ABSTRACT
Retinoblastoma protein (Rb) is a prototypical tumor suppressor that is vital to the negative regulation of the cell cycle and tumor progression. Hypo-phosphorylated Rb is associated with G0/G1 arrest by suppressing E2F transcription factor activity, whereas Rb hyper-phosphorylation allows E2F release and cell cycle progression from G0/G1 to S phase. However, the factors that regulate cyclin-dependent protein kinase (CDK)-dependent hyper-phosphorylation of Rb during the cell cycle remain obscure. In this study, we show that throughout the cell cycle, Rb is specifically small ubiquitin-like modifier (SUMO)ylated at early G1 phase. SUMOylation of Rb stimulates its phosphorylation level by recruiting a SUMO-interaction motif (SIM)-containing kinase CDK2, leading to Rb hyper-phosphorylation and E2F-1 release. In contrast, a SUMO-deficient Rb mutant results in reduced SUMOylation and phosphorylation, weakened CDK2 binding, and attenuated E2F-1 sequestration. Furthermore, we reveal that Rb SUMOylation is required for cell proliferation. Therefore, our study describes a novel mechanism that regulates Rb phosphorylation during cell cycle progression.
Introduction
The accurate control of the G0/G1 to S phase transition during the eukaryotic cell cycle is essential for cell proliferation, and it is controlled by a delicate regulatory network.Citation1,2 One of the key players is Rb, the first identified tumor suppressor.Citation2,3 Its inactivation directly induces the development of retinoblastoma, the most common malignant tumor in children, as well as several other major cancers.Citation3,4 Rb and its homologs p130 and p107 belong to the pocket protein family, which inhibit cell cycle progression by regulating the family of E2F transcription factors, whose activities are essential for the G0/G1 to S transition.Citation4,5 Briefly, in non-cycling, quiescent cells (G0 phase), Rb is present in its hypo-phosphorylated (hypo-Rb), activated form, which is phosphorylated only at a few sites, and hypo-Rb is bound to E2F transcription factors to repress the transcription of genes required for DNA replication and cell division.Citation6-8 As cells progress through G0/G1 toward S phase, Rb becomes hyper-phosphorylated (hyper-pRb) to its inactivated form by cyclins and CDKs, such as cyclin E/CDK2 and cyclin D/CDK4/6, leading to the release of E2F factors and the transcription of S phase genes.Citation9-11 Because it is so important in cell cycle regulation, the phosphorylation of Rb by CDKs during the G0/G1 to S transition has been extensively studied.Citation7,12 However, the factors regulating this process remain unclear.
There has been evidence of phosphorylation cross-talk with SUMOylation.Citation13,14 SUMOylation is a form of post-translational modification using an enzymatic pathway similar to ubiquitination.Citation15,16 It is facilitated by E1 activating enzymes SAE1 and SAE2, the sole E2 conjugating enzyme Ubc9 and various E3 ligases. SUMO comprises 4 distinct proteins in humans (SUMO-1, −2, −3, and −4). SUMO2 and SUMO3 are closely related, whereas SUMO1 shares 50% similarity with either SUMO2 or SUMO3. By the covalent but reversible attachment of SUMO to protein substrates at specific lysine residues, SUMOylation can modulate many cellular processes, such as sub-cellular localization, transcription activation, cell cycle regulation and DNA synthesis and repair.Citation13,17 Besides, it is worth noting that SUMO proteins also have the ability to bind target proteins in a non-covalent manner through the SIM, which can act as a docking site to enhance the interaction between SUMOylated proteins and SIM-containing proteins.
Recent studies indicated a role for SUMOylation in cell cycle regulation as well as cancer development and progression. Furthermore, previous studies suggested that Rb could be SUMOylated on K720in the B domain of the pocket region by both SUMO1 and SUMO2.Citation18,19 However, the role for SUMOylation on Rb function is not clear. Here we show that the SUMOylation of Rb plays an essential role during early G1 phase by increasing its binding with CDK2 through SUMO-SIM interaction and its phosphorylation level, leading to downstream E2F1 transcription factor release and S phase gene expression, which promotes cell cycle progression.
Results
Rb is SUMOylated at early G1 phase
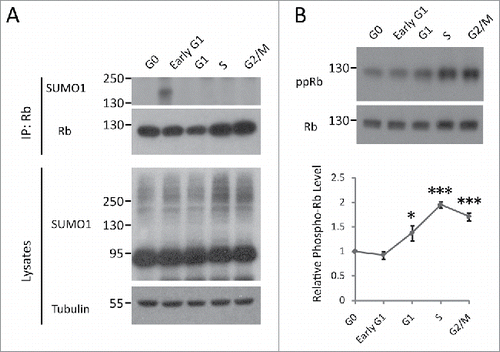
Rb can be SUMOylated, but the functional and physiological relevance are unknown.Citation18,19 To gain insight into the functional role of SUMO conjugation to Rb, we first examined the SUMOylation status of Rb during cell cycle progression. HEK293 cells were synchronized at 5 different stages of the cell cycle (G0, early G1, G1, S and G2/M) as described in the Materials and Methods section. The cells were lysed in the presence of 20 mM N-ethylmaleimide (NEM), a SUMO protease inhibitor, to protect SUMO-conjugated products during the experiments. After immunoprecipitation of endogenous Rb species under denaturing conditions, we detected the presence of the SUMOylated Rb signal using an anti-SUMO1 antibody specifically at early G1 phase, suggesting SUMO1-Rb is involved in G0/G1 to S phase transition ().
Figure 1. Dynamics of Rb SUMOylation and phosphorylation during the cell cycle. (A) Rb is SUMOylated at early G1 phase. HEK293 cells were synchronized at the G0, early G1, G1, S and G2/M phases of the cell cycle, as described in the Materials and Methods section. The lysates in RIPA buffer were subjected to SDS-PAGE or immunoprecipitation using anti-Rb antibody and then blotted with SUMO1 antibody. (B) Rb is gradually phosphorylated after late G1 phase. HEK293 cells were synchronized and lysed as described above, followed by Western blot analysis with phosphorylated-Rb (Ser 807/811) antibody. Quantification of the data are represented as the mean ± the SEM (n = 3).
