ABSTRACT
During S phase, following activation of the S phase CDKs and the DBF4-dependent kinases (DDK), double hexamers of Mcm2-7 at licensed replication origins are activated to form the core replicative helicase. Mcm10 is one of several proteins that have been implicated from work in yeasts to play a role in forming a mature replisome during the initiation process. Mcm10 has also been proposed to play a role in promoting replisome stability after initiation has taken place. The role of Mcm10 is particularly unclear in metazoans, where conflicting data has been presented. Here, we investigate the role and regulation of Mcm10 in Xenopus egg extracts. We show that Xenopus Mcm10 is recruited to chromatin late in the process of replication initiation and this requires prior action of DDKs and CDKs. We also provide evidence that Mcm10 is a CDK substrate but does not need to be phosphorylated in order to associate with chromatin. We show that in extracts depleted of more than 99% of Mcm10, the bulk of DNA replication still occurs, suggesting that Mcm10 is not required for the process of replication initiation. However, in extracts depleted of Mcm10, the replication fork elongation rate is reduced. Furthermore, the absence of Mcm10 or its phosphorylation by CDK results in instability of replisome proteins on DNA, which is particularly important under conditions of replication stress.
Introduction
The process of DNA replication is highly regulated, involving the stepwise recruitment of different factors onto replication origins in an ordered fashion. The primary purpose of this regulation is to ensure complete duplication of genetic material. From late M-phase to early G1, replication origins are licensed for subsequent activity by loading double heterohexamers of Mcm2-7. In S-phase the sequential action of Dbf4-dependent kinase (DDK) and Cyclin dependent kinase (CDK) drives the recruitment of different replication factors onto DNA-bound Mcm2-7 to generate a pre-initiation complex which includes 2 essential components, Cdc45 and the heterotetrameric GINS complex. This allows assembly of the active replicative helicase CMG (Cdc45, Mcm2-7 and GINS) which drives DNA unwinding ahead of the replication fork.
Mcm10 is an evolutionarily-conserved protein that is involved in DNA replication in different eukaryotic systems, though its exact involvement in this process is still unclear.Citation1 Mcm10 was first identified in S. cerevisiae as a gene required for DNA replication.Citation2,3 Studies in different organisms from yeast to humans have shown that Mcm10 can interact with several replication initiation factors including Mcm2-7,Citation2,4-10 Cdc45,Citation11-13 TopBP1Citation14 and RecQ4.Citation15-17
Previous studies in various organisms have implicated Mcm10 in various roles including activating the Mcm2-7 helicase in fission yeast,Citation18 recruiting Cdc45 and the GINS complex to the pre-RC, stabilization of polα in yeast, Xenopus and humansCitation7,15,19-23 and modulating chromatin dynamics in budding yeast and Drosophila.Citation24,25 The prevalent model from in vitro and in vivo studies in budding and fission yeast is that Mcm10 plays a role late in replication initiation where it is required for unwinding of origin DNA and separation of Mcm2-7 double hexamersCitation10,26-29 In addition to its involvement in DNA replication initiation, Mcm10 has also been shown to promote genomic integrity in human cells, as lack of Mcm10 leads to accumulation of DNA damage and cell cycle arrestCitation22,30-32 Budding yeast Mcm10 performs some of its genome protection functions by interactions with 9-1-1 checkpoint clamp and other factors implicated in double strand break repair.Citation33,34
In the current study we show that in Xenopus egg extracts, Mcm10 binds to chromatin at a later stage in process of DNA replication initiation in an S-CDK- and DDK-dependent manner. This is in contrast to a previous study on Xenopus Mcm10,Citation19 but is consistent with results obtained in yeasts and other organisms. We demonstrate that Xenopus Mcm10 is not required for bulk DNA replication but is required for replisome stability, with depleted extracts having reduced rates of replication fork elongation. We also show that the ability of Xenopus Mcm10 to promote replisome stability requires it to undergo a CDK-dependent phosphorylation.
Results
Mcm10 chromatin binding is dependent on S-phase kinases
We raised 2 polyclonal antisera to Xenopus Mcm10, one against the N-terminus of the protein and one against the C-terminus. Both antibodies recognized several bands in whole egg extract, but recognized a common band at ∼100 kDa both in extract and on chromatin, as expected of Xenopus Mcm10 (Fig S1A). The same ∼100 kDa protein was immuno-depleted from extract by both antibodies. Mass spectrometry of immunoprecipitates from extracts and chromatin showed Mcm10 as the most abundant precipitated protein (Fig S1B).
Mcm10 chromatin binding in Xenopus egg extracts was previously reported to be dependent on replication licensing but independent of CDK activity.Citation19 In contrast, recent reports in yeast have demonstrated that Mcm10 is loaded on chromatin at one of the last steps in the assembly of the pre-initiation complex, after both DDK- and CDK- dependent steps have been executed.Citation27-29 In light of these contradictory observations in different organisms, we re-investigated the requirements for Mcm10 chromatin loading in Xenopus egg extracts. Consistent with its playing a role in DNA replication, Xenopus Mcm10 associated with chromatin precisely at the time of replication, matching the binding pattern of Cdc45, Psf2 and PCNA () which all function at active replisomes when DNA synthesis occurs (Fig S2A). As previously reported in Xenopus egg extractCitation19 prior DNA licensing was required for Mcm10 chromatin recruitment as upon Geminin addition, Mcm10 chromatin binding was inhibited ().
Figure 1. Mcm10 chromatin loading requirements. A, Xenopus egg extract was supplemented with demembranated sperm nuclei. After incubation for the indicated times, chromatin was isolated and immunoblotted for Mcm10, Mcm3, Cdc45, Psf2 and PCNA. The lower portion of the gel was stained with Coomassie to visualize histones. (B- F), Interphase (B, D, E, F) or membrane free (C) egg extracts were supplemented with demembranated sperm nuclei and were optionally supplemented with Geminin (B), 50 μM PHA-767491 (D), 100 nM p27kip1 (E), 40 µM aphidicolin or 5 mM Caffeine (F). At the indicated times, chromatin was isolated and immunoblotted for Mcm10, Mcm3, Mcm4, Mcm7, Orc2, Cdc45, Psf2 and PCNA.
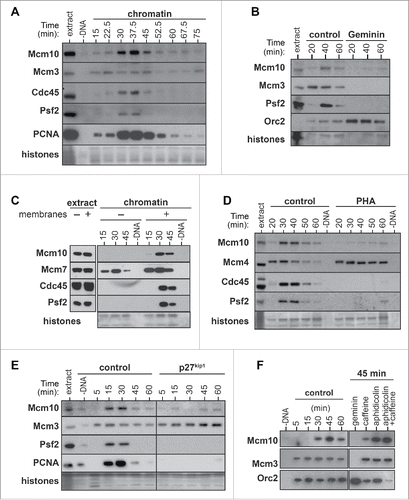
Once origin licensing is complete in Xenopus extracts, chromatin is assembled into interphase nuclei, allowing the nuclear import and activation of the DDK and CDK kinases that are required for the initiation of replication.Citation35-37 When sperm DNA was incubated in extracts lacking nuclear membrane precursors, no Mcm10 chromatin recruitment was observed (). Consistent with this result, when nuclei were assembled in egg extract supplemented with wheat germ agglutinin, a lectin that blocks nuclear pore formation,Citation38,39 Mcm10 did not bind the DNA (Fig. S2B).
