ABSTRACT
The mitotic spindle is made of microtubules (MTs) nucleated through different pathways involving the centrosomes, the chromosomes or the walls of pre-existing MTs. MCRS1 is a RanGTP target that specifically associates with the chromosome-driven MTs protecting them from MT depolymerases. MCRS1 is also needed for the control of kinetochore fiber (K-fiber) MT minus-ends dynamics in metaphase. Here, we investigated the regulation of MCRS1 activity in M-phase. We show that MCRS1 is phosphorylated by the Aurora-A kinase in mitosis on Ser35/36. Although this phosphorylation has no role on MCRS1 localization to chromosomal MTs and K-fiber minus-ends, we show that it regulates MCRS1 activity in mitosis. We conclude that Aurora-A activity is particularly important in the tuning of K-fiber minus-ends dynamics in mitosis.
Introduction
The mitotic spindle is a finely regulated molecular machine that segregates the sister chromatids during cell division. It is mainly composed by microtubules (MTs), themselves formed by protofilaments of α- and β-tubulin heterodimers. The MTs composing the mitotic spindle however differ in function, organization and dynamics.Citation1 The astral MTs anchor the spindle to the cell cortex; the interpolar MTs form the interdigitating antiparallel arrays that constitute the majority of the spindle MTs and provide mechanical support needed for the movement of the chromosomes; the K-fibers are bundles of 20 to 40 MTs that connect each kinetochore to one spindle pole.Citation2 Although they are more stable than the other spindle MTs, K-fiber dynamic properties are essential for chromosome movements, congression and segregation.Citation3 Interestingly, spindle MTs are not only different functionally and mechanically but they also originate through different pathways involving the centrosomes, the chromosomes and pre-existing MTs.Citation4,5 Chromosome-dependent MT assembly relies on a gradient of the GTP-bound form of the small GTPase Ran resulting from the association of its exchange factor RCC1 with the chromatin.Citation6,7 RanGTP releases spindle assembly factors (SAFs) from their inhibitory binding to importins thereby enabling them to promote MT nucleation, stabilization and organization around the chromosomes.Citation8,9 One of these RanGTP regulated SAFs is MCRS1, which is involved in chromosome-dependent MT stabilization and the regulation of K-fiber MT minus-ends dynamics.Citation10
MCRS1 does not associate with the centrosome-driven MTs but specifically with the chromosomal MTs in the M-phase cytoplasm and with the K-fiber MT minus-ends in metaphase.Citation10 MCRS1 function is essential because it protects the chromosome RanGTP-dependent MTs and the K-fibers from MT depolymerase activities. Recently we showed that during mitosis, MCRS1 acts in complex with several members of the NSL complex.Citation11 One of them, KANSL3 is a new MT minus-end binding factor that targets MCRS1 to K-fiber MT minus-ends.Citation11 However, the regulation of MCRS1 mitotic activity and localization to chromosomal MTs and K-fibers is still not understood. Here we investigated the regulation of MCRS1 by phosphorylation. We show that MCRS1 is phosphorylated by the kinase Aurora-A that regulates its function.
Results
MCRS1 is a novel mitotic substrate of Aurora-A
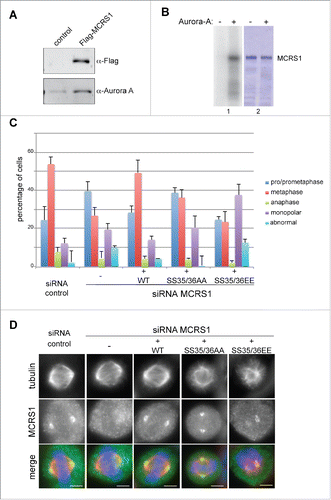
We previously showed that TPX2 co-immunoprecipitates with MCRS1 during mitosis.Citation11 Since TPX2 is a RanGTP-dependent partner of Aurora-A in mitosis,Citation12-16 we investigated whether MCRS1 also co-immunoprecipitates Aurora-A and found that the kinase was present in Flag-MCRS1 pull-downs performed in mitotic HeLa cells (). To determine whether MCRS1 may be a substrate of Aurora-A we first performed phosphorylation assays in vitro. We found that indeed, Aurora-A phosphorylates MCRS1 in vitro () whereas the closely related kinase Aurora-B did not under similar conditions (Suppl. Fig. 1A). We conclude that MCRS1 is a specific substrate of Aurora-A. Mass spectrometry analysis of the MCRS1 phosphorylated in vitro by Aurora-A revealed the incorporation of a phosphate at position Ser35 although it was not possible to discard the possibility of a phosphate addition on the next residue Ser36. To confirm these results, we performed in vitro kinase assays on recombinant MCRS1 in which Ser35 and Ser36 were substituted by alanines by site directed mutagenesis. MCRS1 SS35/36AA was still phosphorylated by Aurora-A in vitro although to a lesser extent (88% of the wild type protein) (Suppl. Fig. 1B), suggesting the existence of additional sites for this kinase at least in vitro. Since the analysis of the MCRS1 sequence did not reveal other putative consensus sites for phosphorylation by Aurora-A (Suppl. Fig. 1C) we repeated the mass spectrometry analysis of the in vitro phosphorylated protein to obtain a better coverage. Indeed, this analysis revealed the incorporation of a phosphate at another site (mapped to Ser85 and/or Ser87) both in the wild type and double alanine mutant of MCRS1. We cannot rule out that additional sites may exist since our mass spectrometry data did not provide a full coverage for the protein sequences. We decided to focus on the phosphorylation of MCRS1 on Ser35/36 for further functional analysis because it has been previously reported in mitotic phosphoproteome analysis.Citation17,18 To determine whether phosphorylation at this site is indeed specific for mitosis, we analyzed this site by mass spectrometry on Flag-MCRS1 pull downs from HeLa cells. The cells were either unsynchronized or synchronized in mitosis by a nocodazole arrest followed by release and shake-off after 1 h. Some of these cells were also incubated with the Aurora-A inhibitor MLN8237. We found that MCRS1 was indeed phosphorylated at Ser35/36 in mitotic cells but not in unsynchronized cells nor in mitotic cells incubated with the Aurora-A inhibitor MLN8237. We conclude that Aurora-A phosphorylates MCRS1 on Ser35/36 specifically during mitosis.