SUMOylation of Rb promotes its phosphorylation
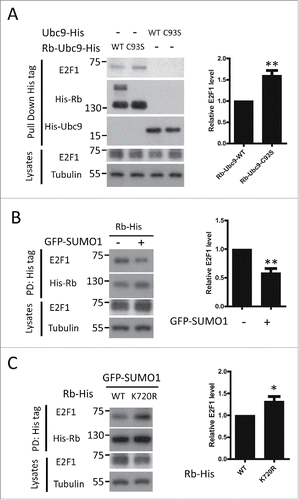
Increasing evidence has shown that SUMOylation and phosphorylation may be associated with one another to fine-tune protein function in a wide variety of cellular processes, e.g., substrate phosphorylation could either facilitate or antagonize the self-SUMOylationCitation20-23; and evidence for SUMOylation-enhanced protein phosphorylation was also found.Citation14 For Rb, research over the past 2 decades has revealed that phosphorylation is crucial for its activity in cell cycle regulation. Thus, we wanted to assess whether there is interplay between the 2 modifications of Rb. To this end, we first examined the Rb phosphorylation level during cell cycle progression. We found that the phosphorylation of Rb was significantly elevated following late G1 phase and increased further as the cell cycle progressed, as measured by immunoblots for 2 major phosphorylation sites of phospho-Rb(807/811,ppRb), which is consistent with previous reports ().Citation3,24 Because SUMO modification of Rb occurred prior to phosphorylation during the cell cycle (), the SUMO conjugation of Rb may regulate its phosphorylation status. To confirm this theory, we generated a constitutive SUMOylated Rb construct by fusing SUMO E2 ligase Ubc9 to its C-terminus, which allowed efficient and selective SUMOylation of Rb ().Citation25 Initially, to identify the Rb-SUMO conjugation caused by Ubc9 fusion-directed SUMOylation(UFDS), we omitted NEM during the cell extract preparation. We discovered that the omission of this reagent led to a loss of Rb-SUMO conjugates, suggesting that this higher molecular weight, NEM-sensitive band was the SUMOylated form of Rb, whereas a Ubc9 defective mutation, C93S, failed to produce this band (). Then, we further confirmed the UFDS of Rb using an anti-SUMO1 antibody (). Ubc9 alone did not cause any SUMO conjugation, further confirming Rb-specific SUMOylation (). We determined that the SUMOylation of Rb led to significantly increased phospho-Rb levels using the phospho-specific Rb antibody (S807/S811, ppRb) (). Moreover, the SUMOylation of Rb resulted in a band that migrated slightly between Rb and Rb-SUMO1 with the most notably increased phosphorylation level (), suggesting that it is the hyper-phosphorylated form. Nevertheless, we also noticed that the SUMO-conjugation led to an increase in phosphorylation of unmodified Rb-Ubc9 (). Then, we further validated this phenomenon by monitoring the change in Rb phosphorylation at various SUMOylation levels by expressing increasing amounts of GFP-SUMO1 (). The phosphorylated Rb showed a significant increase, even when the exogenous SUMOylation just started to accumulate ().
Figure 2. SUMOylation of Rb promotes its phosphorylation. (A) Diagram of the Ubc9 fusion-directed SUMOylation (UFDS) constructs of Rb. (B) Constitutive SUMOylation of Rb caused by UFDS. HEK293 cells transiently transfected with His-tagged UFDS constructs were lysed in RIPA buffer with or without NEM, and then blotted with anti-His antibody. Rb-Ubc9: unmodified Rb-Ubc9; Rb-Ubc9-S: SUMOylated Rb-Ubc9. (C) UFDS of VCP promotes its phosphorylation level. HEK293 cells were co-transfected with indicated constructs. Lysates were incubated with Ni-NTA agarose beads to pull down His-tagged Rb-Ubc9 or Ubc9 (used as negative control), and then immunoblotted with anti-SUMO1, anti-phosphorylated-Rb (Ser 807/811) or anti-His antibodies. Hyper-pRb-Ubc9: hyper-phosphorylated Rb-Ubc9. The relative phosphorylation level of each form of the Rb-Ubc9 fusion proteins were quantified and represented as the mean ± the SEM(n = 4). (D) A mild increase in global SUMO-1 conjugation is sufficient to enhance Rb phosphorylation. The phosphorylation levels of endogenous Rb in HEK293 cells expressing increasing amounts of GFP-SUMO-1 were determined as above. The results represent the mean± the SEM (n = 3). (E) The SUMO-deficient K720R mutation reduces the SUMOylation and phosphorylation of Rb. HEK293 cells were co-transfected with the WT or mutant Rb-His constructs together with SUMO-related plasmids, and analyzed for SUMO conjugation and phosphorylation as described above.
Because SUMO1 is attached to lysine 720 of the Rb protein, we generated a SUMO-deficient mutation by replacing this lysine residue with an arginine (K720R). His-tagged wild type or mutant Rb was co-transfected into HEK293 cells with GFP-SUMO1, followed by analysis of Rb-SUMO1 conjugation capability. Consistent with previous report, the SUMO modification was completely absent in the K720R mutant ().Citation18 Furthermore, we observed that the amount of phosphorylated Rb decreased compared to wild type (). Overall, these results suggest that the SUMOylation of Rb significantly increases its phosphorylation. Moreover, we noticed that the phosphorylation level of the Rb-SUMO1 species is lower than its hyper-phosphorylated form (). This data, combined with the fact that there was a time delay between the 2 Rb modifications during the cell cycle process (), suggests that Rb SUMOylation was a transient, initial step required for its multi-step phosphorylation procedure.