We followed up this result by determining whether Mcm10 chromatin loading in Xenopus extracts depends on either DDK or CDK kinase activity. When replication initiation was prevented by inhibiting DDK activity with PHA-767491Citation40 or by inhibiting CDK with p27kip1, Mcm10 was not loaded onto chromatin (). When replication fork elongation was inhibited with aphidicolin, levels of chromatin-bound Mcm10 remained high (), but when additional dormant origin activation was induced by a combination of aphidicolin and caffeine,Citation41 additional Mcm10 was loaded onto chromatin. These results show that Mcm10 loading occurs during replication initiation downstream of DDK and CDK activity, in agreement with results in S. cerevisiae and S. pombe,Citation27-29 but in disagreement with the earlier study of Mcm10 in Xenopus egg extracts.Citation19
Mcm10 is a CDK substrate
Previous reports have shown that Xenopus Cdc7 executes its essential function early in S-phase before the need for CDKs.Citation40,42,43 Since Mcm10 chromatin association depends on CDK activity, we asked if Mcm10 is a direct CDK-substrate by using mass spectrometry to identify Mcm10 phosphopeptides. To determine whether any observed phosphorylation was CDK-dependent, chromatin was assembled in the presence or absence of the CDK inhibitor, p27kip1, and also examined the effect of aphidicolin and caffeine. Proteins were eluted from chromatin under these different conditions, digested with trypsin and bound to TiO2 beads or a strong cation exchanger (SCX) to enrich mono- and multiply-phosphorylated peptides, respectively. Consistent with Mcm10 chromatin loading being strictly CDK-dependent, no peptides (phosphopeptides or otherwise) were identified in chromatin isolated from p27kip1 samples (, p27kip1). Mcm10 phosphopeptides were identified from all the other conditions with the highest recovery from extracts treated with aphidicolin plus caffeine, where the greatest loading of Mcm10 on chromatin was observed (). Three Mcm10 phosphopeptides were identified across the different samples, each containing a phosphorylated serine followed by a proline (S173, S596 and S630, ), which conforms to the minimal consensus motif for CDK-phosphorylation.
Figure 2. CDK-regulation of Mcm10 chromatin loading. A, B, CDK-dependent phosphopeptides of chromatin bound Mcm10 identified in mid-S-phase chromatin by phosphopeptide enrichment of total proteins (A) or direct immunoprecipitation (B) of Mcm10 followed by mass spectrometric analysis. (A) Heat map for Mcm10 phosphopeptides enrichment observed in control, CDK-inhibited (p27kip1), replication inhibition (aphidicolin, Aph), checkpoint-inhibition (caffeine, Caff) with black showing the presence and white the absence of a peptide. C, Extracts were supplemented with demembranated sperm nuclei with or without the addition of 1 ng/µl wt-rMcm10. Samples containing rMcm10 were optionally supplemented with p27kip1 or Geminin. After incubation for the indicated times, chromatin was isolated and immunoblotted for Mcm10, Mcm7, Cdc45 and Psf2. The lower portion of the gel was stained with Coomassie to visualize histones. D-G, Undepleted or Mcm10 depleted interphase egg extracts were supplemented with demembranated sperm nuclei. After incubation for 35 min, aliquots were optionally supplemented with wt-rMcm10 or undepleted interphase extract with or without p27kip1. At the indicated times, chromatin was isolated and immunoblotted for Mcm10 and/or Mcm7, Cdc45 and PCNA. (D) Cartoon of experimental set-up. (E) rMcm10 was added to undepleted extracts at 35 in presence of p27kip1, aphidicolin or both. (F, G) rMcm10 or undepleted extract was added to Mcm10 depleted extract at 35 min in presence of p27kip1. H, Control (nonimmune IgG), RecQ4, TopBP1 or Treslin depleted extract were supplemented with demembranated sperm nuclei. After incubation for the indicated times, chromatin was isolated and immunoblotted for Mcm10.
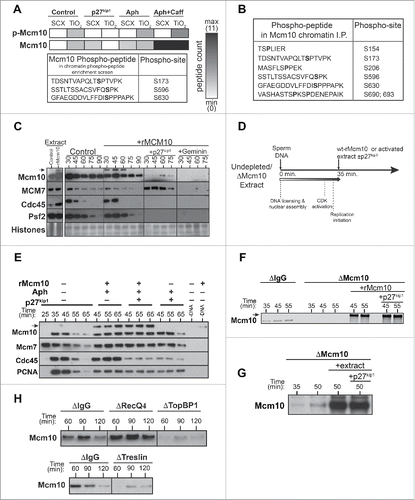
In an alternative approach, Mcm10 protein was immunoprecipitated from chromatin assembled in extract that was optionally treated with p27kip1 and the immunoprecipitates were then analyzed by mass-spectrometry. Six different CDK-dependent phosphopeptides were identified (), including the 3 identified in the general phosphopeptide enrichment screen. CDK-dependent phosphorylation of Mcm10 appears to occur only when Mcm10 is bound to chromatin as no Mcm10 phosphopeptides were observed when Mcm10 was immunoprecipitated from egg extract without DNA (data no shown).
Mcm10 phosphorylation is not required for chromatin loading
When recombinant GST-tagged Mcm10 protein (rMcm10) expressed in E. coli was added to Xenopus extract, it was loaded onto chromatin in a manner that resembled endogenous Mcm10 (). The peak of rMcm10 chromatin binding occurred during DNA replication, coinciding with the binding of Cdc45 and Psf2. Moreover no rMcm10 binding took place when licensing was inhibited with geminin or when CDK activity was inhibited with p27kip1. We next addressed whether CDK-phosphorylation is a pre-requisite for Mcm10 chromatin binding with experiments that are outlined in . In the first set of experiments, rMcm10 was added to extracts in S-phase with or without p27kip1; extract was also supplemented with aphidicolin at this time to prevent the termination of replication forks already active at the time of p27kip1 addition. shows that inhibiting CDK activity with p27kip1 did not block the chromatin loading of rMcm10. This suggests that prior phosphorylation of Mcm10 protein by CDKs is not essential for is chromatin binding.
