-R. (B) Histogram shows cell cycle distribution in control and S. typhimurium A1-R-treated cultures. Scale bar: 500 mm. (C) S. typhimurium A1-R stimulated cell-cycle transit from G0/G1 to S/G2 phase in quiescent tumor spheres formed from MKN45-FUCCI cells in vitro. Representative images of control tumor spheres and and tumor spheres treated with S. typhimurium A1-R. (D) Histogram shows cell-cycle distribution in control and S. typhimurium A1-R-treated tumor spheres. (E) S. typhimurium A1-R decoyed the cell-cycle transit of quiescent cancer cells in MKN45-FUCCI tumors in vivo. Representative images of cross sections of FUCCI-expressing MKN45 tumor xenografts treated with S. typhimurium A1-R or untreated control. (F) Histograms show the cell-cycle phase distribution of FUCCI-expressing cells within the tumors treated with S. typhimurium A1-R or untreated control. The cells in G0/G1, S, or G2/M phases appear red, yellow, or green, respectively.
Recombinant methionine (rMETase) trap of cancer cells in S/G2 visualized by FUCCI imaging
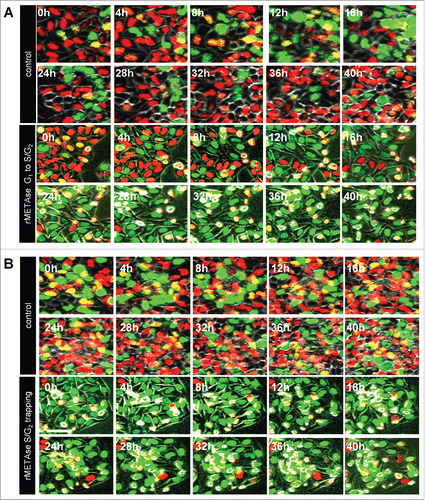
Control HeLa cells in vitro continue to divide. In contrast, rMETase trapped HeLa-FUCCI cells in S/G2 phase before cell division (). rMETase continued to trap HeLa-FUCCI cells in S/G2 phase over time without entry into mitosis ().
Figure 2. rMETase traps cancer cells in S/G2 phase. Time-course imaging of HeLa-FUCCI cells treated with rMETase. After seeding on 35 mm glass dishes and culture overnight, HeLa-FUCCI cells were treated with rMETase at a dose of 1.0 unit/ml. (A) Kinetics of rMETase trapping of cells in S/G2. (B) Maintenance of rMETase trap in S/G2 over time. All images were acquired with the FV1000 confocal microscope (Olympus, Tokyo, Japan).Citation81 The cells in G0/G1, S, or G2/M phases appear red, yellow, or green, respectively.
Decoy, trap and shoot chemotherapy with CDDP
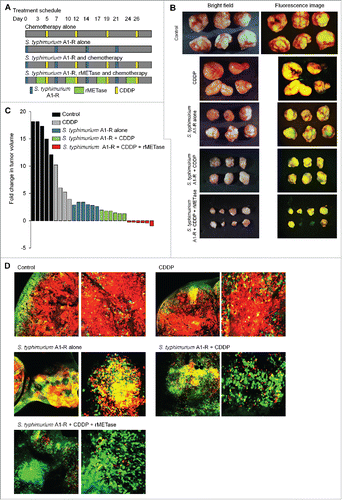
MKN45 tumor-bearing mice were treated with CDDP; or S. typhimurium A1-R; or S. typhimurium A1-R and CDDP or S. typhimurium A1-R, rMETase and CDDP. CDDP inhibited tumor growth (p < 0.01). S. typhimurium A1-R inhibited tumor growth more than CDDP (p < 0.01). S. typhimurium A1-R and CDDP combined had a greater inhibition of tumor growth (p < 0.01). The sequential combination of S. typhimurium A1-R, rMETase and CDDP prevented or regressed tumor growth more than S. typhimurium A1-R or CDDP alone or the combination of these two agents (p < 0.01) ().
Figure 3. Decoy, trap and shoot chemotherapy with CDDP. (A) Treatment schedule. FUCCI-expressing MKN45 cells (5 × 106 cells/mouse) were injected subcutaneously into the left flank of nude mice. When the tumors reached approximately 8 mm in diameter (tumor volume, 300 mm3), mice were administered iv S. typhimurium A1-R alone (5 × 107 CFU/mouse, iv, qW × 4); or cisplatinum (CDDP) alone (5 mg/kg, ip, q3d); or S. typhimurium A1-R followed by CDDP; or S. typhimurium A1-R, rMETase (200 units/mouse, ip, q d for 3 d × 4) and CDDP in that order. (B) Macroscopic photographs of FUCCI-expressing tumors: untreated control; S. typhimurium A1-R-treated; CDDP-treated; S. typhimurium A1-R and CDDP-treated; or treated with the sequential combination of S. typhimurium A1-R, rMETase and CDDP. (C) Waterfall plot indicating fold change in tumor volume: untreated control; CDDP-treated; S. typhimurium A1-R-treated; S. typhimurium A1-R and CDDP-treated; or treated with the sequential combination of S. typhimurium A1-R, rMETase and CDDP. (D) Representative images of cross-sections of FUCCI-expressing MKN45 subcutaneous tumors: untreated control; S. typhimurium A1-R-treated; CDDP-treated; S. typhimurium A1-R and CDDP-treated; or treated with the sequential combination of S. typhimurium A1-R, rMETase and CDDP.