CDK2 is required for SUMOylation-enhanced phosphorylation of Rb through its SIM
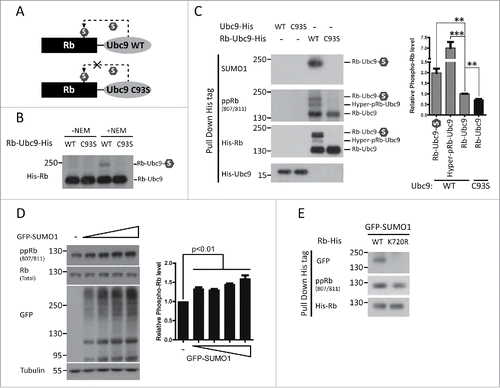
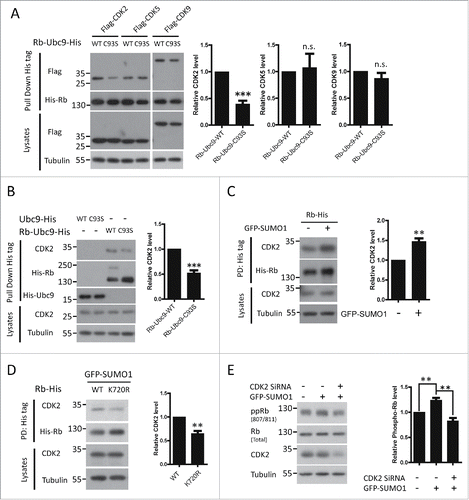
Rb phosphorylation is directly mediated by several CDKs. More specifically, CDK2 and CDK4 regulate G1/S transition during cell cycle progressionCitation11; CDK5 phosphorylates Rb in neurons, leading to cell cycle reentry and neuronal deathCitation26; and CDK9 also binds Rb both in vitro and in vivo to phosphorylate Rb.Citation27 As SUMOylation may affect protein interactions, we examined whether Rb SUMOylation promoted its interaction with CDK2/5/9.By co-transfecting HEK293 cells with His-tagged Rb-Ubc9 (wild type (WT) /C93S) fusion constructs and Flag-tagged CDK2/5/9, we found significantly increased binding of Rb to CDK2 after its constitutive SUMOylation (). In contrast, the interactions between Rb and CDK5 and CDK9, 2 CDKs not involved in cell cycle regulation, were not affected by the status of Rb SUMOylation (). Furthermore, we observed that UFDS of Rb similarly enhanced the binding of Rb to endogenous CDK2 (). Then, we confirmed this observation by stimulating Rb SUMOylation through direct over-expression of GFP-SUMO1 in HEK293 cells (). In contrast, the Rb SUMO-deficient mutant, K720R, exhibited reduced CDK2 binding capacity (). Therefore, increased SUMOylation of Rb leads to enhanced binding to CDK2.
Figure 3. CDK2 is required for SUMOylation-enhanced phosphorylation of Rb. (A) UFDS of Rb stimulates its binding with CDK2. HEK293 cells were co-transfected with His-tagged Rb-Ubc9 together with Flag-tagged CDKs. The binding capability of Rb-Ubc9 with each CDK was analyzed by pull down assay 72 h post-transfection. Data are shown as the mean± the SEM (n = 3). (B, C) Improved binding of Rb to endogenous CDK2 caused by UFDS (B) or enhanced of global SUMOylation (C). HEK293 cells were transfected as indicated. The binding of Rb to CDK2 was determined as described above. These results are represented as the mean± the SEM (n = 3). PD: pull down. (D) The SUMO-deficient K720R mutation exhibited reduced binding to CDK2. HEK293 cells were co-transfected with the WT or mutant Rb-His constructs together with GFP-SUMO1, and analyzed for CDK2 binding as described above. PD: pull down. (E) Inhibition of CDK2 by siRNA blocked the SUMOylation-enhanced Rb phosphorylation. HEK293 cells were first transfected with GFP-SUMO1, and they were then transfected with control or CDK2 siRNAas indicated. Rb phosphorylation was determined using an antibody against Rb S807/S811 (n = 4 independent experiments), and the values represent the mean± the SEM.
Next, we examined whether CDK2 played a role in this SUMO-enhanced phosphorylation of Rb. After knockdown of CDK2 by siRNA, SUMO-stimulated phosphorylation was blocked, indicating that CDK2 is required for the SUMOylation-enhanced phosphorylation of Rb ().
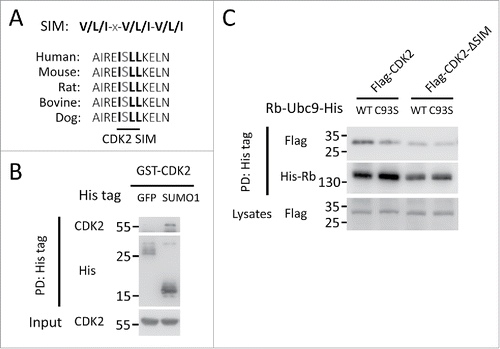
As SUMOylated protein could recruit SIM-containing proteins,Citation28 we set to examine whether the SUMOylation-enhanced CDK2 recruitment of Rb is through the SUMO-SIM interaction. To address this question, we firstly analyzed the amino acid sequence of human CDK2 and found the residues 53I-S-L-L56 corresponded to the reported SIM consensus sequence,Citation29,30 which is evolutionally conserved among mammals (). Consistently, we conducted an in vitro pull down experiment using purified recombinant proteins and confirmed the directly interaction between CDK2 and SUMO1 (). To investigate the functional relevance of the SIM, we generated a CDK2 mutant by deleting this region between 53 and 56. The in vivo binding assay revealed that the deletion of the SIM completely abolished the recruitment of CDK2 upon Rb SUMOylation (). Thus, these data suggest that CDK2 could directly bind SUMOylated Rb through its SIM, leading to increased Rb phosphorylation.
Figure 4. The recruitment of CDK2 to SUMOylated Rb is mediated by a functional SIM. (A) The alignment of the sequences corresponding to the putative CDK2 SIM in various species with the consensus SIM site. x stands for any amino acid. (B) CDK2 binds non-covalently to SUMO1 in vitro. Recombinant GST-tagged SUMO1 were incubated with His-tagged CDK2 and control GFP, followed by affinity pull down with Ni-NTA beads and Western blot. PD: Pull down. (C) The CDK2 SIM is required for its enhanced recruitment to SUMOylated Rb. HEK293 cells were co-transfected with His-tagged Rb-Ubc9 together with Flag-tagged wild-type CDK2 and a mutant lacking the SIM (ΔSIM) for the binding capacity assay as described in .
SUMOylation of Rb disrupts the E2F1-Rb interaction
Rb is an anti-oncoprotein that binds E2F, leading to E2F transcriptional inhibition and cell cycle arrest. To investigate the effect of Rb SUMOylation on E2F binding, we first compared the E2F1 binding capability between the SUMO-enhanced and SUMO-defective Rb-Ubc9 fusion proteins. The Rb-Ubc9(WT) protein, the one with enhanced Rb SUMOylation, showed weakened E2F binding compared to Rb-Ubc9(C93S), which is consistent with its elevated phosphorylation level (). We also observed decreased E2F1-Rb binding following GFP-SUMO1 overexpression (). In contrast, when Rb SUMOylation was blocked by the K720R mutant, its binding to E2F1 was significantly increased (). These findings indicate that the SUMOylation of Rb disrupts E2F1-Rb binding.