Xenopus Mcm10 has the potential to self-associate via its N-terminal coiled-coil domainCitation44 and human Mcm10 has also the potential to form a homo-hexameric structure.Citation45 We therefore used Mcm10-depleted extracts to rule out the possibility that interactions with endogenous Mcm10 already bound to chromatin at the time of addition might allow unphosphorylated rMcm10 to bind chromatin. DNA was incubated in Mcm10-depleted extract and at 35 minutes rMcm10 was added with or without p27kip1 shows that rMcm10 was loaded onto chromatin regardless of the co-addition of p27kip1.
In order to rule out the possibility that an unusual feature of rMcm10 not present on the endogenous protein makes it able to bind chromatin without being phosphorylated by CDKs, we performed similar experiments with endogenous Mcm10 in place of rMcm10 (). DNA was incubated for 35 min in extract immunodepleted of Mcm10, at which time the depleted extract was supplemented with an aliquot of undepleted extract with or without p27kip1 shows that endogenous Mcm10 was loaded onto chromatin regardless of the co-addition of p27kip1. Together, these results suggest that although the association of Mcm10 with chromatin requires CDK activity, Mcm10 itself does not need to be phosphorylated by CDKs to allow this to happen.
We next examined which of the known replication initiation factors are required for Mcm10 chromatin loading, by examining the binding of Mcm10 to chromatin in extracts depleted of TopBP1, Treslin or RecQ4. shows that Mcm10 chromatin loading requires TopBP1 and Treslin, but does not require RecQ4. Given that in Xenopus egg extracts RecQ4 is required late in the initiation process, after Cdc45 and GINS recruitment,Citation46-48 this suggests that Mcm10 is also loaded onto chromatin at this late stage.
Mcm10 is required for replication fork elongation and stability
To understand the function of Mcm10 in DNA replication, we removed >99% Mcm10 from interphase Xenopus egg extracts by immunodepletion using antibodies raised against the N- or C-terminus of the protein individually (Supp. Fig S1A) or when mixed together (). Mcm10 immunodepletion did not prevent nuclear assembly (data not shown). No Mcm10 could be observed on chromatin in the immunodepleted extracts ( and Supp. Fig S1A). Unlike the previous report on Xenopus Mcm10,Citation19 extracts depleted of Mcm10 with our 2 antibodies (used together) still loaded Cdc45, GINS, Polα and PCNA onto DNA (). However the quantity of these proteins bound to chromatin was reproducibly reduced to ∼50% of control levels in Mcm10-depleted extracts ( and Supp. Fig S3C). Similarly, although Mcm10-depleted extracts could still replicate DNA, they did so at a slower rate and less efficiently than control-depleted extracts ( and Supp. Fig S3A and S3B). Since Mcm10-depleted extract still managed to replicate the bulk of input DNA, this suggests that Mcm10 is either not required for initiation in Xenopus extracts or performs a function that can be taken over by another protein. A modest rescue in the amount of replication was observed upon addition of wt-Mcm10 to Mcm10 depleted extract (Supp. Fig S3D). However, the consistently slower replication rate and reduction in replication factors on chromatin in the absence of Mcm10 suggests that there could be problems with the elongation or stability of replisomes.
Figure 3. Lack of Mcm10 leads to reduction in chromatin bound replisome proteins. A, Egg extract was immunodepleted with either nonimmune IgG or Mcm10 antibodies (raised against N- or C-terminus of the protein). So that the efficiency of depletion could be estimated, 0.5 µl of each of the depleted extracts and known amounts of nonimmune-depleted extract was immunoblotted for Mcm10 using 2 different antibodies. B, Control (nonimmune IgG) and Mcm10 depleted extract were supplemented with demembranated sperm nuclei. After incubation for the indicated times, chromatin was isolated and immunoblotted for Mcm10, Mcm4, Cdc45, Psf2, Pol α and PCNA. The lower portion of the gel was stained with Coomassie to visualize histones. C, Quantitation of Cdc45, Pol α, PCNA and Psf2 bound to chromatin at 60 min in control and Mcm10 depleted extracts. Mean and SEM of 3 independent experiments is shown. D, E, Control and Mcm10 depleted extracts were supplemented with 3 ng DNA/µl and [α-32 P]dATP; total DNA synthesis was determined at the indicated times. (E) Mean incorporation of [α-32 P]dATP at 90 and 120 min. and SEM of 3 independent experiments is shown.
![Figure 3. Lack of Mcm10 leads to reduction in chromatin bound replisome proteins. A, Egg extract was immunodepleted with either nonimmune IgG or Mcm10 antibodies (raised against N- or C-terminus of the protein). So that the efficiency of depletion could be estimated, 0.5 µl of each of the depleted extracts and known amounts of nonimmune-depleted extract was immunoblotted for Mcm10 using 2 different antibodies. B, Control (nonimmune IgG) and Mcm10 depleted extract were supplemented with demembranated sperm nuclei. After incubation for the indicated times, chromatin was isolated and immunoblotted for Mcm10, Mcm4, Cdc45, Psf2, Pol α and PCNA. The lower portion of the gel was stained with Coomassie to visualize histones. C, Quantitation of Cdc45, Pol α, PCNA and Psf2 bound to chromatin at 60 min in control and Mcm10 depleted extracts. Mean and SEM of 3 independent experiments is shown. D, E, Control and Mcm10 depleted extracts were supplemented with 3 ng DNA/µl and [α-32 P]dATP; total DNA synthesis was determined at the indicated times. (E) Mean incorporation of [α-32 P]dATP at 90 and 120 min. and SEM of 3 independent experiments is shown.](/cms/asset/0fc4fa6c-66f9-445b-b219-ae8d58d347f7/kccy_a_1199305_f0003_b.gif)
We directly addressed whether Mcm10 is required for fork elongation with the experiment outlined in .Citation40,49-51 Sperm nuclei were incubated in Mcm10-depleted extract in the presence of 100 µM aphidicolin, so that forks could initiate but were unable to move away from replication origins. After 60 min, chromatin lacking bound-Mcm10 was isolated and transferred to either non-immune depleted or Mcm10-depleted extract containing p27kip1 to prevent any further initiation events from occurring. Replication fork progression rate was measured as the [α-32P]dATP incorporated over next 10 minutes. Forks in Mcm10 depleted extract, lacking chromatin bound Mcm10 (), progressed at a slower rate than in non-immune-depleted extract (). Measuring the overall rate of replication in 8 independent experiments gave a mean reduction in the replication rate of 0.81±0.05 (mean ± s.e.m.) in Mcm10-depleted extract compared to the non-immune-depleted controls. This suggests that the reduced rate of bulk DNA replication observed in Mcm10 depleted extract results from a reduced rate of fork elongation.