rMETase induces mitotic catastrophe after late-S/G2 trap visualized by FUCCI imaging
HeLa-FUCCI cells were treated with rMETase for more than 80 hours. HeLa-FUCCI cells trapped in late-S/G2 phase did not divide and their nuclei turned red, after which they died (). These results showed that methionine was indispensable for cell division, and therefore rMETase induced mitotic catastrophe.Citation34-36
Figure 4. Prolonged administration of rMETase induced mitotic catastrophe after late S/G2 phase blocking. (A) Time-lapse imaging of HeLa-FUCCI cells treated with rMETase. After seeding on 35 mm glass dishes and culture overnight, HeLa-FUCCI cells were treated with rMETase at a dose of 1.0 unit/ml for 80 hours. All images were acquired with the FV1000 confocal microscope (Olympus, Tokyo, Japan). The cells in G0/G1, S, or G2/M phases appear red, yellow, or green, respectively. (B) High magnificent image of A. Arrowheads refer to a cell dying from mitotic catastrophe.
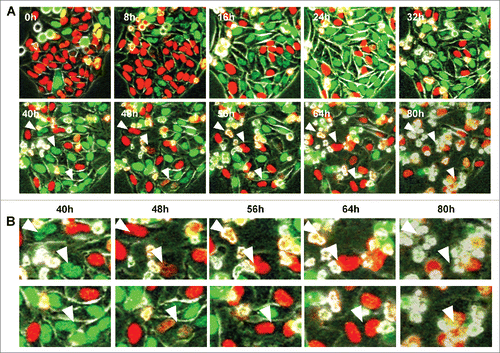
Figure 5. Decoy, trap and shoot chemotherapy with PTX. A). Treatment schedule. FUCCI-expressing MKN45 cells (5 × 106 cells/mouse) were injected subcutaneously into the left flank of nude mice. When the tumors reached approximately 8 mm in diameter (tumor volume, 300 mm3), mice were administered S. typhimurium A1-R alone (5 × 107 CFU/mouse, iv, qW × 4), or PTX alone (6 mg/kg, ip, q3d × 4); or S. typhimurium A1-R followed by PTX, or S. typhimurium A1-R, rMETase (200 units/mouse, ip, q d for 3 d × 4) and PTX sequentially. (B) Macroscopic photographs of FUCCI-expressing tumors: untreated control; S. typhimurium A1-R-treated; PTX-treated; S. typhimurium A1-R in combination with PTX-treated; or treated with the sequential combination of S. typhimurium A1-R, rMETase and PTX. Scale bars, 10 mm. (C) Waterfall plot indicating fold change in tumor volume: untreated control; PTX-treated; S. typhimurium A1-R-treated; S. typhimurium A1-R in combination with PTX-treated; or treated with the sequential combination of S. typhimurium A1-R, rMETase and PTX. (D) Representative images of cross-sections of FUCCI-expressing MKN45 subcutaneous tumors: untreated control; S. typhimurium A1-R-treated; PTX-treated; S. typhimurium A1-R in combination with PTX-treated; or treated with the sequential combination of S. typhimurium A1-R, rMETase and PTX.
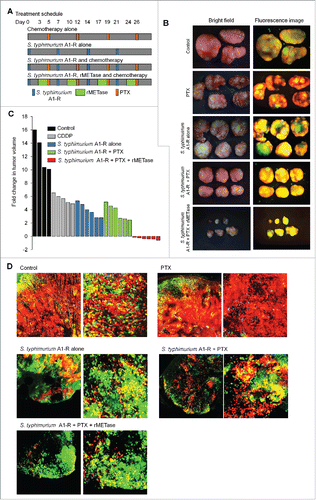
Decoy, trap and shoot chemotherapy with mitotic inhibitor PTX paclitaxel (PTX)
Based on rMETase prevention of cell division, we tested decoy, trap and shoot chemotherapy on MKN45 tumor bearing mice with paclitaxel (PTX). PTX alone and S. typhimurium A1-R alone significantly inhibited tumor growth (p < 0.05). S. typhimurium A1-R combined with PTX had a similar inhibition of tumor growth compared with PTX alone or S. typhimurium A1-R alone. The sequential combination of S. typhimurium A1-R, rMETase and PTX prevented or regressed tumor growth more than S. typhimurium A1-R or PTX alone or the combination of these two agents (p < 0.05).