Figure 1. MCRS1 is phosphorylated by Aurora-A in vitro and in cells. (A) Western-blot of anti-Flag pulldowns from untransfected (control) or Flag-MCRS1 expressing mitotic HeLa cells probed with anti-Flag and anti-Aurora-A antibodies, as indicated. Flag-MCRS1 specifically pulls down Aurora-A in mitotic HeLa cells. (B) In vitro kinase assay. Recombinant MBP-tagged human MCRS1 was incubated (+) or not (−) with purified human Aurora-A and P32-ATP. The autoradiography (1) and the corresponding Coomassie blue stained gel (2) are shown. Aurora-A phosphorylates MCRS1 in vitro. (C) Characterization of the mitotic cells in MCRS1-silenced cells expressing the MCRS1 phospho-variants on Ser35/36. MCRS1 silencing results in an accumulation of cells in prometaphase and cells with spindle organization defects (monopolar or abnormal). Expression of MCRS1 WT rescues these defects whereas expression of MCRS1 SS35/36AA or MCRS1 SS35/36EE does not. The graph shows the percentage of cells in each category, as indicated. Mean of 3 independent experiments counting at least 150 cells per condition and per experiment. Bars: standard deviation. (D) Immunofluorescence images showing the localization of the MCRS1 proteins in CTRL or MCRS1-silenced cells, expressing the different MCRS1 phospho-variants, as indicated. Tubulin is shown in red, MCRS1 in green and DNA in blue. Scale bars 5 µm.
MCRS1 phosphorylation by Aurora-A on Ser35/36 is required for mitotic progression
In order to study the function of MCRS1 phosphorylation on Ser35/36 during mitosis, we prepared different stable HeLa-FRT isogenic cell lines expressing in a tetracycline-inducible manner the different phospho-variants of siRNA-resistant Flag-tagged MCRS1: wild-type MCRS1 (WT), the phospho-null MCRS1 with Ser35 and Ser36 substituted by alanines (MCRS1 SS35/36AA), or the phospho-mimicking variant MCRS1 with Ser35 and Ser36 substituted to either glutamic acids or aspartic acids (MCRS1 SS35/36EE and MCRS1 SS35/36DD). All the cell lines expressed homogenously the corresponding MCRS1 WT and phospho-variants at endogenous levels upon silencing of endogenous MCRS1 (Suppl. Fig. 2A) for 48 h and induction of exogenous expression with tetracycline at 0,1µg/ml overnight (Suppl. Fig. 2B). For simplicity we present here the results obtained with one version of the phospho-mimicking variants, MCRS1 SS35/36EE, as we obtained similar results with cells expressing the other variant MCRS1 SS35/36DD (data not shown).
We first determined whether phosphorylation could regulate some of the known interactions of MCRS1 during mitosis. Pull-down experiments of the different phospho-variants of MCRS1 showed that in all cases the interactions with TPX2, KANSL3 and Aurora-A itself were maintained Citation10,11 (Suppl. Fig. 3A).