Figure 4. Mcm10 loss affects replication fork elongation and replisome stability. A-C, Mcm10-depleted interphase egg extracts were supplemented with demembranated sperm nuclei and 100 µM aphidicolin. After 60 min incubation, chromatin was isolated. Chromatin was then incubated in non-immune or Mcm10 depleted extract supplemented with 100 nM p27kip1 and optionally with [α-32 P]dATP. (A) Cartoon of experimental set-up. (B) Chromatin isolated after incubation in the first (lane 1) and second extracts (lane 2, 3) was immunoblotted for Mcm10 and histone H3. (C) At the indicated times, incorporation of [α-32 P]dATP into nascent DNA strand was determined. (D) Control (nonimmune IgG) and Mcm10 depleted extract were supplemented with 15 ng DNA/µl. At 120 mins [α-32 P]dATP was added for 15 seconds, and then DNA was isolated and separated on alkaline sucrose gradients. The 32 P content of fractions was determined by scintillation counting. The black arrowhead shows the migration of tRNA in a parallel neutral sucrose gradient. (E) Non-immune and Mcm10 depleted extract were supplemented with demembranated sperm nuclei and optionally supplemented with 50 µM camptothecin. At 60 min., chromatin was isolated and immunoblotted for Mcm10 and γH2A-X.
![Figure 4. Mcm10 loss affects replication fork elongation and replisome stability. A-C, Mcm10-depleted interphase egg extracts were supplemented with demembranated sperm nuclei and 100 µM aphidicolin. After 60 min incubation, chromatin was isolated. Chromatin was then incubated in non-immune or Mcm10 depleted extract supplemented with 100 nM p27kip1 and optionally with [α-32 P]dATP. (A) Cartoon of experimental set-up. (B) Chromatin isolated after incubation in the first (lane 1) and second extracts (lane 2, 3) was immunoblotted for Mcm10 and histone H3. (C) At the indicated times, incorporation of [α-32 P]dATP into nascent DNA strand was determined. (D) Control (nonimmune IgG) and Mcm10 depleted extract were supplemented with 15 ng DNA/µl. At 120 mins [α-32 P]dATP was added for 15 seconds, and then DNA was isolated and separated on alkaline sucrose gradients. The 32 P content of fractions was determined by scintillation counting. The black arrowhead shows the migration of tRNA in a parallel neutral sucrose gradient. (E) Non-immune and Mcm10 depleted extract were supplemented with demembranated sperm nuclei and optionally supplemented with 50 µM camptothecin. At 60 min., chromatin was isolated and immunoblotted for Mcm10 and γH2A-X.](/cms/asset/27aa596a-f05d-4dd7-9fd7-3a0b6ed3aefa/kccy_a_1199305_f0004_b.gif)
In order to examine the effect of Mcm10 depletion we pulse-labeled nascent DNA very briefly (15 seconds) with 32P-dATP and then separated nascent DNA on a denaturing alkaline sucrose gradient. This shows 32P-dATP incorporation into both high molecular weight and Okazaki-fragment-sized DNA; if the 32P-dATP is chased with unlabelled dATP, the Okazaki fragment-sized DNA is rapidly ligated into high molecular weight DNA, demonstrating semi-discontinuous DNA synthesis in Xenopus egg extracts.Citation52 After a 15 second pulse of 32P-dATP in extract immunodepleted of Mcm10, there was a relative lack of label in high molecular weight DNA (). This suggests that a lack of Mcm10 causes abnormal processing of nascent DNA at the replication fork, either due to defects in leading strand synthesis or in the ligation of Okazaki fragments.
In order to determine effect of Mcm10 on the stability of replication forks, we challenged DNA replication by incubating sperm chromatin in control and Mcm10-depleted extracts supplemented with 50 µM camptothecin, a topoisomerase I inhibitor. Camptothecin stabilizes complexes consisting of topoisomerase-I covalently linked to DNA, thereby providing an impediment to replication forks which can result in double strand DNA breaks and checkpoint activation. When DNA was replicated in Mcm10-depleted extracts there was a very large increase in γH2AX on the chromatin, suggesting the formation of a large number of double strand DNA breaks ().
shows that Mcm10 is phosphorylated by CDKs, but this is not essential for its chromatin loading. To investigate a potential function for Mcm10 phosphorylation by CDKs we aligned the Mcm10 protein sequence from various vertebrates. Interestingly among the different phosphorylation sites we identified on Xenopus Mcm10 () only S630 was well conserved among Xenopus, zebrafish, mouse and human (). We therefore synthesized recombinant Mcm10 with serine 630 replaced by alanine (Mcm10 S630A). No major effect on bulk DNA synthesis was observed when wild-type or S630A rMcm10 was added to Xenopus extract (). However, addition of recombinant S630A Mcm10 to extracts caused a marked reduction in the abundance of CMG components (Cdc45 and Psf2) and DNA polymerase-α on chromatin (). This reduction was similar in magnitude to the effect of depleting Mcm10 from extract ().
Figure 5. CDK-dependent Mcm10 phosphorylation is important for its function. A, Comparison of a segment of Mcm10 protein sequence alignment from Xenopus laevis, Xenopus tropicalis, Dana rerio, Mus musculus and Homo sapiens. B, C, Interphase extract was supplemented with demembranated sperm nuclei and optionally supplemented with [α-32 P]dATP, wt-Mcm10 or S630A-Mcm10. (B) At the indicated times, DNA synthesis was determined by measuring [α-32 P]dATP incorporation. A representative of 3 independent experiment is shown. (C) Chromatin was isolated at indicated times and immunoblotted for Mcm10, Mcm7, Cdc45, Psf2, Polα, or PCNA. The lower portion of the gel was stained with Coomassie to visualize histones. (D) Interphase extract was supplemented with demembranated sperm nuclei and optionally supplemented with wt-Mcm10, S630A-Mcm10 or 50 µM camptothecin. After incubation for the indicated times, chromatin was isolated and immunoblotted for Mcm10 and γH2A-X. (E) Interphase extract was supplemented with demembranated sperm nuclei and optionally supplemented with 50 µM camptothecin. At 45 min. chromatin Mcm10 was immunoprecipitated using antibodies against Mcm10 and samples were immunoblotted for Mcm10, ATR, ATM, Nbs1, Mre11 and Rad17.