Previously-developed concepts and strategies of highly selective tumor-targetingCitation37-48 can take advantage of bacterial cell-cycle decoy and rMETase cell-cycle trap described in the present and previous reports.Citation1,49,50
Excess thymidine or its analogs have also been used to arrest cancer cells in S-phase, where they are sensitized to S-phase drugs, and after the release of the block, the cancer cells are sensitive to M-Phase drugs.Citation51-53
Cytosine arabinoside, methotrexate and hydroxyorea have been used to block cancer cells in S-phase which can sensitize them to an M-phase drug administered after the S-phase block is lifted.Citation54-58
Mibefradil, a calcium channel blocker, has been used to synchronize glioblastoma cells at the G1/S checkpoint sensitizing them to temozolomide.Citation59 Lovastatin can be used to synchronize cancer cells in G1.Citation60,61 The cancer cells can be effectively treated with an S-phase drug after the block is lifted.
PDO332991, a pyridopyrimidine, inhibits cyclin-dependent kinases 4 and 6 and induced early-G1 arrest in myeloma cells in vitro and in vivo where they become sensitive to cytotoxic drugs.Citation62 RO-3306, another cyclin-kinase inhibitor, arrests cancer cells in G2 phase which become sensitive to M-phase drugs after the block is lifted.Citation63 EGF, G-CSF, and IL-6 can stimulate cancer cell out of G0 and can sensitize them to cytotoxic chemotherapy.Citation64-66 Reviews on cell synchronization are available.Citation67-70
The critical advantage of S. typhimurium A1-decoy and rMETase trapping is that both are tumor specific, unlike the methods listed above, and can overcome tumor intrinsic chemoresistance (TIC).Citation27,28,32,71-79
Materials and methods
FUCCI (Fluorescence ubiquitination cell cycle indicator)
The FUCCI probe was generated by fusing mKO2 (monomeric kusabira orange2) and mAG (monomeric azami green) to the ubiquitination domains of human Cdt1 and geminin, respectively. These 2 chimeric proteins, mKO2-hCdt1and mAG-hGem, accumulate reciprocally in the nuclei of transfected cells during the cell cycle, labeling the nuclei of G1 phase cells orange and nuclei of cells in S/G2/M phase green.Citation1 Plasmids expressing mKO2-hCdt1 (orange fluorescent protein) or mAG-hGem (green fluorescent protein) were obtained from the Medical and Biological Laboratory. Plasmids expressing mAG-hGem were transfected into MKN45 cells using Lipofectamine™ LTX (Invitrogen). The cells were incubated for 48 h after transfection and were then trypsinized and seeded in 96-well plates at a density of 10 cells/well. In the first step, cells were sorted into green (S, G2, and M phase) cells using a cell sorter. The first-step-sorted green-fluorescent cells were then re-transfected with mKO2-hCdt1 (orange) and then sorted by orange fluorescence.Citation1,2
Cells
MKN45 human stomach cancer cells were grown in RPMI 1640 medium with 10% fetal bovine serum and penicillin/streptomycin.Citation1 HeLa cells were grown in DMEM supplemented with 10% fetal bovine serum and penicillin/streptomycin.Citation32
Mice
Athymic nu/nu nude mice (AntiCancer, Inc., San Diego, CA) were maintained in a barrier facility under HEPA filtration and fed with autoclaved laboratory rodent diet (Teklad LM-485; Harlan). All animal procedures were performed under anesthesia using s.c. administration of a ketamine mixture (10 μl ketamine HCl, 7.6 μl xylazine, 2.4 μl acepromazine maleate, and 10 μl PBS) (Henry-Schein). FUCCI-expressing MKN45 cells were harvested from monolayer culture by brief trypsinization. Single-cell suspensions were prepared at a final concentration of 5 × 106 cells and injected subcutaneously in the left flank of nude mice. All animal studies were conducted in accordance with the principles and procedures outlined in the National Institute of Health Guide for the Care and Use of Animals under Assurance Number A3873–1.Citation19
Recombinant methioninase (rMETase)
Recombinant L-methionine α-deamino-γ- mercaptomethane lyase (methioninase, METase) [EC 4.4.1.11] from Pseudomonas putida has been previously cloned and was produced in Escherichia coli (AntiCancer, Inc.,). rMETase is a homotetrameric PLP enzyme of 172-kDa molecular mass.Citation30,80
Decoy, trap and shoot chemotherapy
When the tumors reached approximately 8 mm in diameter (tumor volume, 300 mmCitation3), mice were administered iv S. typhimurium A1-R (5 × 107 CFU/mouse, iv, qW × 4) alone or in combination with cisplatinum (CDDP) (5 mg/kg ip) or paclitaxel (PTX) (6 mg/kg ip) q 3 d × 5 or the combination of S. typhimurium A1-R and either CDDP or PTX, or these combinations with rMETase (200 units/mouse).Citation19 Please see text and figure legends for dosing schedules.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Dedication
This paper is dedicated to the memory of A.R. Moossa, MD, and Sun Lee, MD.