We then examined whether MCRS1 phosphorylation may be required for mitosis. We quantified the distribution of the mitotic cells in the different mitotic phases in control and MCRS1-silenced cells expressing or not the different MCRS1 phospho-variants. As we described previously,Citation10 MCRS1-silenced cells showed an increased proportion of prometaphase-like figures (70%) including monopolar spindles and abnormal disorganized mitotic figures. Concomitantly the number of cells in metaphase and anaphase decreased. Expression of WT-MCRS1 fully rescued this phenotype as expected (). Consistently, immunofluorescence analysis showed that the exogenously expressed recombinant MCRS1 localized to the spindle poles like the endogenous protein (). In contrast, the expression of the MCRS1 SS35/36AA phospho-null variant in the silenced cells did not rescue the mitotic defects. Indeed these cells showed a similar phenotype to the silenced cells with an accumulation of prometaphase-like figures including monopolar spindles and abnormal mitotic figures (). However, immunofluorescence analysis showed that the MCRS1 SS35/36AA protein localized to the spindle poles similarly to the endogenous and WT proteins (). These data suggested that Aurora-A dependent MCRS1 phosphorylation on Ser35/36 does not regulate MCRS1 localization. However it regulates MCRS1 function in spindle assembly and dynamics.
We then examined MCRS1-silenced cells expressing the phospho-mimicking variant MCRS1 SS35/36EE. Surprisingly, not only the mitotic defects were not rescued but the cells showed more dramatic mitotic defects than the silenced cells with a significant accumulation of monopolar and abnormal spindles (50%) (). This dominant-negative phenotype underscores the importance of phosphorylation of MCRS1 on Ser35/36 during mitosis. However, MCRS1 SS35/36EE did localize to the poles or to focus points of MTs in the abnormal spindle-like structures ().
The different MCRS1 phospho-variants appeared to retain a localization similar to the endogenous protein. We therefore checked by immunofluorescence analysis in Nuf2-silenced cells whether their localization is K-fiber-dependent like it is for the endogenous protein.Citation10 Indeed, we found that all the phospho-variants failed to localize in cells lacking K-fibers (Suppl. Fig. 3B).
These results indicate that phosphorylation of MCRS1 by Aurora-A on Ser35/36 plays no role in its localization. However it is required for MCRS1 function and mitosis. Altogether, these data indicate that the function of MCRS1 goes beyond its targeting to the spindle poles in mitosis and suggest that its phosphorylation by Aurora-A on Ser35/36 provides a dynamic regulation of its activity.
MCRS1 phosphorylation by Aurora-A on Ser35/36 is not required for chromosome-induced MT aster formation
We previously showed that MCRS1 is essential for the assembly of acentrosomal MTs in the mitotic cell.Citation10 Since the expression of the MCRS1 phospho-variants did not rescue the mitotic phenotype induced by MCRS1 silencing, we then investigated whether phosphorylation could be required for chromosome-induced MT assembly. We performed MT regrowth assays in MCRS1-silenced cells expressing the different phospho-variants and quantified the number of MT asters in mitotic cells fixed 5 min after nocodazole washout. Expression of the WT-MCRS1 rescued MT asters assembly to almost control levels (). Surprisingly, the expression of the MCRS1 SS35/36AA phospho-null mutant also rescued the silencing phenotype and restored the number of MT asters to control levels. Immunofluorescence analysis showed that the phospho-null protein localized specifically to the chromosome-dependent MT asters and not to the centrosomes like the endogenous and WT proteins (). These results suggest that although phosphorylation is required for spindle assembly it is not required for the localization and function of MCRS1 at the level of chromosome-dependent MT assembly.
Figure 2. Role of MCRS1 phosphorylation at Ser35/36 in chromosomal MT assembly. (A) MT regrowth experiment. Data obtained from 3 independent experiments counting the number of MT asters in more than 60 cells per condition and per experiment. Box-and-whisker plot: boxes show the upper and lower quartiles (25–75%) with a line at the median, whiskers extend from the 10th to the 90th percentile and dots correspond to outliers. Expression of MCRS1 WT or MCRS1 SS35/36AA rescues the silencing phenotype but not SS35/36EE. t test, *** corresponds to p <0,05. (B) Immunofluorescence images of cells quantified in A showing the localization of the MCRS1 variants during MT regrowth. Cells were fixed 5 min after nocodazole washout and then stained for tubulin (red), MCRS1 (green) and DNA (blue). Maximum projections of confocal images are presented. All the MCRS1 versions localize to the chromosomal MT asters and not to the centrosomal ones. Scale bars 5 µm.
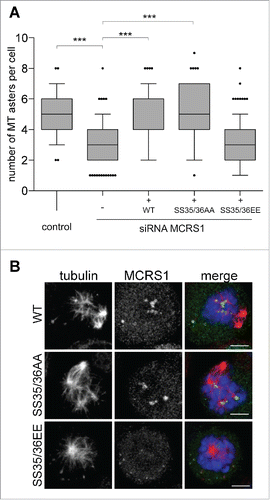
By contrast, the expression of the phospho-mimicking MCRS1 SS35/36EE in the silenced cells did not restore MT assembly around the mitotic chromosomes (). MCRS1 SS35/36EE did not localize in most cases () although it did localize to the center of chromosome-induced asters when present (Suppl. Fig. 3C). This result indicated that MCRS1 SS35/36EE is not functional and may have a dominant-negative effect on chromosome-induced MT assembly that could explain, at least in part, the strong phenotype on spindle assembly reported above.