Figure 5. SUMOylation of Rb disrupts the E2F1-Rb interaction. (A) The constitutive SUMOylated Rb construct shows reduced interaction with E2F1. HEK293 cells were transfected with His-tagged Rb-Ubc9 or Ubc9, followed by pull-down experiments. The amount of Rb-bound E2F1 was determined by immunoblotting. Quantification of the data is shown as the mean± the SEM (n = 4). (B) Elevated global SUMOylation causes decreased association between Rb and E2F1. HEK293 cells transfected as indicated were lysed and subjected to pull down assay. Rb and E2F1 were detected by Western blot protein gel blot analysis using the indicated antibodies. The mean (n = 3) with the SEM values for the amount of Rb-bound E2F1 are shown. PD: Pull down. (C) Defective SUMOylation of Rb leads to increased sequestration of E2F1. WT and K720 Rb were precipitated by Ni-NTA using lysates from HEK293 cells transfected with the indicated constructs, and the amount of associated E2F1 was determined by immunoblotting. Data are expressed as the mean ± the SEM (n = 3). PD: pull down.
The role of Rb SUMOylation in cell proliferation
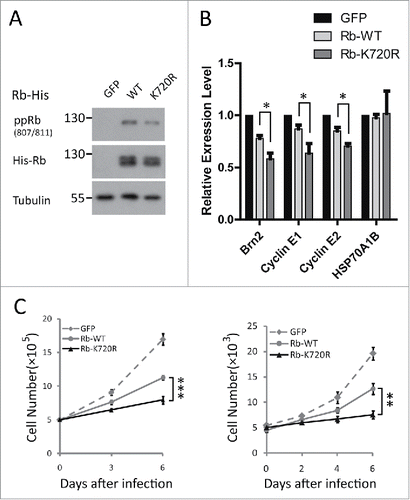
A previous study indicated that the restoration of Rb to Rb-deficient Y79 retinoblastoma cells could inhibit E2F target cell cycle gene expression and cell proliferation.Citation31 To investigate the functional consequence of Rb SUMOylation, we compared the effect between Rb and the SUMO-defective K720R Rb mutant on the proliferation of Y79 retinoblastoma cells, where the disturbance arising from endogenous Rb was preventable. After transduction of Y79 with Rb constructs, we first validated the decreased phosphorylation level of Rb K720R (). Then, we performed quantitative reverse transcription (qRT)-PCR analyses to examine the effect of WT or K720R Rb on the mRNA level of 3 known E2F-response genes, Brn2, Cyclin E1 and Cyclin E2, and a negative control gene, HSP70A1B.Citation31 As shown in , both Rb species could specifically reduce the expression levels of the E2F target genes. Furthermore, compared with wild type Rb, the inhibition effect of the SUMO-defective Rb mutation was stronger, suggesting that Rb SUMOylation is required for E2F target cell cycle gene expression. For further confirmation, we analyzed the effect of WT and K720R Rb on Y79 cell proliferation. We found that the restored K720R mutant inhibited Y79 cell proliferation compared with WT Rb, indicating that SUMO conjugation of Rb is essential for cell growth (). Collectively, our results suggest that the SUMOylation of Rb is required for cell cycle gene expression and cell proliferation.
Figure 6. Rb SUMOylation is required for cell proliferation. (A) The comparable expression level of restored WT and K720R Rb via lentiviral transduction of Y79 cells at days 3 and 6 was verified by Western blot. (B) Enhanced inhibitory effect of Rb K720R on E2F target cell cycle gene expression by Rb lentiviral infection in Y79 cells. The mRNA levels of the cell cycle-related genes (Brn2, cyclin E1, cyclin E2) and a negative control gene (HSP70A1B) were measured by qRT-PCR in Y79 cells 3 d after infection. (C) After restoration in Y79 cells, Rb promoted cell growth compared with its SUMO-deficient mutant. Y79 cells expressing exogenous WT and K720R Rb were cultured for 6 d, and the cell numbers were calculated by counting cell numbers (left panel) at days 3 and 6 and by CCK8 assay (right panel) every 48 h.
Discussion
Rb is a nuclear protein that undergoes cell cycle dependent phosphorylation to regulate cell proliferation by controlling the expression of E2F-dependent genes. Here we report a novel positive regulation of Rb phosphorylation. In cells at early G1 phase, Rb is SUMOylated prior to its phosphorylation. The SUMOylation of Rb results in an increased binding capacity for CDK2 through SUMO-SIM interaction, as well as Rb hyper-phosphorylation, which is essential for E2F factor release and S phase gene expression. Therefore, our study highlights SUMOylation as a molecular switch to control the phosphorylation and function of an essential cell cycle regulator.
Mounting evidence has suggested that SUMOylation is involved in cell proliferation control and cancer development. For example, genome-wide RNAi screens revealed that several key proteins in the SUMOylation pathway are essential for cell proliferation.Citation32 Furthermore, the loss of the SUMO pathway significantly inhibited cell proliferation, whereas the growth of cells overexpressing SUMO2 was markedly promoted.Citation33,34 Additionally, the global SUMOylation is elevated in patient-derived tumor tissues compared with normal tissues.Citation35 Consistent with these findings, we have observed increased global SUMOylation during cell cycle progress in the S/M phase (). Furthermore, although Rb has been reported to be modified by SUMO1 and SUMO2, the SUMOylation of endogenous Rb as well as the functional and physiological consequence of this modification have not yet been identified.Citation18,19, 36 The results presented herein validated the SUMOylation of endogenous Rb at early G1 phase in particular of the cell cycle and identified SUMOylation-regulated Rb activity, suggesting that in addition to global SUMOylation change in S/M phase, the SUMOylation of specific substrates is also important for regulation of the G0/G1 to S transition. These data are corroborated by studies revealing that the loss of SUMO pathway function resulted in G1/S phase cell cycle arrest in both Drosophila and human cells, further emphasizing the importance of SUMO modification in accurate cell cycle progression control.Citation37,38 Importantly, Rb-SUMO1 exists only for a very short window of time, consistent with the concept that SUMOylation is a highly dynamic modification and suggesting that this modification must be tightly regulated. Thus, determining the regulatory pathway that stimulates and attenuates Rb SUMOylation at the beginning of G1 phase of the cell cycle will be of great interest.