Funding
This study was supported in part by the National Cancer Institute grants CA13297 and CA142669. This study was also supported by grants from the Ministry of Health, Labour, and Welfare, Japan (to T. Fujiwara; No. 10103827, No. 13801426, No. 14525167) and grants from the Ministry of Education, Culture, Sports, Science and Technology, Japan (to T. Fujiwara; No. 25293283).
References
- Yano S, Zhang Y, Miwa S, Tome Y, Hiroshima Y, Uehara F, Yamamoto M, Suetsugu A, Kishimoto H, Tazawa H, et al. Spatial-temporal FUCCI imaging of each cell in a tumor demonstrates locational dependence of cell cycle dynamics and chemoresponsiveness. Cell Cycle 2014; 13:2110-9; PMID:24811200; http://dx.doi.org/10.4161/cc.29156
- Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, Kashiwagi S, Fukami K, Miyata T, Miyoshi H, et al. Visualizing spatiotemporal dynamics of multicellular cell cycle progression. Cell 2008; 132:487-98; PMID:18267078; http://dx.doi.org/10.1016/j.cell.2007.12.033
- Zhang Y, Zhang N, Zhao M, Hoffman RM. Comparison of the selective targeting efficacy of Salmonella typhimurium A1-R and VNP20009 on the Lewis lung carcinoma in nude mice. Oncotarget 2015; 6:14625-31; PMID:25714030; http://dx.doi.org/10.18632/oncotarget.3342
- Hoffman RM, editor. Bacterial Therapy of Cancer: Methods and Protocols. Methods in Molecular Biology 1409. Walker John M, series ed. Humana Press (Springer Science + Business Media New York), 2016
- Zhao M, Yang M, Li XM, Jiang P, Baranov E, Li S, Xu M, Penman S, Hoffman RM. Tumor-targeting bacterial therapy with amino acid auxotrophs of GFP-expressing Salmonella typhimurium. Proc Natl Acad Sci USA 2005; 102:755-60; PMID:15644448; http://dx.doi.org/10.1073/pnas.0408422102
- Zhao M, Geller J, Ma H, Yang M, Penman S, Hoffman RM. Monotherapy with a tumor-targeting mutant of Salmonella typhimurium cures orthotopic metastatic mouse models of human prostate cancer. Proc Natl Acad Sci USA 2007; 104:10170-4; PMID:17548809; http://dx.doi.org/10.1073/pnas.0703867104
- Zhao M, Yang M, Ma H, Li X, Tan X, Li S, Yang Z, Hoffman RM. Targeted therapy with a Salmonella typhimurium leucine-arginine auxotroph cures orthotopic human breast tumors in nude mice. Cancer Res 2006; 66:7647-52; PMID:16885365; http://dx.doi.org/10.1158/0008-5472.CAN-06-0716
- Zhang Y, Tome Y, Suetsugu A, Zhang L, Zhang N, Hoffman RM, Zhao M. Determination of the optimal route of administration of Salmonella typhimurium A1-R to target breast cancer in nude mice. Anticancer Res 2012; 32:2501-8; PMID:22753706
- Zhang Y, Miwa S, Zhang N, Hoffman RM, Zhao M. Tumor-targeting Salmonella typhimurium A1-R arrests growth of breast-cancer brain metastasis. Oncotarget 2015; 6:2615-22; PMID:25575815; http://dx.doi.org/10.18632/oncotarget.2811
- Uchugonova A, Zhao M, Zhang Y, Weinigel M, König K, Hoffman RM. Cancer-cell killing by engineered Salmonella imaged by multiphoton tomography in live mice. Anticancer Res 2012; 32:4331-7; PMID:23060555
- Liu F, Zhang L, Hoffman RM, Zhao M. Vessel destruction by tumor-targeting Salmonella typhimurium A1-R is enhanced by high tumor vascularity. Cell Cycle 2010; 9:4518-24; PMID:21135579; http://dx.doi.org/10.4161/cc.9.22.13744
- Nagakura C, Hayashi K, Zhao M, Yamauchi K, Yamamoto N, Tsuchiya H, Tomita K, Bouvet M, Hoffman RM. Efficacy of a genetically-modified Salmonella typhimurium in an orthotopic human pancreatic cancer in nude mice. Anticancer Res 2009; 29:1873-8; PMID:19528442
- Yam C, Zhao M, Hayashi K, Ma H, Kishimoto H, McElroy M, Bouvet M, Hoffman RM. Monotherapy with a tumor-targeting mutant of S. typhimurium inhibits liver metastasis in a mouse model of pancreatic cancer. J Surg Res 2010; 164:248-55; PMID:19766244; http://dx.doi.org/10.1016/j.jss.2009.02.023