![Figure 5. CDK-dependent Mcm10 phosphorylation is important for its function. A, Comparison of a segment of Mcm10 protein sequence alignment from Xenopus laevis, Xenopus tropicalis, Dana rerio, Mus musculus and Homo sapiens. B, C, Interphase extract was supplemented with demembranated sperm nuclei and optionally supplemented with [α-32 P]dATP, wt-Mcm10 or S630A-Mcm10. (B) At the indicated times, DNA synthesis was determined by measuring [α-32 P]dATP incorporation. A representative of 3 independent experiment is shown. (C) Chromatin was isolated at indicated times and immunoblotted for Mcm10, Mcm7, Cdc45, Psf2, Polα, or PCNA. The lower portion of the gel was stained with Coomassie to visualize histones. (D) Interphase extract was supplemented with demembranated sperm nuclei and optionally supplemented with wt-Mcm10, S630A-Mcm10 or 50 µM camptothecin. After incubation for the indicated times, chromatin was isolated and immunoblotted for Mcm10 and γH2A-X. (E) Interphase extract was supplemented with demembranated sperm nuclei and optionally supplemented with 50 µM camptothecin. At 45 min. chromatin Mcm10 was immunoprecipitated using antibodies against Mcm10 and samples were immunoblotted for Mcm10, ATR, ATM, Nbs1, Mre11 and Rad17.](/cms/asset/94a3ce8e-d729-4d70-8769-47a43a8356a2/kccy_a_1199305_f0005_c.gif)
We therefore asked whether S630A-Mcm10 caused the loss of replisome proteins from replication forks when challenged by replication stress. Replication forks were allowed to initiate in Xenopus extracts in the presence of camptothecin and rMcm10 proteins. shows that addition of S630A-Mcm10 led to an increase in the amount of γH2AX on DNA that was induced by camptothecin. Remarkably, addition of wt-Mcm10 reduced the amount of γH2AX relative to the amount generated in untreated extract.
These results show that although Mcm10 is not essential for bulk DNA replication in Xenopus egg extracts, it plays an important role in protecting replisome stability and preventing the generation of double strand DNA breaks during replication. This is consistent with work in other organisms which have implicated Mcm10 in preventing DNA damage during S phase and stabilizing replisomes.Citation20,22,30,33,53,54 We therefore examined whether we could detect interactions between Mcm10 and DNA repair factors at active replication forks in the Xenopus system. In immunoprecipitates of Mcm10 from replicating chromatin we identified interactions with the different subunits of MRN complex (Mre11, Rad17 and Nbs1) as ATM and ATR (). Treatment with camptothecin did not result in further increase in interaction between Mcm10 and MRN proteins. These results are consistent with Mcm10 working with the MRN complex and checkpoint kinases to respond to problems occurring during replisome progression that could lead to double strand break formation.Citation34
Discussion
Mcm10 is one of several replication factors that form the replisome and it plays a role in ensuring the accurate replication of the genome. Despite intensive study, the precise role of Mcm10 in DNA replication still remains somewhat mysterious, with apparently conflicting results in different model systems.Citation1 In the current study we have used Xenopus egg extracts to demonstrate that Mcm10 chromatin binding is a late event in the process of replication initiation and requires prior action of DDK and CDK. We also show that Mcm10 is a CDK substrate that requires binding of other CDK-dependent replisome factors for its chromatin association. We show that lack of Mcm10 or its phosphorylation by CDK results in instability of replication fork proteins on DNA, which is crucial particularly under conditions of replication stress.
Mcm10 chromatin association
Previous work in S. cerevisiae and S. pombe has shown that the CMG complex assembles in the absence of Mcm10,Citation20,26-29 and that Mcm10 loading occurs only after CMG assembly, thus post DDK and CDK activities.Citation55 However, a previous study of DNA replication in Xenopus egg extracts showed Mcm10 loading to be independent of CDK or DDK activities and to be required for Cdc45 loading.Citation19 We therefore revisited the kinetics and timing of Mcm10 chromatin association in Xenopus cell free extracts. We showed that Mcm10 loading onto chromatin depends on origin licensing, nuclear assembly and both DDK and CDK activities. We also observed increased Mcm10 loading upon hyper-activation of dormant origins by simultaneous treatment with both aphidicolin and caffeine.Citation41 These results show that while Mcm10 chromatin binding occurs at the time of replication initiation, it is downstream of DDK and CDK activities. CDK- and DDK-dependent association of Mcm10 with replication origins has recently been reported in human cells,Citation56 and we conclude that this is likely to be a conserved feature of Mcm10 regulation in eukaryotes.
Requirement for Mcm10 in replication initiation
Mcm10 has been implicated in several replication-related functions, but the prevailing model in budding yeast suggests that the essential function of Mcm10 is at the step of DNA unwinding to allow RPA loading.Citation26-29 However, we show that Xenopus extracts depleted of Mcm10 can load RPA and support bulk DNA replication at an efficiency of ∼80% of that of control extracts. Although impossible to completely rule out, we think it is very unlikely that this DNA replication depends on trace amounts of Mcm10 remaining in the depleted extracts. Immunodepletion removed >99% of total Mcm10 from the extract, and no Mcm10 was detectable on chromatin assembled in the depleted extract. Our results are consistent with studies in other metazoan cell types, where knockdown of Mcm10 by RNAi in C. elegans,Citation57 Drosophila KC cellsCitation11 and Hela cellsCitation30 resulted in no major block to bulk DNA replication. More dramatically, Drosophila Mcm10 mutants were not defective for adult cell proliferation, though they were essential for embryo viability and tissue types, like compound eye, for replication and differentiation.Citation58
CDK phosphorylation of Mcm10
We have provided evidence that Mcm10 is a direct substrate of S-phase CDKs. We show that CDK phosphorylation of Mcm10 is not a pre-requisite for its loading onto chromatin but only occurs once Mcm10 is on chromatin. This is in apparent contrast to the human Mcm10 protein, which is phosphorylated at the onset of metaphase to promote its release from the chromatin.Citation5 While investigating CDK regulation of Mcm10 we identified, among number of phosphorylations sites, S630 as crucial for Mcm10 function. The potential importance of S630 is indicated by its conservation in vertebrates through Xenopus to humans. When added to extract, a non-phosphorylatable S630A mutant of Mcm10 resulted in a phenotype that showed similarities with Mcm10 depletion: reduced recruitment of replisome proteins (Cdc45, Psf2, PCNA, polα) and a strongly enhanced γ-H2AX signal when extracts were challenged with camptothecin.
Mcm10 and replisome stability
Although Mcm10-depleted Xenopus extract supported bulk DNA replication, we observed that replication was unusual in several ways. In Mcm10-depleted extracts, the replication was slightly slower and less extensive than normal, the rate of replication fork elongation was reduced, the chromatin recruitment of replisome proteins (Cdc45, Psf2, PCNA, polα) was reduced and there was a strongly enhanced chromatin recruitment of γ-H2AX when extracts were further challenged with camptothecin. Similar defects were seen in the non-phosphorylatable S360A mutant of Mcm10. These results are also consistent with the effect of Mcm10 knockdown in Drosophila KC cellsCitation11 and Hela cellsCitation22,30,31 where various features of aberrant replication were detected.