Altogether, these data suggest that not only MCRS1 phosphorylation by Aurora-A on Ser35/36 is not required for chromosome-dependent MT assembly but it is detrimental for this process.
MCRS1 phosphorylation by Aurora-A on Ser35/36 regulates K-fiber formation and dynamics
We previously showed that MCRS1 acts at K-fiber minus-ends protecting them from the activity of MT depolymerases, playing an essential role in defining K-fiber length and dynamics.Citation10 We therefore examined whether the Aurora-A dependent phosphorylation of MCRS1 at Ser35/36 is important for this function. We first measured the K-fiber length () in the MCRS1-silenced cells expressing the different MCRS1 phospho-variants. To facilitate these measurements, cells were first incubated in monastrol to promote the formation of monopolar spindles and then placed on ice for 10 min to induce the depolymerization of the more unstable astral MTs while maintaining the K-fibers.Citation10 As we previously described, MCRS1-silenced cells had significantly shorter K-fibers than control cellsCitation10 (). Expression of WT–MCRS1 in the silenced cells efficiently rescued K-fiber length to control levels. In contrast, the expression of the MCRS1 SS35/36AA phospho-null variant in the silenced cells did not (). These defects may explain why these cells have difficulties in progressing through mitosis (see above). Similarly, the expression of the MCRS1 SS35/36EE phospho-mimicking variant in the silenced cells failed to rescue the K-fiber length to control values. Since the cells expressing this protein are defective for chromosome-induced MT assembly (see above), the assembly of shorter K-fibers may be a direct consequence of these earlier defects rather than a direct dominant-negative effect on K-fiber assembly.
Figure 3. MCRS1 phosphorylation by Aurora-A regulates its function at K-fiber minus-ends. (A) K-fiber length in monastrol incubated control and MCRS1-silenced cells expressing the different MCRS1 phospho-variants as indicated. Data from 3 independent experiments monitoring at least 40 cells per condition and experiment. Box-and-whisker plot: boxes show the upper and lower quartiles (25–75%) with a line at the median, whiskers extend from the 10th to the 90th percentile and dots correspond to outliers. Neither MCRS1 SS35/36AA nor MCRS1 SS35/36EE rescues the MCRS1 silencing phenotype. A representative picture of a monastrol-incubated control cell fixed and stained for tubulin is shown. t test, *** corresponds to p <0,05. (B) K-fiber stability in control and MCRS1-silenced cells expressing the different MCRS1 phospho-variants as indicated. The presence of K-fibers (intact K-fibers), K-fibers remnants or the absence of MTs (no K-fibers) was monitored in cells exposed to cold-induced MT depolymerization for different times as indicated. The graph shows the percentage of cells in each category as indicated. At least 140 cells were quantified per experiment and condition. One representative out of 3 independent experiments is shown. Expression of MCRS1 SS35/36AA or MCRS1 SS35/36EE mutants does not rescue the MCRS1 silencing phenotype.
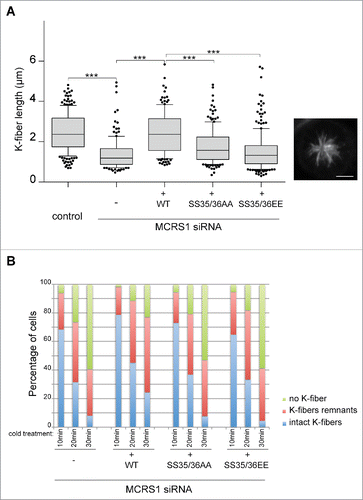
We then examined K-fiber stability under the different conditions. MCRS1-silenced cells expressing the different MCRS1 phospho-variants were fixed after different times of incubation on ice to monitor MT depolymerization over time and therefore K-fiber stability, as previously described.Citation10 K-fiber stability was compromised to a similar extent in MCRS1-silenced cells and in silenced cells expressing any of the 2 MCRS1 phospho-variants compared to MCRS1-silenced cells expressing WT-MCRS1 ().
Overall these results show that the phosphorylation of MCRS1 at Ser35/36 is important for K-fiber assembly. They suggest the existence of a complex regulatory function for Aurora-A on the function of MCRS1 that requires a balance between the phosphorylated and unphosphorylated forms of MCRS1 to promote K-fiber formation and spindle assembly.
Discussion
In this work we have uncovered an important regulatory function for the Aurora-A dependent phosphorylation of MCRS1 on Ser35/36. This phosphorylation occurs specifically during mitosis when MCRS1 stabilizes and regulates the dynamics of the chromosome-driven MT and the K-fibers, performing an essential function in spindle assembly and cell division.