- Hiroshima Y, Zhao M, Zhang Y, Maawy A, Hassanein MK, Uehara F, Miwa S, Yano S, Momiyama M, Suetsugu A, et al. Comparison of efficacy of Salmonella typhimurium A1-R and chemotherapy on stem-like and non-stem human pancreatic cancer cells. Cell Cycle 2013; 12:2774-80; PMID:23966167; http://dx.doi.org/10.4161/cc.25872
- Hiroshima Y, Zhao M, Maawy A, Zhang Y, Katz MH, Fleming JB, Uehara F, Miwa S, Yano S, Momiyama M, et al. Efficacy of Salmonella typhimurium A1-R versus chemotherapy on a pancreatic cancer patient-derived orthotopic xenograft (PDOX). J Cell Biochem 2014; 115:1254-61; PMID:24435915; http://dx.doi.org/10.1002/jcb.24769
- Hiroshima Y, Zhang Y, Murakami T, Maawy AA, Miwa S, Yamamoto M, Yano S, Sato S, Momiyama M, Mori R, et al. Efficacy of tumor-targeting Salmonella typhimurium A1-R in combination with anti-angiogenesis therapy on a pancreatic cancer patient-derived orthotopic xenograph (PDOX) and cell line mouse models. Oncotarget 2014; 5:12346-57; PMID:25402324; http://dx.doi.org/10.18632/oncotarget.2641
- Matsumoto Y, Miwa S, Zhang Y, Hiroshima Y, Yano S, Uehara F, Yamamoto M, Toneri M, Bouvet M, Matsubara H, et al. Efficacy of tumor-targeting Salmonella typhimurium A1-R on nude mouse models of metastatic and disseminated human ovarian cancer. J Cell Biochem 2014; 115:1996-2003; PMID:24924355
- Matsumoto Y, Miwa S, Zhang Y, Zhao M, Yano S, Uehara F, Yamamoto M, Hiroshima Y, Toneri M, Bouvet M, et al. Intraperitoneal administration of tumor-targeting Salmonella typhimurium A1-R inhibits disseminated human ovarian cancer and extends survival in nude mice. Oncotarget 2015; 6:11369-77; PMID:25957417; http://dx.doi.org/10.18632/oncotarget.3607
- Yano S, Zhang Y, Zhao M, Hiroshima Y, Miwa S, Uehara F, Kishimoto H, Tazawa H, Bouvet M, Fujiwara T, et al. Tumor-targeting Salmonella typhimurium A1-R decoys quiescent cancer cells to cycle as visualized by FUCCI imaging and become sensitive to chemotherapy. Cell Cycle 2014; 13:3958-63; PMID:25483077; http://dx.doi.org/10.4161/15384101.2014.964115
- Hiroshima Y, Zhang Y, Zhao M, Zhang N, Murakami T, Maawy A, Mii S, Uehara F, Yamamoto M, Miwa S, et al. Tumor-targeting Salmonella typhimurium A1-R in combination with Trastuzumab eradicates HER-2-positive cervical cancer cells in patient-derived mouse models. PLoS One 2015; 10:e0120358; PMID:26047477; http://dx.doi.org/10.1371/journal.pone.0120358
- Hayashi K, Zhao M, Yamauchi K, Yamamoto N, Tsuchiya H, Tomita K, Hoffman RM. Cancer metastasis directly eradicated by targeted therapy with a modified Salmonella typhimurium. J Cell Biochem 2009; 106:992-8; PMID:19199339; http://dx.doi.org/10.1002/jcb.22078
- Hayashi K, Zhao M, Yamauchi K, Yamamoto N, Tsuchiya H, Tomita K, Kishimoto H, Bouvet M, Hoffman RM. Systemic targeting of primary bone tumor and lung metastasis of high-grade osteosarcoma in nude mice with a tumor-selective strain of Salmonella typhimurium. Cell Cycle 2009; 8:870-5; PMID:19221501; http://dx.doi.org/10.4161/cc.8.6.7891
- Miwa S, Zhang Y, Baek K-E, Uehara F, Yano S, Yamamoto M, Hiroshima Y, Matsumoto Y, Kimura H, Hayashi K, et al. Inhibition of spontaneous and experimental lung metastasis of soft-tissue sarcoma by tumor-targeting Salmonella typhimurium A1-R. Oncotarget 2014; 5:12849-61; PMID:25528763; http://dx.doi.org/10.18632/oncotarget.2561
- Murakami T, DeLong J, Eilber FC, Zhao M, Zhang Y, Zhang N, Singh A, Russell T, Deng S, Reynoso J, et al. Tumor-targeting Salmonella typhimurium A1-R in combination with doxorubicin eradicate soft tissue sarcoma in a patient-derived orthotopic xenograft PDOX model. Oncotarget, 2016; 7:12783-90.