Our report shows that SUMO modification results in Rb hyper-phosphorylation and increased binding capacity for CDK2. Rb harbors 16 consensus CDK phosphorylation sites, which are mediated by the CDK/cyclin complex in a cell cycle dependent manner.Citation7,39 Although it is widely accepted that sequential hyper-phosphorylation is required for Rb inactivation, the exact mechanism regulating Rb phosphorylation is not completely understood. Previous studies have shown that the initial phosphorylation events in early G1phase are catalyzed by CDK4/6, whereas CDK2 is responsible for further Rb phosphorylation in middle to late G1 phase.Citation11,12 However, our data suggested that CDK2 is involved in the regulation of SUMO-enhanced Rb phosphorylation in early G1 phase. Because CDK2 is crucial for the inactivation of Rb and the release of E2F factors, it is possible that Rb SUMOylation plays an essential role in modulating the Rb-CDK2 interaction.Citation40,41 Furthermore, we found that CDK2 containing a conserved SIM, which is responsable for the enhanced recruitment upon Rb SUMOylation through non-covalent SUMO-SIM interactions. However, based on the observation that Rb SUMOylation is only a transient, initial step of its sequential phosphorylation (), the possibility that kinases other than CDK2 (e.g., CDK4/6) may participate the SUMO-enhanced phosphorylation process has not been fully ruled out in this report and needs further exploration. In addition to being phosphorylated and SUMOylated, Rb can also be ubiquitylated, acetylated and methylated.Citation4,39,42 Mdm2 binds Rb and promotes its ubiquitination and subsequent degradation.Citation43 Acetylation occurs at Lys873 and Lys874 of Rb, increasing its affinity for Mdm2 and resulting in reduced phosphorylation.Citation44 Acetylated Rb maybe induced by DNA damage and plays a role in cell differentiation.Citation45,46 The methylation of Rb K810 by the methyltransferase Set7/9 exerts a negative effect on both Rb phosphorylation and cell growth.Citation47 As growing evidence demonstrates the functional interplay between SUMOylation and other post-translational modifications, it remains to be determined whether SUMOylation has a modulatory effect on any of these modifications.
Additionally, by restoring exogenous WT and K720R Rb in Rb-deficient Y79 retinoblastoma cells, we discovered that the loss of Rb SUMOylation enhanced its ability to inhibit both E2F gene expression and cell proliferation. This is consistent with a previous study where a SUMO-deficient Rb mutant exhibited increased repressive activity on an E2F-responsive reporter gene.Citation18 However, due to the proliferation-promoting effect derived from Ubc9 itself,Citation48-50 we failed to detect whether SUMO modificaiton of Rb is sufficient to initiate the cell cycle and activate cell proliferation by Rb-Ubc9 fusion protein (data not shown). This possibility surely warrant further investigation. Thus, our data suggest that the SUMOylation of Rb is required for cell cycle gene expression and cell proliferation. In summary, our study adds to the growing evidence that posttranslational modifications play a major role in Rb activity regulation and provides a previously unknown mechanism that stimulates Rb phosphorylation.
Materials and methods
Plasmids, siRNA and reagents
cDNAs for human Rb and CDK2/5/9 were cloned, sequenced, and then sub-cloned into a pcDNA3.1-based expression vector with appropriate tags (His and Flag, respectively). Site-directed mutagenesis was performed to generate the Rb K720R mutation. Both Rb species were cloned into lentiviral vector plv-GFP, kindly provided by Dr. Jianwei Jiao, by replacing GFP with Rb. The Rb and Ubc9 cDNA were cloned into pcDNA3.1 to generate Rb-Ubc9 (WT) and Rb-Ubc9 (C93S) fusion constructs as previously reported.Citation25 CDK2 siRNA was chemically synthesized (GenePharma, Shanghai, China) and the sequence (sense strand) was 5′-AAGGUGGUGGCGCUUAAGAAA-3′.Citation51 Recombinant proteins including GST- SUMO1, His-GFP and His-CDK2 were ordered from Abcam.
Antibodies
The primary antibodies used were: mouse anti-Rb antibody (9309, Cell Signaling Technology); rabbit anti-phospho-Rb (S807/S811) antibody (9308, Cell Signaling Technology); mouse anti-SUMO1 antibody (33-2400, Invitrogen); mouse anti-tubulin antibody (M30109, Abmart); mouse anti-Flag tag (DYKDDDDK-Tag) antibody (M20008, Abmart); mouse anti-His tag antibody (M30111, Abmart); mouse anti-GFP antibody (M20004, Abmart); rabbit anti-CDK2 antibody (10122-1-AP, Proteintech); and rabbit anti-E2F1 antibody (12171-1-AP, Proteintech).
Cell culture and Synchronization
The HEK293 and 293T cells were cultured in DMEM medium (Gibco) supplemented with 10% FBS (Gibco) and the appropriate antibiotic. The human Y79 retinoblastoma cells were kindly provided by Dr. Peiquan Zhao (Shanghai Jiao Tong University) and were grown in RPMI-1640 medium (Gibco) supplemented with 10% FBS (Gibco) and antibiotic. The cells were grown at 37°C with 5% CO2 in a humidified incubator. The HEK293 cells were synchronized at different cell cycle phases as described previously.Citation52,53 The cells were incubated in DMEM medium without serum for 72 h to induce G0 arrest. For the G1 phase, the cells were collected 0.5 (early G1) or 2 (G1) h after the addition of serum to the G0 arrested cells. To obtain cells synchronized at the beginning of S phase, the cells were synchronized by a double thymidine block. Briefly, the cells were treated with 2.5 mM thymidine (T1895, Sigma) for 18 h. Then, they were washed and supplied fresh media for 14 h, followed by a second treatment with 2.5 mM thymidine for 18 h. To arrest the cells in the G2/M phase, the cells were incubated in medium containing 400ng/ml nocodazole (M1404, Sigma) for 16 h.