Our data show that phosphorylation does not play any role in the precise localization of MCRS1 to the MT minus-ends during mitosis nor in its specificity for the chromosome-dependent MTs and the K-fibers. This suggests that MCRS1 mitotic localization relies on another mechanism. We recently showed that it indeed requires another protein, KANSL3, which binds directly to the MT minus-ends.Citation11 Consistently, we found here that phosphorylation on Ser35/36 has no impact on the interaction between the 2 proteins. Since we however found that phosphorylation on Ser35/36 has important functional implications, this suggests that it has a regulatory role that goes beyond MCRS1 localization. On the other hand it also suggests that the targeting of the MCRS1 complex to the MT minus-ends is not sufficient for its function.
Interestingly, our data show that abolishing or mimicking phosphorylation on MCRS1 Ser35/36 strongly interferes with its function and spindle assembly. The phenotypes are however different for the phospho-null and the phospho-mimicking variants. While cells expressing MCRS1 SS35/36AA support chromosome-dependent MT assembly and only later show defects in K-fiber assembly and dynamics, those expressing MCRS1 SS35/36EE do not assemble chromosome-dependent MTs. It is therefore difficult to interpret the spindle phenotypes observed in these cells as they may well be a consequence of the absence of the chromosome-derived MTs. However, it is interesting to note that the spindle phenotype is stronger in these cells than in MCRS1-silenced cells, suggesting that the phospho-mimicking form of MCRS1 acts as a dominant-negative mutant promoting major spindle assembly defects.
In addition, the assembly of the chromosomal MTs (in cells expressing the AA mutant) is not sufficient to support the formation of functional K-fibers. These results altogether indicate that the whole process is highly complex and that phosphorylation on MCRS1 Ser35/36 plays a key role for defining its dynamics.
Our results suggest the existence of a 2-step process (). First, chromosome-dependent MT nucleation relies on the activity of a complex including the γTURC,Citation15,19,20 therefore generating MTs with capped minus-ends. At this step unphosphorylated MCRS1 is required and its phosphorylation on Ser35/36 has a negative effect on its stabilizing activity. It is also possible that Aurora-A may not be able to phosphorylate MCRS1 at this early mitosis step. Later, as the spindle organizes and the K-fibers form, MCRS1 becomes specifically associated to their MT minus-ends that are under continuous depolymerization.Citation21,22 Our data suggest that this dynamic association with the depolymerizing minus-ends and the stabilizing activity of MCRS1 require its phosphorylation on Ser35/36 by Aurora-A ().
Figure 4. Model. (A) During spindle assembly, MCRS1 protects the chromosomal MTs against the action of MT depolymerases like MCAK. To be functional and support chromosome-dependent MT assembly in these early steps, MCRS1 has to be unphosphorylated on Ser35/36. (B) In metaphase, Aurora-A phosphorylation of MCRS1 on its S35/36 is required for its function at the minus-ends of the K-fibers establishing their proper dynamics by counteracting MCAK-like MT depolymerase activities.
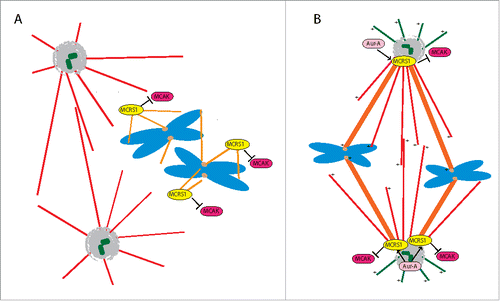
Although the mechanism by which this regulates the MT minus-end depolymerization rate is still unclear, it may involve the activity of other proteins to provide the transition from MTs capped by the γTURC at their minus-ends to MTs with free depolymerizing minus-ends in the K-Fibers. The 2 sides of MCRS1 function, in the initial phases of MT assembly and in the context of the formed spindle, suppose a tight regulation of the balance between MCRS1 phosphorylation and dephosphorylation. This would most probably involve the coordinated activity of a phosphatase that still remains to be identified.
The regulation of K-fiber dynamics at their minus-ends is essential to ensure the constant length of the K-fibers during metaphase, to control the tubulin poleward flux,Citation22-24 and also probably to trigger the fast shortening of the K-fibers that occurs in anaphase.Citation22,25,26 Here, we show that Aurora-A, by phosphorylating the specific K-fiber minus-end binding protein MCRS1, may provide a key regulatory mechanism for the control of K-fiber dynamic properties. Aurora-A promotes MT nucleation and stabilization in mitosisCitation15,27-29 and this is compatible with our data on the defects in K-fiber assembly and stability in cells expressing MCRS1 SS35/36AA. Our work therefore suggests yet another role for Aurora-A during spindle assembly, controlling K-fiber dynamics.