- Kimura H, Zhang L, Zhao M, Hayashi K, Tsuchiya H, Tomita K, Bouvet M, Wessels J, Hoffman RM. Targeted therapy of spinal cord glioma with a genetically-modified Salmonella typhimurium. Cell Proliferation 2010; 43:41-48; PMID:19922490; http://dx.doi.org/10.1111/j.1365-2184.2009.00652.x
- Momiyama M, Zhao M, Kimura H, Tran B, Chishima T, Bouvet M, Endo I, Hoffman RM. Inhibition and eradication of human glioma with tumor-targeting Salmonella typhimurium in an orthotopic nude-mouse model. Cell Cycle 2012; 11:628-32; PMID:22274398; http://dx.doi.org/10.4161/cc.11.3.19116
- Hoffman RM, Jacobsen SJ. Reversible growth arrest in simian virus 40-transformed human fibroblasts. Proc Natl Acad Sci USA 1980; 77:7306-10; PMID:6261250; http://dx.doi.org/10.1073/pnas.77.12.7306
- Stern PH, Hoffman RM. Enhanced in vitro selective toxicity of chemotherapeutic agents for human cancer cells based on a metabolic defect. J Natl Cancer Inst 1986; 76:629-39; PMID:3457200
- Guo H, Lishko VK, Herrera H, Groce A, Kubota T, Hoffman RM. Therapeutic tumorspecific cell cycle block induced by methionine starvation in vivo. Cancer Res 1993; 53:5676-9; PMID:8242623
- Yano S, Li S, Han Q, Tan Y, Bouvet M, Fujiwara T, Hoffman RM. Selective methioninase-induced trap of cancer cells in S/G2 phase visualized by FUCCI imaging confers chemosensitivity. Oncotarget 2014; 5:8729-36; PMID:25238266; http://dx.doi.org/10.18632/oncotarget.2369
- Mecham JO, Rowitch D, Wallace CD, Stern PH, Hoffman RM. The metabolic defect of methionine dependence occurs frequently in human tumor cell lines. Biochem Biophys Res Commun 1983; 117:429-34; PMID:6661235; http://dx.doi.org/10.1016/0006-291X(83)91218-4
- Tan Y, Xu M, Hoffman RM. Broad selective efficacy of recombinant methioninase and polyethylene glycol-modified recombinant methioninase on cancer cells in vitro. Anticancer Res 2010; 30:1041-6; PMID:20530407
- Hoffman RM. Development of recombinant methioninase to target the general cancer-specific metabolic defect of methionine dependence: a 40-year odyssey. Expert Opin Biol Ther 2015; 15:21-31; PMID:25439528; http://dx.doi.org/10.1517/14712598.2015.963050
- Castedo M, Perfettini JL, Roumier T, Andreau K, Medema R, Kroemer G. Cell death by mitotic catastrophe: A molecular definition. Oncogene 2004; 23:2825-37; PMID:15077146; http://dx.doi.org/10.1038/sj.onc.1207528
- Chow JPH, Poon RYC. Mitotic catastrophe. In: Greg Enders, editor. Cell cycle deregulation in cancer. Springer, New York. 2010; 79-96.
- Vakifahmetoglu H, Olsson M, Zhivotovsky B. 2008. Death through a tragedy: Mitotic catastrophe. Cell Death Differ 2008; 15:1153-62; PMID:18404154; http://dx.doi.org/10.1038/cdd.2008.47
- Blagosklonny MV. How cancer could be cured by 2015. Cell Cycle 2005; 4:269-78; PMID:15655345
- Blagosklonny MV. Tissue-selective therapy of cancer. Br J Cancer 2003; 89:1147-51; PMID:14520435; http://dx.doi.org/10.1038/sj.bjc.6601256
- Blagosklonny MV. Matching targets for selective cancer therapy. Drug Discov Today 2003; 8:1104-7; PMID:14678733; http://dx.doi.org/10.1016/S1359-6446(03)02806-X
- Blagosklonny MV. “Targeting the absence” and therapeutic engineering for cancer therapy. Cell Cycle 2008; 7:1307-12; PMID:18487952; http://dx.doi.org/10.4161/cc.7.10.6250
- Blagosklonny MV. Teratogens as anti-cancer drugs. Cell Cycle 2005; 4:1518-21; PMID:16258270; http://dx.doi.org/10.4161/cc.4.11.2208
- Blagosklonny MV. Treatment with inhibitors of caspases, that are substrates of drug transporters, selectively permits chemotherapy-induced apoptosis in multidrug-resistant cells but protects normal cells. Leukemia 2001; 15:936-41; PMID:11417480; http://dx.doi.org/10.1038/sj.leu.2402127
- Blagosklonny MV. Target for cancer therapy: proliferating cells or stem cells. Leukemia 2006; 20:385-91; PMID:16357832; http://dx.doi.org/10.1038/sj.leu.2404075
- Blagosklonny MV. Cancer stem cell and cancer stemloids: from biology to therapy. Cancer Biol Ther 2007; 6:1684-90; PMID:18344680; http://dx.doi.org/10.4161/cbt.6.11.5167
- Apontes P, Leontieva OV, Demidenko ZN, Li F, Blagosklonny MV. Exploring long-term protection of normal human fibroblasts and epithelial cells from chemotherapy in cell culture. Oncotarget 2011; 2:222-33; PMID:21447859; http://dx.doi.org/10.18632/oncotarget.248
- Rao B, van Leeuwen IM, Higgins M, Campbel J, Thompson AM, Lane DP, Lain S. Evaluation of an Actinomycin D/VX-680 aurora kinase inhibitor combination in p53-based cyclotherapy. Oncotarget 2010; 1:639-50; PMID:21317459; http://dx.doi.org/10.18632/oncotarget.198
- Blagosklonny MV. NCI's provocative questions on cancer: some answers to ignite discussion. Oncotarget 2011; 2:1352-67; PMID:22267462; http://dx.doi.org/10.18632/oncotarget.432