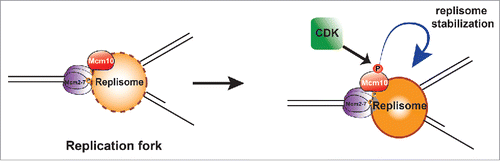
The ∼20% reduction in fork elongation rate in Xenopus extracts lacking Mcm10 is similar to the reduction in replication of bulk DNA in the extracts, consistent with the idea that the primary defect is in replisome progression. Since Mcm10 has been shown to interact with multiple replisome proteinsCitation4-17 and is a component of moving replisomes,Citation59 the slowed fork elongation rate in Mcm10-depleted extracts could likely result from a reduced stability of replisome proteins at the fork. This is consistent with the significant reduction in chromatin-bound replisome proteins that we observed in Mcm10-depleted extracts. The fact that the large reduction in replisome proteins was associated with only a modest reduction in replication rate could be due to the presence of a large number of licensed but dormant origins which can be activated if replication forks stall.Citation41,60 Taken together, these observations are consistent with the primary role of Mcm10 in metazoans being to maintain or stabilizing replisomes once they have initiated ().
Figure 6. The role and regulation of Mcm10 in Xenopus egg extracts. Replication forks are shown, with DNA in black, the CMG helicase in purple, Mcm10 in red, and other replisome proteins in orange. Mcm10 is required for stability of the other replisome proteins, and this effect is enhanced when Mcm10 is phosphorylated by CDK.
Mcm10 has been shown to interact with DNA damage repair proteins which could also explain its role in stabilizing replisomes.Citation1,33,61 Alver et al.Citation33 have shown that N-terminus of Mcm10 plays an important role in resisting the DNA damage induced by different genomic insults including treatment with camptothecin in budding yeast. We did not find any Mcm10 interaction with 9-1-1 complex but instead showed that Xenopus Mcm10 interacts with the different subunits of the MRN complex (Mre11, Rad17 and Nbs1) as well as ATM and ATR. We therefore conclude that although Mcm10 is not an essential factor for DNA replication initiation in Xenopus, it is an S-CDK substrate required for stabilizing other protein components at the replication fork.
Materials and methods
Xenopus egg extract and DNA templates
Metaphase-arrested Xenopus laevis egg extract and demembranated Xenopus sperm nuclei were prepared as described.Citation62 Extracts were supplemented with 250 μg/ml cycloheximide, 25 mM phosphocreatine and 15 μg/ml creatine phosphokinase and incubated with 0.3 mM CaCl2 for 15 minutes to release from metaphase arrest. For DNA synthesis reactions, sperm nuclei were incubated at 6–10 ng DNA/μl in extract. DNA synthesis was assayed by measuring incorporation of [α-32 P]dATP into acid-insoluble material followed by scintillation counting, as described.Citation62 All incubations were carried out at 23°C.
Chromatin isolation from egg extract
Chromatin isolation for immunoblotting was carried out as described.Citation62 Briefly, extract was diluted with ice-cold NIB (50 mM KCl, 50 mM mM HEPES-KOH pH 7.6, 5 mM MgCl2, 0.5 mM spermidine, 0.15 mM spermine, 2 mM DTT) containing phosphatase inhibitors, under-laid with NIB + 20% sucrose (w/v) and centrifuged in a swinging bucket rotor at 2100 g, 5 min, 4°C. Following a cushion wash, chromatin was compacted by spinning at 13000 g, 2 min in a fixed angle rotor. The resulting pellet was resuspended in SDS loading buffer.
Isolation of intact nuclei for transfer experiments for fork-elongation assay was carried out as described.Citation40 Extracts were diluted as before but in NIB buffer where Triton X-100 was omitted, and were then under-laid with a double cushion of NIB + 20% sucrose and NIB + 30% glycerol (v:v in NIB) and centrifuged in a swinging bucket rotor. Following a cushion wash, nuclei were resuspended in the glycerol cushion and added to the second extract at a final concentration of 10 ng DNA/µl.
Immunoblotting
For immunoblotting, samples were separated on 4–12% Bis-Tris gradient gels (Invitrogen). Proteins were transferred onto PVDF membranes (GE Healthcare, RPN303F) using a wet transfer system, blocked in PBS with 0.2% Tween-20 and 5% non-fat milk. After incubation with primary and secondary antibodies, membranes were developed using enhanced chemiluminescence detection (SuperSignal® West Pico Chemiluminescent; Thermo Scientific, 34087) or signals were acquired using Li-COR Odyssey bio-systems where Li-Cor secondary antibodies were used. The lower portion of each gel was typically cut and treated with Coomassie stain to visualize histones. Band intensities were quantified using GelEval (FrogDance Software).
Immunoprecipitation and immunodepletion
Chromatin-bound Mcm10 from Xenopus egg extract was isolated as described.Citation62,63 Briefly, chromatin was isolated in the middle of S-phase and resuspended in NIB+20% sucrose. To release proteins DNA was digested with 2 units/µl Benzonase (Novagen) for 10min. Samples were sonicated (Bioruptor, Diagenode) for 5 min (10 s sonication, 45 s break at medium intensity), and centrifuged 20,000 g, 4°C. The supernatant was used as an input for immunoprecipitation with Mcm10 or preimmune sheep IgG (Sigma) antibody previously bound to protein G-Dynabeads (Invitrogen) for 1 h at 4°C. For immunoprecipitation of Mcm10 in Xenopus egg extracts (not supplemented with DNA), interphase extracts were diluted (5-fold) in LFB 1/50 (40 mM HEPES-KOH pH 8.0, 20 mM K2HPO4/KH2PO4 pH 8.0, 2 mM MgCl2, 1 mM EGTA, 2 mM dithiothreitol, 10% (w/v) sucrose, 1 μg/ml each of leupeptin, pepstatin and aprotinin; supplemented with 50 mM KCl) and incubated with antibody conjugated beads as described above. The beads were then washed (5x) with filtered PBS before resuspension in loading buffer.
Immunodepletion of Mcm10 from Xenopus egg extract was performed as described.Citation62 Briefly, metaphase arrested extract was activated to bring it into interphase. Subsequently three successive rounds of incubations of 1 volume extract with 0.2 volumes of protein G-Dynabeads beads conjugated to anti-Mcm10 antibody for 30 minutes at 4°C were carried out.
Phosphopeptide enrichment and mass spectrometry
To enrich chromatin bound phosphoproteins, chromatin was isolated in middle of S-phase and resuspended in NIB+20% sucrose.Citation63 To release proteins, DNA was digested with 2 units/µl Benzonase (Novagen) for 10min. Samples were sonicated (Bioruptor, Diagenode) for 5 min (10 s sonication, 45 s break at medium intensity), centrifuged 20,000 g, 4°C. Supernatant proteins were reduced upon incubation with 10 mM DTT at 50°C for 15 min followed by 15 min incubation with 20 mM iodoacetamide in the dark. Proteins were digested overnight with trypsin (1:100 w/w enzyme to substrate ratio) at 37°C. Samples were cleaned over ZipTip (C18 material) before sequential incubation with strong cation exchange resin (SCX) and TiO2 beads to enrich multiply or mono phosphorylated peptides. The extracted peptide solutions were analyzed using nano LC-MS/MS on an LTQ Orbitrap Velos (ThermoFisher, San Jose, CA).