Transfection and lentiviral infection
The plasmids and siRNA were introduced into HEK293 cells using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. The total amount of plasmid DNA was adjusted to 4 μg per 35mm dish or 20 μg per 10 cm dish with an empty vector or GFP expression plasmid, respectively. Cells were harvested 36-48 h post-transfection for immunoblotting. For the preparation of lentivirus, the 293T cells were co-transfected using Lipofectamine 2000 with viral vectors PSPAX2 and PMD2.G. Cell supernatants were harvested 48 h after transfection, and they were collected and passed through 0.45-μm cellulose acetate filters. For infections, Y79 cells were plated at 5 × 106 cells per 35mm dish and supplemented with 500μl virus-containing cell supernatants (titer: 2∼3 × 106 virus/ml) and 6μg/ml polybrene.
SUMOylation analysis, pull down and immunoprecipitation
For analysis of Rb SUMOylation, HEK293 cells (106) were plated on a 10-cm dish and synchronized as described above. The cells were lysed in denaturing RIPA (50 mM Tris, pH 8.0; 150 mM NaCl; 1% NP-40; 1% sodium deoxycholate; 0.1% SDS) lysis buffer supplemented with 20 mM N-ethylmaleimide (E3876, Sigma) as well as protease and phosphatase inhibitors (Roche). Extracts of the cell lysates (1 mg) were incubated with anti-Rb antibodies for 16 h at 4°C with gentle inversion mixing. Then, protein A/G Sepharose (A10001, Abmart) was added and incubated for 3 h. The beads were collected and washed 4 times with lysis buffer, and the immunoprecipitated proteins were eluted by 1×SDS-sample buffer, immunoblotted and probed with anti-SUMO antibody. For the Rb pull down, the lysates were incubated with Ni-NTA agarose beads (30230, Qiagen) for 3h at 4°C. The beads were washed 5 times with washing buffer containing 20 mM imidazole, and they were eluted with 40μl of elution buffer containing 250 mM imidazole. For protein interaction analysis, HEK293 cells were lysed in non-denaturing NP-40 buffer (50 mM Tris, pH 8.0; 150 mM NaCl; 1% NP-40), followed by immunoprecipitation or pull down assay. For the in vitro pull down assay, 2μg of GST-tagged SUMO1 were incubated with the same anount of His-tagged GFP or CDK2 together with Ni-NTA beads for 4 h at 4°C. After extensive washing, eluates were further analyzed by SDS-PAGE and immunoblotting.
Gene expression and proliferation assay
The total RNA was extracted from Y79 cells using TRIzol reagent (Invitrogen) 72 h after infection. The cDNA was synthesized using 1 mg of RNA in a reverse transcription reaction (Takara). The expression levels of Brn2, CyclinE1 and CyclinE2 mRNA were normalized to the GAPDH mRNA levels and represented relative to the expression in GFP control cells (mean and SEM from PCR triplicates of 3 independent experiments). The PCR primers were designed as previously reported Citation31: Brn2, 5′-CAGAGAGATGGCAAGCACTG-3′ and 5′-TCAGGAAGCTGCATTTTGTG-3′; cyclin E1, 5′-CGTGCGTTTGCTTTTACAGA-3′ and 5′-AGCACCTTCCATAGCAGCAT-3′; cyclin E2, 5′-CCTCCATTGTGAGATAAGGACA and GCCTATGTACAGCAAGTTTTCA-3′; HSP70A1B, 5′-CCGAGAAGGACGAGTTTGAG-3′ and 5′-GCAGCAAAGTCCTTGAGTCC-3′. To examine the proliferation in the Y79 retinoblastoma cell line, the cells were infected and seeded into 6-well plates (500,000 cells/well) or 96-well plates (5,000 cells/well). Cell numbers for each group were determined by Countess Automated Cell Counter (Invitrogen), and by a cell counting kit-8 assay (CCK-8; Dojindo Laboratories).
Statistics
Band intensity in the Western blot was determined using the BIO-RAD Quantity One software. All quantification data are presented as the mean ± SEM. The statistical significance was analyzed using the student t-test in all experiments. (***, P < 0.001; **, P < 0.01; *, P < 0.05).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgment
We want to thank Dr. Jianwei Jiao for lentiviral vectors, and Dr. Peiquan Zhao for Y79 retinoblastoma cell line.
Funding
This study was supported by grants from the Science and Technology Commission of Shanghai (Grant No. 14411961800).
References
- Massague J. G1 cell-cycle control and cancer. Nature 2004; 432:298-306; PMID:15549091; http://dx.doi.org/10.1038/nature03094
- Bertoli C, Skotheim JM, de Bruin RA. Control of cell cycle transcription during G1 and S phases. Nat Rev Mol Cell Biol 2013; 14:518-28; PMID:23877564; http://dx.doi.org/10.1038/nrm3629
- Weinberg RA. The retinoblastoma protein and cell cycle control. Cell 1995; 81:323-30; PMID:7736585; http://dx.doi.org/10.1016/0092-8674(95)90385-2
- Burkhart DL, Sage J. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat Rev Cancer 2008; 8:671-82; PMID:18650841; http://dx.doi.org/10.1038/nrc2399
- Dyson N. The regulation of E2F by pRB-family proteins. Gen Dev 1998; 12:2245-62; PMID:9694791; http://dx.doi.org/10.1101/gad.12.15.2245
- Ezhevsky SA, Ho A, Becker-Hapak M, Davis PK, Dowdy SF. Differential regulation of retinoblastoma tumor suppressor protein by G(1) cyclin-dependent kinase complexes in vivo. Mol Cellular Biol 2001; 21:4773-84; PMID:11416152; http://dx.doi.org/10.1128/MCB.21.14.4773-4784.2001
- Giacinti C, Giordano A. RB and cell cycle progression. Oncogene 2006; 25:5220-7; PMID:16936740; http://dx.doi.org/10.1038/sj.onc.1209615