- Blagosklonny MV. Antagonistic drug combinations that select against drug resistance: from bacteria to cancer. Cancer Biol Ther 2007; 6:1013-4; PMID:17646740; http://dx.doi.org/10.4161/cbt.6.7.4340
- Yano S, Miwa S, Mii S, Hiroshima Y, Uehara F, Yamamoto M, Kishimoto H, Tazawa H, Bouvet M, Fujiwara T, et al. Invading cancer cells are predominantly in G0/G1 resulting in chemoresistance demonstrated by real-time FUCCI imaging. Cell Cycle 2014; 13:953-960; PMID:24552821; http://dx.doi.org/10.4161/cc.27818
- Yano S, Tazawa H, Hashimoto Y, Shirakawa Y, Kuroda S, Nishizaki M, Kishimoto H, Uno F, Nagasaka T, Urata Y, et al. A genetically engineered oncolytic adenovirus decoys and lethally traps quiescent cancer stem-like cells into S/G2/M phases. Clin Cancer Res 2013; 19:6495-505; PMID:24081978; http://dx.doi.org/10.1158/1078-0432.CCR-13-0742
- Wang X, Pan L, Mao N, Sun L, Qin X, Yin J. Cell-cycle synchronization reverses Taxol resistance of human ovarian cancer cell lines. Cancer Cell Int 2013; 13:77; PMID:23899403; http://dx.doi.org/10.1186/1475-2867-13-77
- Chandrasekaran B, Kute TE, Duch DS. Synchronization of cells in the S phase of the cell cycle by 3′-azido-3′-deoxythymidine: implications for cell cytotoxicity. Cancer Chemother Pharmacol 1995; 35:489-95; PMID:7882457; http://dx.doi.org/10.1007/BF00686833
- Kufe DW, Egan EM, Rosowsky A, Ensminger W, Frei E 3rd. Thymidine arrest and synchrony of cellular growth in vivo. Cancer Treat Rep 1980; 64:1307-17; PMID:7471121
- Vogler WR, Kremer WB, Knospe WH, Omura GA, Tornyos K. Synchronization with phase-specific agents in leukemia and correlation with clinical response to chemotherapy. Cancer Treat Rep 1976; 60:1845-59; PMID:1026339
- Morris CM, Fitzgerald PH. An evaluation of high resolution chromosome banding of hematologic cells by methotrexate synchronization and thymidine release. Cancer Genet Cytogenet 1985; 14:275-84; PMID:3967208; http://dx.doi.org/10.1016/0165-4608(85)90193-1
- Moran RE, Straus MJ. Synchronization of L1210 leukemia with hydroxyurea infusion and the effect of subsequent pulse dose chemotherapy. Cancer Treat Rep 1980; 64:81-6; PMID:7379059
- Dethlefsen LA, Sorensen SP, Riley RM. Effects of double and multiple doses of hydroxyurea on mouse duodenum and mammary tumors. Cancer Res 1975; 35:694-9; PMID:1116130
- Finzi L, Kraemer A, Capron C, Noullet S, Goere D, Penna C, Nordlinger B, Legagneux J, Emile JF, Malafosse R. Improved retroviral suicide gene transfer in colon cancer cell lines after cell synchronization with methotrexate. J Exp Clin Cancer Res 2011; 30:92; PMID:21970612
- Keir ST, Friedman HS, Reardon DA, Bigner DD, Gray LA. Mibefradil, a novel therapy for glioblastoma multiforme: cell cycle synchronization and interlaced therapy in a murine model. J Neurooncol 2013; 111:97-102; PMID:23086436; http://dx.doi.org/10.1007/s11060-012-0995-0
- Keyomarsi K, Sandoval L, Band V, Pardee AB. Synchronization of tumor and normal cells from G1 to multiple cell cycles by lovastatin. Cancer Res 1991; 51:3602-9; PMID:1711413
- Javanmoghadam-Kamrani S, Keyomarsi K. Synchronization of the cell cycle using lovastatin. Cell Cycle 2008; 7:2434-40; PMID:18677105; http://dx.doi.org/10.4161/cc.6364
- Huang X, Di Liberto M, Jayabalan D, Liang J, Ely S, Bretz J, Shaffer AL 3rd, Louie T, Chen I, Randolph S, et al. Prolonged early G(1) arrest by selective CDK4/CDK6 inhibition sensitizes myeloma cells to cytotoxic killing through cell cycle-coupled loss of IRF4. Blood 2012; 120:1095-106; PMID:22718837; http://dx.doi.org/10.1182/blood-2012-03-415984
- Vassilev LT. Cell cycle synchronization at the G2/M phase border by reversible inhibition of CDK1. Cell Cycle 2006; 5:2555-6; PMID:17172841; http://dx.doi.org/10.4161/cc.5.22.3463
- Dong XF, Berthois Y, Dussert C, Isnardon D, Palmari J, Martin PM. Mode of EGF action on cell cycle kinetics in human breast cancer cell line MCF-7: some evidence that EGF acts as a “progression factor”. Anticancer Res 1992; 12:2085-92; PMID:1295452
- Hambek M, Werner C, Baghi M, Gstöttner W, Knecht R. Enhancement of docetaxel efficacy in head and neck cancer treatment by G0 cell stimulation. Eur J Cancer 2007; 43:1502-7; PMID:17524637; http://dx.doi.org/10.1016/j.ejca.2005.09.037
- Hambek M, Werner C, Baghi M, Gstöttner W, Knecht R. Prestimulation of head and neck cancer cells with growth factors enhances treatment efficacy. Anticancer Res 2006; 26:1091-5; PMID:16619511
- Grdina DJ, Meistrich ML, Meyn RE. Cell synchrony techniques. A comparison of methods. In: Techniques in Cell Cycle Analysis. Gray JW and Darzynkiewicz A (eds). Humana Press Inc., Clifton, NJ, 1987; 367-403.