Recombinant proteins, reagents and antibodies
Geminin was produced as previously described.Citation64 PHA-767491 and full-length p27kip1 were procured and used as described.Citation40 Full length Xenopus Mcm10 was expressed from a pGEX-Mcm10 plasmidCitation19 (a gift of J. Walter, Harvard Medical School) and purified from Rosetta(DE3)pLysS cells (Novagen) using glutathione–Sepharose. The purified protein was dialysed against LFB1/50 buffer. To determine protein concentration, different amounts of protein alongside known amounts of BSA were resolved by SDS-PAGE and band intensities were quantitated. Mcm10 serine to alanine point mutants were prepared from pGEX-Mcm10 using QuickChange Site-Directed Mutagenesis kit from Agilent Technologies; protein was expressed and purified as described above. Antibody against PCNA was from Santa Cruz Biotechnology. Mcm3, Mcm4, Mcm7 and Orc2 antibodies were as previously described.Citation65 Cdc45, Psf2 and Polα antibodies were as previously described.Citation63 RPA34 antibody was a gift from Marcel Mechali (Institute of Human Genetics, CNRS, Montpellier, France).Citation66 The Mcm10 antibody was raised in sheep against a bacterially-expressed immunogen consisting of the previously described His6-tagged C-terminal (278–860 aa)Citation63 and His6-tagged N-terminal (1–445 aa) fragments of Xenopus Mcm10 expressed and purified from Escherichia coli (RosettaTM (DE3)pLysS, Novagen) using Ni2+-NTA affinity chromatography. Mcm10-C antibody was used for Western blotting unless stated otherwise. The antiserum was affinity purified prior to use (see Fig. S1). Antibodies were used for immunoblotting at 1:1000 dilutions. Sheep polyclonal antisera were raised against synthetic peptides corresponding to sequences of ATR (MATDPGLEMASMIPALREC), Nbs1 (CEDLFRYNPKPSKRRR), Rad17 (KIEEYDSDCKIEEYDSD), ATM (DEVDLNATLGGDDPE) and Mre11 (MSSSSSSLDDEDTFKC). Immunization and antibody purification were performed as described.Citation67 Blots of these antibodies against whole Xenopus egg extract or replicating chromatin isolated from egg extract is shown in Supplementary Figure S4.
Alkaline sucrose gradients
Alkaline sucrose gradients were performed as described.Citation52 Sperm nuclei were incubated for 120 min at 15 ng DNA/µl in 80 µl of either non-immune-depleted or Mcm10-depleted extract. The extract was then supplemented with 5 μl [α-32 P]dATP (10 mCi/ml). The pulse was stopped by the addition of 1 ml Buffer A (60 mM KCl; 15 mM Tris-HCI, pH 7.4; 15 mM NaCI,;1 mM β-mercaptoethanol; 0.5 mM spermidine: and 0.15 mM spermine) at 0°C. Nuclei were centrifuged at 2500 x g for 5 min, and then resuspended in 190 μl 0.5% SDS, 20 mM Tris- HCI (pH 7.4), and 20 mM EDTA. The DNA was denatured at 20°C by the addition of 10 μl 10 M NaOH. Samples were layered on top of a 5-20% sucrose gradient in 0.5 M NaCl, 0.25 M NaOH, 1 mM EDTA, and centrifuged at 55,000 rpm in an SW60Ti rotor at 20°C for 5 hr. Fractions were collected onto Whatman GF/C glass fiber filters previously soaked in saturated Na4P207, and 10 μg/ml calf thymus DNA. After drying, filters were TCA precipitated, washed in ethanol, dried again, and subjected to scintillation counting.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Author contributions
GSC performed all experiments; JJB managed the study; GSC and JJB designed and wrote the manuscript; AG raised the antibodies against Mcm10; PJG raised the antibodies against ATM, ATR, Mre11, Rad17 and Nbs1.
Supplemental Files
Download PDF (2 MB)Acknowledgements
We would like to thank Robert Alver for helpful comments on the manuscript.
Funding
This work was supported by CR-UK program grant C303/A14301. We also acknowledge the Dundee Proteomics Facility, supported by Wellcome Trust award 097945/B/11/Z.
References
- Thu YM, Bielinsky AK. Enigmatic roles of Mcm10 in DNA replication. Trends Biochem Sci 2013; 38:184-94; PMID:23332289; http://dx.doi.org/10.1016/j.tibs.2012.12.003.
- Merchant AM, Kawasaki Y, Chen Y, Lei M, Tye BK. A lesion in the DNA replication initiation factor Mcm10 induces pausing of elongation forks through chromosomal replication origins in Saccharomyces cerevisiae. Mol Cell Biol 1997; 17:3261-71; PMID:9154825; http://dx.doi.org/10.1128/MCB.17.6.3261.
- Solomon NA, Wright MB, Chang S, Buckley AM, Dumas LB, Gaber RF. Genetic and molecular analysis of DNA43 and DNA52: two new cell-cycle genes in Saccharomyces cerevisiae. Yeast 1992; 8:273-89; PMID:1514326; http://dx.doi.org/10.1002/yea.320080405.
- Homesley L, Lei M, Kawasaki Y, Sawyer S, Christensen T, Tye BK. Mcm10 and the MCM2-7 complex interact to initiate DNA synthesis and to release replication factors from origins. Genes Dev 2000; 14:913-26; PMID:10783164.
- Izumi M, Yatagai F, Hanaoka F. Cell cycle-dependent proteolysis and phosphorylation of human Mcm10. J Biol Chem 2001; 276:48526-31; PMID:11602595; http://dx.doi.org/10.1074/jbc.M101463200.
- Hart EA, Bryant JA, Moore K, Aves SJ. Fission yeast Cdc23 interactions with DNA replication initiation proteins. Curr Genet 2002; 41:342-8; PMID:12185500; http://dx.doi.org/10.1007/s00294-002-0316-9.
- Zhu W, Ukomadu C, Jha S, Senga T, Dhar SK, Wohlschlegel JA, Nutt LK, Kornbluth S, Dutta A. Mcm10 and And-1/CTF4 recruit DNA polymerase α to chromatin for initiation of DNA replication. Genes Dev 2007; 21:2288-99; PMID:17761813; http://dx.doi.org/10.1101/gad.1585607.
- Lee C, Liachko I, Bouten R, Kelman Z, Tye BK. Alternative mechanisms for coordinating polymerase α and MCM helicase. Mol Cell Biol 2010; 30:423-35; PMID:19917723; http://dx.doi.org/10.1128/MCB.01240-09.