Material and methods
Antibodies
The anti α-tubulin DM1A antibody (Sigma T9026) was used at 1:1000 in immunofluorescence; the anti-human MCRS1 antibody was produced in the labCitation10 and used at 1:100 in immunofluorescence. The home made anti-TPX2 and Aurora-A antibodies were produced in the Vernos lab as previously described.Citation11,30 They were used at 1µg/ml in western-blot. Anti-KANSL3 antibody was used as previously described.Citation11 Secondary antibodies were anti-rabbit and anti-mouse antibodies conjugated to Alexa-488, 568, 680 or 800 (Molecular Probes) and were used at 1:1000 for immunofluorescence and 1:10000 for western-blot.
In vitro kinase assay
Expression and purification of recombinant Aurora-A and MCRS1 were previously described.Citation10,29 5 μM of MBP alone, GST alone, MBP-MCRS1 or GST-MCRS1 were incubated with 0.1 μM of purified His-hAurora-A in kinase buffer (20 mM HEPES, 0.2 M KCl, 5 mM MgCl2, 0.5 mM EGTA, 0.05 % Triton X-100, 0.1 mM ATP) in the presence of [γ-32P]-ATP. The reactions were incubated at 30°C for 10 to 15 min and stopped by addition of SDS-PAGE loading buffer. Proteins were resolved by SDS-PAGE and visualized by Coomassie blue staining. Autoradiographies were obtained by exposing the gel to an Imaging Plate (Fuji Film) that was later scanned with a Typhoon Trio Imager (Amersham Biosciences).
Cell culture, inhibitor, siRNA and plasmid transfections
HeLa cells were grown in DMEM, 10% fetal calf serum and 1X penicillin and streptomycin (Life Technologies). To generate cell lines expressing the WT, SS35/36AA and SS35/36EE mutants, a siRNA-resistant ORF of MCRS1 was cloned in pcDNA5/FRT/TO (Invitrogen). Mutations of S35/36 in A and in E were then generated by site directed mutagenesis. These plasmids were transfected into a HeLa-FRT cell line (gift from J. Pines) and stable cell lines were generated using the FLIP-in system (Invitrogen). Positive clones were selected using Hygromycin B (5 mg/ml, Invitrogen). To induce protein expression from the inducible promoter, cells were incubated with tetracyclin (Calbiochem) at 0.1µg/ml.
MLN8237 (Aurora-A inhibitor, Sigma) was used at 250nM, for 2 h before fixing the cells.
Plasmid transfection was carried out using 10µg of DNA in a 10 cm cell culture dish with x-tremeGENE 9 DNA transfection reagent (Roche) following manufacturer's protocol.
siRNA targeting MCRS1, NUF2 and scrambled siRNA were previously described.Citation10 They were transfected using Lipofectamine 2000 (Invitrogen) using 100 pmol per well in 6-well plates according to the manufacturer's protocol and analyzed 48 h after transfection.
Cell synchronization and anti-flag pull-downs
HeLa cells were synchronized in mitosis by incubating them for 16 h in 3µM nocodazole (Sigma). Nocodazole was then washed out with PBS (X4) and cells were collected by shake-off after 1 h of nocodazole release.
Cells expressing Flag-MCRS1 (WT or mutant forms) synchronized in mitosis were collected and resuspended in lysis buffer (50 mM Tris/HCl pH7.4, 150 mM NaCl, 1 mM EDTA, 5 mM NaF, 100µM orthovanadate, 20 mM β-glycerophosphate, 1%NP40) and incubated for 30 min on ice. The lysate was then cleared by centrifugation at 12000 g (4°C). Anti-Flag magnetic beads (Sigma) were incubated with the lysate for 1 h at 4°C following manufacturer's protocol. After 3 washes with lysis buffer, the beads were then subjected to elution with 400µg/ml of Flag-peptide (Sigma). The eluted fraction was then loaded on SDS-PAGE before western-blot analysis.
Mass spectrometry
Immunoprecipitated proteins were reduced with dithiothreitol (14 mM, 1 h, 37°C ), alkylated in the dark with iodoacetamide (28 mM, 30 min, 25°C) and digested overnight at 37°C with trypsin (Promega). Samples were analyzed by LC-MSMS using an LTQ-Orbitrap Velos mass spectrometer (Thermo Fisher Scientific) coupled to a Proxeon EasyLC (Thermo Fisher Scientific). Peptide mixtures were loaded directly onto the analytical column and they were separated by C18 reversed-phase chromatography. The Mascot search engine was used for peptide identification with the swissprot human database. The precursor ion mass tolerance was set 7 ppm whereas the fragment ion mass tolerance was set to 0.5 Da. Oxidation of methionine, and N-terminal protein acetylation were defined as variable modifications whereas carbamidomethylation on cysteines was set as fixed modifications. Peptides were filtered by Mascot score higher than 20.