- Ezhevsky SA, Nagahara H, Vocero-Akbani AM, Gius DR, Wei MC, Dowdy SF. Hypo-phosphorylation of the retinoblastoma protein (pRb) by cyclin D:Cdk4/6 complexes results in active pRb. Proc Natl Acad Sci U S A 1997; 94:10699-704; PMID:9380698; http://dx.doi.org/10.1073/pnas.94.20.10699
- Zarkowska T, Mittnacht S. Differential phosphorylation of the retinoblastoma protein by G1/S cyclin-dependent kinases. J Biol Chem 1997; 272:12738-46; PMID:9139732; http://dx.doi.org/10.1074/jbc.272.19.12738
- Stevens C, La Thangue NB. E2F and cell cycle control: a double-edged sword. Archives of biochemistry and biophysics 2003; 412:157-69; PMID:12667479; http://dx.doi.org/10.1016/S0003-9861(03)00054-7
- Harbour JW, Luo RX, Dei Santi A, Postigo AA, Dean DC. Cdk phosphorylation triggers sequential intramolecular interactions that progressively block Rb functions as cells move through G1. Cell 1999; 98:859-69; PMID:10499802; http://dx.doi.org/10.1016/S0092-8674(00)81519-6
- Harbour JW, Dean DC. The Rb/E2F pathway: expanding roles and emerging paradigms. Gen Dev 2000; 14:2393-409; PMID:11018009; http://dx.doi.org/10.1101/gad.813200
- Gareau JR, Lima CD. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol 2010; 11:861-71; PMID:21102611; http://dx.doi.org/10.1038/nrm3011
- Yao Q, Li H, Liu BQ, Huang XY, Guo L. SUMOylation-regulated protein phosphorylation, evidence from quantitative phosphoproteomics analyses. J Biol Chem 2011; 286:27342-9; PMID:21685386; http://dx.doi.org/10.1074/jbc.M111.220848
- Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol 2007; 8:947-56; PMID:18000527; http://dx.doi.org/10.1038/nrm2293
- Hay RT. SUMO: a history of modification. Mol Cell 2005; 18:1-12; PMID:15808504; http://dx.doi.org/10.1016/j.molcel.2005.03.012
- Hickey CM, Wilson NR, Hochstrasser M. Function and regulation of SUMO proteases. Nat Rev Mol Cell Biol 2012; 13:755-66; PMID:23175280; http://dx.doi.org/10.1038/nrm3478
- Ledl A, Schmidt D, Muller S. Viral oncoproteins E1A and E7 and cellular LxCxE proteins repress SUMO modification of the retinoblastoma tumor suppressor. Oncogene 2005; 24:3810-8; PMID:15806172; http://dx.doi.org/10.1038/sj.onc.1208539
- Li T, Santockyte R, Shen RF, Tekle E, Wang G, Yang DC, Chock PB. Expression of SUMO-2/3 induced senescence through p53- and pRB-mediated pathways. J Biol Chem 2006; 281:36221-7; PMID:17012228; http://dx.doi.org/10.1074/jbc.M608236200
- Muller S, Berger M, Lehembre F, Seeler JS, Haupt Y, Dejean A. c-Jun and p53 activity is modulated by SUMO-1 modification. J Biol Chem 2000; 275:13321-9; PMID:10788439; http://dx.doi.org/10.1074/jbc.275.18.13321
- Tan JA, Song J, Chen Y, Durrin LK. Phosphorylation-dependent interaction of SATB1 and PIAS1 directs SUMO-regulated caspase cleavage of SATB1. Mol Cellular Biol 2010; 30:2823-36; PMID:20351170; http://dx.doi.org/10.1128/MCB.01603-09
- Gresko E, Ritterhoff S, Sevilla-Perez J, Roscic A, Frobius K, Kotevic I, Vichalkovski A, Hess D, Hemmings BA, Schmitz ML. PML tumor suppressor is regulated by HIPK2-mediated phosphorylation in response to DNA damage. Oncogene 2009; 28:698-708; PMID:19015637; http://dx.doi.org/10.1038/onc.2008.420
- Gupta P, Ho PC, Huq MM, Ha SG, Park SW, Khan AA, Tsai NP, Wei LN. Retinoic acid-stimulated sequential phosphorylation, PML recruitment, and SUMOylation of nuclear receptor TR2 to suppress Oct4 expression. Proc Natl Acad Sci U S A 2008; 105:11424-9; PMID:18682553; http://dx.doi.org/10.1073/pnas.0710561105
- Mittnacht S. Control of pRB phosphorylation. Curr Opin Genet Dev 1998; 8:21-7; PMID:9529601; http://dx.doi.org/10.1016/S0959-437X(98)80057-9
- Jakobs A, Koehnke J, Himstedt F, Funk M, Korn B, Gaestel M, Niedenthal R. Ubc9 fusion-directed SUMOylation (UFDS): a method to analyze function of protein SUMOylation. Nat Methods 2007; 4:245-50; PMID:17277783; http://dx.doi.org/10.1038/nmeth1006
- Hamdane M, Bretteville A, Sambo AV, Schindowski K, Begard S, Delacourte A, Bertrand P, Buee L. p25/Cdk5-mediated retinoblastoma phosphorylation is an early event in neuronal cell death. J Cell Sci 2005; 118:1291-8; PMID:15741232; http://dx.doi.org/10.1242/jcs.01724
- Simone C, Bagella L, Bellan C, Giordano A. Physical interaction between pRb and cdk9/cyclinT2 complex. Oncogene 2002; 21:4158-65; PMID:12037672; http://dx.doi.org/10.1038/sj.onc.1205511
- Kerscher O. SUMO junction-what's your function? New insights through SUMO-interacting motifs. EMBO reports 2007; 8:550-5; PMID:17545995; http://dx.doi.org/10.1038/sj.embor.7400980
- Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc Natl Acad Sci U S A 2004; 101:14373-8; PMID:15388847; http://dx.doi.org/10.1073/pnas.0403498101
- Zhao Q, Xie Y, Zheng Y, Jiang S, Liu W, Mu W, Liu Z, Zhao Y, Xue Y, Ren J. GPS-SUMO: a tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res 2014; 42:W325-30; PMID:24880689; http://dx.doi.org/10.1093/nar/gku383
- Cobrinik D, Francis RO, Abramson DH, Lee TC. Rb induces a proliferative arrest and curtails Brn-2 expression in retinoblastoma cells. Mol Cancer 2006; 5:72; PMID:17163992; http://dx.doi.org/10.1186/1476-4598-5-72