- Davis PK, Ho A, Dowdy SF. Biological methods for cell-cycle synchronization of mammalian cells. Biotechniques 2001; 30:1322-6, 1328, 1330-1.
- Merril GF. Cell synchronization. Methods Cell Biol 1998; 57:229-249; PMID:9648108; http://dx.doi.org/10.1016/S0091-679X(08)61582-4
- Amon A. Synchronization procedures. Methods Enzymol 2002; 351:457-67; PMID:12073363; http://dx.doi.org/10.1016/S0076-6879(02)51864-4
- Hoffman RM. Altered methionine metabolism, DNA methylation and oncogene expression in carcinogenesis: a review and synthesis. Biochim Biophys Acta Reviews on Cancer 1984; 738:49-87; http://dx.doi.org/10.1016/0304-419X(84)90019-2
- Hoffman RM, Erbe RW. High in vivo rates of methionine biosynthesis in transformed human and malignant rat cells auxotrophic for methionine. Proc Natl Acad Sci USA 1976; 73:1523-7; PMID:179090; http://dx.doi.org/10.1073/pnas.73.5.1523
- Hoffman RM, Jacobsen SJ, Erbe RW. Reversion to methionine independence by malignant rat and SV40-transformed human fibroblasts. Biochem Biophys Res Commun 1978; 82:228-34; PMID:208554; http://dx.doi.org/10.1016/0006-291X(78)90600-9
- Hoffman RM, Jacobsen SJ, Erbe RW. Reversion to methionine independence in simian virus 40-transformed human and malignant rat fibroblasts is associated with altered ploidy and altered properties of transformation. Proc NatlAcad Sci USA 1979; 76:1313-7; http://dx.doi.org/10.1073/pnas.76.3.1313
- Coalson DW, Mecham JO, Stem PH, Hoffman RM. Reduced availability of endogenously synthesized methionine for S-adenosylmethionine formation in methionine-dependent cancer cells. Proc Natl Acad Sci USA 1982; 79:4248-51; PMID:6289297; http://dx.doi.org/10.1073/pnas.79.14.4248
- Stem PH, Mecham JO, Wallace CD, Hoffman RM. Reduced free-methionine in methionine-dependent SV40-transformed human fibroblasts synthesizing apparently normal amounts of methionine. J Cell Physiol l983; 117:9-14.
- Mecham JO, Rowitch D, Wallace CD, Stem PH, Hoffman RM. The metabolic defect of methionine dependence occurs frequently in human tumor cell lines. Biochem Biophys Res Commun 1983; 117:429-34; PMID:6661235; http://dx.doi.org/10.1016/0006-291X(83)91218-4
- Stem PH, Wallace CD, Hoffman RM. Altered methionine metabolism occurs in all members of a set of diverse human tumor cell lines. J Cell Physiol 1984; 9:29-34.
- Stem PH, Hoffman RM. Elevated overall rates of transmethylation in cell lines from diverse human tumors. In Vitro 1984; 20:663-70; PMID:6500606; http://dx.doi.org/10.1007/BF02619617
- Tan Y, Xu M, Tan X-Z, Tan X-Y, Wang, X, Saikawa Y, Nagahama T, Sun X, Lenz M, Hoffman RM. Overexpression and large-scale production of recombinant l-methionine-a-deamino-g-mercaptomethane-lyase for novel anticancer therapy. Prot Exp Purif 1997; 9:233-45; http://dx.doi.org/10.1006/prep.1996.0700
- Uchugonova A, Duong J, Zhang N, König K, Hoffman RM. The bulge area is the origin of nestin-expressing pluripotent stem cells of the hair follicle. J Cell Biochem 2011; 112:2046-50; PMID:21465525; http://dx.doi.org/10.1002/jcb.23122