Immunofluorescence microscopy
Cells were grown on coverslips and were fixed in −20°C methanol for 10 min. Blocking and antibody dilution buffer was 0.5% BSA (Sigma), 0.1% TritonX100 (Sigma). Fixed cells were visualized with an x63 objective on an inverted DMI-6000 Leica wide-field fluorescent microscope. Confocal images were taken in 0.4µm slices with a x63 oil immersion 1.4 numerical aperture objective lens on a Leica TCS SP5 confocal microscope. Pictures were acquired with the Leica Application Suite software. Images were processed with Image J or Photoshop (Adobe) and mounted in figures using Illustrator (Adobe).
Microtubule regrowth and cold stable assays
These assays were performed as previously described.Citation10 Briefly, for MT regrowth, cells were plated 2 d before the experiment. 2 µM nocodazole was added to the medium for 3 h and washed out 4 times with PBS and once with medium at 37°C. Cells were fixed in methanol at −20°C for 5 min after nocodazole release. Results were quantified by counting the number of MT asters in at least 60 cells per condition and per experiment.
Cold-stable assays were used to quantify K-fiber stability. Cells were washed once with PBS and incubated on ice for 10 to 30 min in L15 medium (Sigma) supplemented with 20 mM HEPES at pH 7.3, washed once in PBS and fixed in methanol at −20°C for 5 min. To quantify the phenotype more than 140 cells were classified into the different categories for each experimental condition.
K-fiber length
To measure K-fiber length, cells were grown on coverslips and treated for 4 h with 40µM monastrol (Sigma), and then subjected to 10 min cold treatment to depolymerize spindle microtubules but not K-fibers. After 1 PBS wash, cells were fixed for 10 min in −20°C methanol. All the K-fibers with clearly identifiable ends were measured in at least 40 monopolar spindles (in total more than 100 per experiment) using Image J in each condition.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
1187342_Supplemental_Material.zip
Download Zip (4.8 MB)Acknowledgments
We thank Núria Mallol for excellent technical assistance. We thank Jonathon Pines and Asifa Akhtar for the gift of reagents.
Funding
Work in the Vernos lab is supported by the Spanish Ministry of Economy and Competitiveness grants BFU2012-37163. We acknowledge support of the Spanish Ministry of Economy and Competitiveness, “Centro de Excelencia Severo Ochoa 2013–2017”, SEV-2012-0208.
References
- Meunier S, Vernos I. Microtubule assembly during mitosis - from distinct origins to distinct functions? J Cell Sci 2012; 125:2805-14; PMID:22736044; http://dx.doi.org/10.1242/jcs.092429
- Rieder CL. The structure of the cold-stable kinetochore fiber in metaphase PtK1 cells. Chromosoma 1981; 84:145-58; PMID:7297248; http://dx.doi.org/10.1007/BF00293368
- Rieder CL. Kinetochore fiber formation in animal somatic cells: dueling mechanisms come to a draw. Chromosoma 2005; 114:310-8; PMID:16270218; http://dx.doi.org/10.1007/s00412-005-0028-2
- Meunier S, Vernos I. Acentrosomal Microtubule Assembly in Mitosis: The Where, When, and How. Trends Cell Biol 2016; 26(2):80-7. doi: 10.1016/j.tcb.2015.09.001
- Helmke KJ, Heald R, Wilbur JD. Interplay between spindle architecture and function. Int Rev Cell Mol Biol 2013; 306:83-125; PMID:24016524; http://dx.doi.org/10.1016/B978-0-12-407694-5.00003-1
- Kalab P, Pralle A, Isacoff EY, Heald R, Weis K. Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature 2006; 440:697-701; PMID:16572176; http://dx.doi.org/10.1038/nature04589
- Kalab P, Weis K, Heald R. Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science 2002; 295:2452-6; PMID:11923538; http://dx.doi.org/10.1126/science.1068798
- Kalab P, Heald R. The RanGTP gradient - a GPS for the mitotic spindle. J Cell Sci 2008; 121:1577-86; PMID:18469014; http://dx.doi.org/10.1242/jcs.005959
- Karsenti E, Vernos I. The mitotic spindle: a self-made machine. Science 2001; 294:543-7; PMID:11641489; http://dx.doi.org/10.1126/science.1063488
- Meunier S, Vernos I. K-fibre minus-ends are stabilized by a RanGTP-dependent mechanism essential for functional spindle assembly. Nat Cell Biol 2011; 13:1406-14; PMID:22081094; http://dx.doi.org/10.1038/ncb2372