- Schlabach MR, Luo J, Solimini NL, Hu G, Xu Q, Li MZ, Zhao Z, Smogorzewska A, Sowa ME, Ang XL, et al. Cancer proliferation gene discovery through functional genomics. Science 2008; 319:620-4; PMID:18239126; http://dx.doi.org/10.1126/science.1149200
- Chen LZ, Li XY, Huang H, Xing W, Guo W, He J, Sun ZY, Luo AX, Liang HP, Hu J, et al. SUMO-2 promotes mRNA translation by enhancing interaction between eIF4E and eIF4G. PloS one 2014; 9:e100457; PMID:24971752; http://dx.doi.org/10.1371/journal.pone.0100457
- He X, Riceberg J, Pulukuri SM, Grossman S, Shinde V, Shah P, Brownell JE, Dick L, Newcomb J, Bence N. Characterization of the loss of SUMO pathway function on cancer cells and tumor proliferation. PloS one 2015; 10:e0123882; PMID:25860128; http://dx.doi.org/10.1371/journal.pone.0123882
- Bellail AC, Olson JJ, Hao C. SUMO1 modification stabilizes CDK6 protein and drives the cell cycle and glioblastoma progression. Nat Commun 2014; 5:4234; PMID:24953629; http://dx.doi.org/10.1038/ncomms5234
- Bischof O, Schwamborn K, Martin N, Werner A, Sustmann C, Grosschedl R, Dejean A. The E3 SUMO ligase PIASy is a regulator of cellular senescence and apoptosis. Mol cell 2006; 22:783-94; PMID:16793547; http://dx.doi.org/10.1016/j.molcel.2006.05.016
- Ni HJ, Chang YN, Kao PH, Chai SP, Hsieh YH, Wang DH, Fong JC. Depletion of SUMO ligase hMMS21 impairs G1 to S transition in MCF-7 breast cancer cells. Biochimica et biophysica acta 2012; 1820:1893-900; PMID:22906975; http://dx.doi.org/10.1016/j.bbagen.2012.08.002
- Nie M, Xie Y, Loo JA, Courey AJ. Genetic and proteomic evidence for roles of Drosophila SUMO in cell cycle control, Ras signaling, and early pattern formation. PloS one 2009; 4:e5905; PMID:19529778; http://dx.doi.org/10.1371/journal.pone.0005905
- Macdonald JI, Dick FA. Posttranslational modifications of the retinoblastoma tumor suppressor protein as determinants of function. Genes & cancer 2012; 3:619-33; PMID:23634251; http://dx.doi.org/10.1177/1947601912473305
- De Luca A, MacLachlan TK, Bagella L, Dean C, Howard CM, Claudio PP, Baldi A, Khalili K, Giordano A. A unique domain of pRb2/p130 acts as an inhibitor of Cdk2 kinase activity. J Biol Chem 1997; 272:20971-4; PMID:9261093; http://dx.doi.org/10.1074/jbc.272.34.20971
- Zhang HS, Gavin M, Dahiya A, Postigo AA, Ma D, Luo RX, Harbour JW, Dean DC. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell 2000; 101:79-89; PMID:10778858; http://dx.doi.org/10.1016/S0092-8674(00)80625-X
- Dick FA, Rubin SM. Molecular mechanisms underlying RB protein function. Nat Rev Mol Cell Biol 2013; 14:297-306; PMID:23594950; http://dx.doi.org/10.1038/nrm3567
- Uchida C, Miwa S, Kitagawa K, Hattori T, Isobe T, Otani S, Oda T, Sugimura H, Kamijo T, Ookawa K, et al. Enhanced Mdm2 activity inhibits pRB function via ubiquitin-dependent degradation. EMBO J 2005; 24:160-9; PMID:15577944; http://dx.doi.org/10.1038/sj.emboj.7600486
- Chan HM, Krstic-Demonacos M, Smith L, Demonacos C, La Thangue NB. Acetylation control of the retinoblastoma tumour-suppressor protein. Nat Cell Biol 2001; 3:667-74; PMID:11433299; http://dx.doi.org/10.1038/35083062
- Markham D, Munro S, Soloway J, O'Connor DP, La Thangue NB. DNA-damage-responsive acetylation of pRb regulates binding to E2F-1. EMBO reports 2006; 7:192-8; PMID:16374512; http://dx.doi.org/10.1038/sj.embor.7400591
- Nguyen DX, Baglia LA, Huang SM, Baker CM, McCance DJ. Acetylation regulates the differentiation-specific functions of the retinoblastoma protein. EMBO J 2004; 23:1609-18; PMID:15044952; http://dx.doi.org/10.1038/sj.emboj.7600176
- Carr SM, Munro S, Kessler B, Oppermann U, La Thangue NB. Interplay between lysine methylation and Cdk phosphorylation in growth control by the retinoblastoma protein. EMBO J 2011; 30:317-27; PMID:21119616; http://dx.doi.org/10.1038/emboj.2010.311
- Dong M, Pang X, Xu Y, Wen F, Zhang Y. Ubiquitin-conjugating enzyme 9 promotes epithelial ovarian cancer cell proliferation in vitro. Int J Mol Sci 2013; 14:11061-71; PMID:23708104; http://dx.doi.org/10.3390/ijms140611061
- Li F, Li X, Kou L, Li Y, Meng F, Ma F. SUMO-conjugating enzyme UBC9 promotes proliferation and migration of fibroblast-like synoviocytes in rheumatoid arthritis. Inflammation 2014; 37:1134-41; PMID:24531852; http://dx.doi.org/10.1007/s10753-014-9837-x
- Zhao Z, Tan X, Zhao A, Zhu L, Yin B, Yuan J, Qiang B, Peng X. microRNA-214-mediated UBC9 expression in glioma. BMB reports 2012; 45:641-6; PMID:23187003; http://dx.doi.org/10.5483/BMBRep.2012.45.11.097
- Zhang GJ, Safran M, Wei W, Sorensen E, Lassota P, Zhelev N, Neuberg DS, Shapiro G, Kaelin WG, Jr. Bioluminescent imaging of Cdk2 inhibition in vivo. Nat Med 2004; 10:643-8; PMID:15122251; http://dx.doi.org/10.1038/nm1047
- Pabla N, Bhatt K, Dong Z. Checkpoint kinase 1 (Chk1)-short is a splice variant and endogenous inhibitor of Chk1 that regulates cell cycle and DNA damage checkpoints. Proc Natl Acad Sci U S A 2012; 109:197-202; PMID:22184239; http://dx.doi.org/10.1073/pnas.110-4767109
- Singhmar P, Kumar A. Angelman syndrome protein UBE3A interacts with primary microcephaly protein ASPM, localizes to centrosomes and regulates chromosome segregation. PloS one 2011; 6:e20397; PMID:21633703; http://dx.doi.org/10.1371/journal.pone.0020397