- Meunier S, Shvedunova M, Van Nguyen N, Avila L, Vernos I, Akhtar A. An epigenetic regulator emerges as microtubule minus-end binding and stabilizing factor in mitosis. Nat Commun 2015; 6:7889; PMID:26243146; http://dx.doi.org/10.1038/ncomms8889
- Bayliss R, Sardon T, Vernos I, Conti E. Structural basis of Aurora-A activation by TPX2 at the mitotic spindle. Mol Cell 2003; 12:851-62; PMID:14580337; http://dx.doi.org/10.1016/S1097-2765(03)00392-7
- Eyers PA, Erikson E, Chen LG, Maller JL. A novel mechanism for activation of the protein kinase Aurora A. Curr Biol 2003; 13:691-7; PMID:12699628; http://dx.doi.org/10.1016/S0960-9822(03)00166-0
- Sardon T, Peset I, Petrova B, Vernos I. Dissecting the role of Aurora A during spindle assembly. EMBO J 2008; 27:2567-79; PMID:18756265; http://dx.doi.org/10.1038/emboj.2008.173
- Scrofani J, Sardon T, Meunier S, Vernos I. Microtubule nucleation in mitosis by a RanGTP-dependent protein complex. Curr Biol 2015; 25:131-40; PMID:25532896; http://dx.doi.org/10.1016/j.cub.2014.11.025
- Tsai MY, Wiese C, Cao K, Martin O, Donovan P, Ruderman J, Prigent C, Zheng Y. A Ran signalling pathway mediated by the mitotic kinase Aurora A in spindle assembly. Nat Cell Biol 2003; 5:242-8; PMID:12577065; http://dx.doi.org/10.1038/ncb936
- Malik R, Lenobel R, Santamaria A, Ries A, Nigg EA, Korner R. Quantitative analysis of the human spindle phosphoproteome at distinct mitotic stages. J Proteome Res 2009; 8:4553-63; PMID:19691289; http://dx.doi.org/10.1021/pr9003773
- Nousiainen M, Sillje HH, Sauer G, Nigg EA, Korner R. Phosphoproteome analysis of the human mitotic spindle. Proc Natl Acad Sci U S A 2006; 103:5391-6; PMID:16565220; http://dx.doi.org/10.1073/pnas.0507066103
- Groen AC, Cameron LA, Coughlin M, Miyamoto DT, Mitchison TJ, Ohi R. XRHAMM functions in ran-dependent microtubule nucleation and pole formation during anastral spindle assembly. Curr Biol 2004; 14:1801-11; PMID:15498487; http://dx.doi.org/10.1016/j.cub.2004.10.002
- Luders J, Patel UK, Stearns T. GCP-WD is a gamma-tubulin targeting factor required for centrosomal and chromatin-mediated microtubule nucleation. Nat Cell Biol 2006; 8:137-47; PMID:16378099; http://dx.doi.org/10.1038/ncb1349
- Ganem NJ, Compton DA. Functional roles of poleward microtubule flux during mitosis. Cell Cycle 2006; 5:481-5; PMID:16552178; http://dx.doi.org/10.4161/cc.5.5.2519
- Waters JC, Mitchison TJ, Rieder CL, Salmon ED. The kinetochore microtubule minus-end disassembly associated with poleward flux produces a force that can do work. Mol Biol Cell 1996; 7:1547-58; PMID:8898361; http://dx.doi.org/10.1091/mbc.7.10.1547
- Mitchison T, Evans L, Schulze E, Kirschner M. Sites of microtubule assembly and disassembly in the mitotic spindle. Cell 1986; 45:515-27; PMID:3708686; http://dx.doi.org/10.1016/0092-8674(86)90283-7
- Rogers GC, Rogers SL, Schwimmer TA, Ems-McClung SC, Walczak CE, Vale RD, Scholey JM, Sharp DJ. Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature 2004; 427:364-70; PMID:14681690; http://dx.doi.org/10.1038/nature02256
- Chen W, Zhang D. Kinetochore fibre dynamics outside the context of the spindle during anaphase. Nat Cell Biol 2004; 6:227-31; PMID:15039774
- VandenBeldt KJ, Barnard RM, Hergert PJ, Meng X, Maiato H, McEwen BF. Kinetochores use a novel mechanism for coordinating the dynamics of individual microtubules. Curr Biol 2006; 16:1217-23; PMID:16782013; http://dx.doi.org/10.1016/j.cub.2006.04.046
- Barr AR, Gergely F. Aurora-A: the maker and breaker of spindle poles. J Cell Sci 2007; 120:2987-96; PMID:17715155; http://dx.doi.org/10.1242/jcs.013136
- Peset I, Seiler J, Sardon T, Bejarano LA, Rybina S, Vernos I. Function and regulation of Maskin, a TACC family protein, in microtubule growth during mitosis. J Cell Biol 2005; 170:1057-66; PMID:16172207; http://dx.doi.org/10.1083/jcb.200504037
- Pinyol R, Scrofani J, Vernos I. The role of NEDD1 phosphorylation by Aurora A in chromosomal microtubule nucleation and spindle function. Curr Biol 2013; 23:143-9; PMID:23273898; http://dx.doi.org/10.1016/j.cub.2012.11.046
- Lioutas A, Vernos I. Aurora A kinase and its substrate TACC3 are required for central spindle assembly. EMBO Rep 2013; 14:829-36; PMID:23887685; http://dx.doi.org/10.1038/embor.2013.109