- Douglas ME, Diffley JF. Recruitment of Mcm10 to Sites of Replication Initiation Requires Direct Binding to the MCM Complex. J Biol Chem 2016; 291(11):5879-88; PMID: 26719337; http://dx.doi.org/10.1074/jbc.M115.707802
- Quan Y, Xia Y, Liu L, Cui J, Li Z, Cao Q, Chen XS, Campbell JL, Lou H. Cell-Cycle-Regulated Interaction between Mcm10 and Double Hexameric Mcm2-7 Is Required for Helicase Splitting and Activation during S Phase. Cell Rep 2015; 13:2576-86; PMID:26686640; http://dx.doi.org/10.1016/j.celrep.2015.11.018.
- Christensen TW, Tye BK. Drosophila MCM10 interacts with members of the prereplication complex and is required for proper chromosome condensation. Mol Biol Cell 2003; 14:2206-15; PMID:12808023; http://dx.doi.org/10.1091/mbc.E02-11-0706.
- Sawyer SL, Cheng IH, Chai W, Tye BK. Mcm10 and Cdc45 cooperate in origin activation in Saccharomyces cerevisiae. J Mol Biol 2004; 340:195-202; PMID:15201046; http://dx.doi.org/10.1016/j.jmb.2004.04.066.
- Di Perna R, Aria V, De Falco M, Sannino V, Okorokov AL, Pisani FM, De Felice M. The physical interaction of Mcm10 with Cdc45 modulates their DNA-binding properties. Biochem J 2013; 454:333-43; PMID:23750504; http://dx.doi.org/10.1042/BJ20130059.
- Taylor M, Moore K, Murray J, Aves SJ, Price C. Mcm10 interacts with Rad4/Cut5(TopBP1) and its association with origins of DNA replication is dependent on Rad4/Cut5(TopBP1). DNA Repair (Amst) 2011; 10:1154-63; PMID:21945095; http://dx.doi.org/10.1016/j.dnarep.2011.09.001.
- Im JS, Ki SH, Farina A, Jung DS, Hurwitz J, Lee JK. Assembly of the Cdc45-Mcm2-7-GINS complex in human cells requires the Ctf4/And-1, RecQL4, and Mcm10 proteins. Proc Natl Acad Sci U S A 2009; 106:15628-32; PMID:19805216; http://dx.doi.org/10.1073/pnas.0908039106.
- Kliszczak M, Sedlackova H, Pitchai GP, Streicher WW, Krejci L, Hickson ID. Interaction of RECQ4 and MCM10 is important for efficient DNA replication origin firing in human cells. Oncotarget 2015; 6:40464-79; PMID:26588054.
- Xu X, Rochette PJ, Feyissa EA, Su TV, Liu Y. MCM10 mediates RECQ4 association with MCM2-7 helicase complex during DNA replication. EMBO J 2009; 28:3005-14; PMID:19696745; http://dx.doi.org/10.1038/emboj.2009.235.
- Lee JK, Seo YS, Hurwitz J. The Cdc23 (Mcm10) protein is required for the phosphorylation of minichromosome maintenance complex by the Dfp1-Hsk1 kinase. Proc Natl Acad Sci U S A 2003; 100:2334-9; PMID:12604790; http://dx.doi.org/10.1073/pnas.0237384100.
- Wohlschlegel JA, Dhar SK, Prokhorova TA, Dutta A, Walter JC. Xenopus Mcm10 binds to origins of DNA replication after Mcm2-7 and stimulates origin binding of Cdc45. Mol Cell 2002; 9:233-40; PMID:11864598; http://dx.doi.org/10.1016/S1097-2765(02)00456-2.
- Ricke RM, Bielinsky AK. Mcm10 regulates the stability and chromatin association of DNA polymerase-α. Mol Cell 2004; 16:173-85; PMID:15494305; http://dx.doi.org/10.1016/j.molcel.2004.09.017.
- Ricke RM, Bielinsky AK. A conserved Hsp10-like domain in Mcm10 is required to stabilize the catalytic subunit of DNA polymerase-α in budding yeast. J Biol Chem 2006; 281:18414-25; PMID:16675460; http://dx.doi.org/10.1074/jbc.M513551200.
- Chattopadhyay S, Bielinsky AK. Human Mcm10 regulates the catalytic subunit of DNA polymerase-α and prevents DNA damage during replication. Mol Biol Cell 2007; 18:4085-95; PMID:17699597; http://dx.doi.org/10.1091/mbc.E06-12-1148.
- Perez-Arnaiz P, Bruck I, Kaplan DL. Mcm10 coordinates the timely assembly and activation of the replication fork helicase. Nucleic Acids Res 2016; 44:315-29; PMID:26582917; http://dx.doi.org/10.1093/nar/gkv1260.
- Apger J, Reubens M, Henderson L, Gouge CA, Ilic N, Zhou HH, Christensen TW. Multiple functions for Drosophila Mcm10 suggested through analysis of two Mcm10 mutant alleles. Genetics 2010; 185:1151-65; PMID:20498296; http://dx.doi.org/10.1534/genetics.110.117234.
- Liachko I, Tye BK. Mcm10 mediates the interaction between DNA replication and silencing machineries. Genetics 2009; 181:379-91; PMID:19064704; http://dx.doi.org/10.1534/genetics.108.099101.
- Heller RC, Kang S, Lam WM, Chen S, Chan CS, Bell SP. Eukaryotic origin-dependent DNA replication in vitro reveals sequential action of DDK and S-CDK kinases. Cell 2011; 146:80-91; PMID:21729781; http://dx.doi.org/10.1016/j.cell.2011.06.012.
- Kanke M, Kodama Y, Takahashi TS, Nakagawa T, Masukata H. Mcm10 plays an essential role in origin DNA unwinding after loading of the CMG components. EMBO J 2012; 31:2182-94; PMID:22433840; http://dx.doi.org/10.1038/emboj.2012.68.
- van Deursen F, Sengupta S, De Piccoli G, Sanchez-Diaz A, Labib K. Mcm10 associates with the loaded DNA helicase at replication origins and defines a novel step in its activation. EMBO J 2012; 31:2195-206; PMID:22433841; http://dx.doi.org/10.1038/emboj.2012.69.
- Watase G, Takisawa H, Kanemaki MT. Mcm10 plays a role in functioning of the eukaryotic replicative DNA helicase, Cdc45-Mcm-GINS. Curr Biol 2012; 22:343-9; PMID:22285032; http://dx.doi.org/10.1016/j.cub.2012.01.023.
- Park JH, Bang SW, Jeon Y, Kang S, Hwang DS. Knockdown of human MCM10 exhibits delayed and incomplete chromosome replication. Biochem Biophys Res Commun 2008; 365:575-82; PMID:17997977; http://dx.doi.org/10.1016/j.bbrc.2007.11.003.
- Park JH, Bang SW, Kim SH, Hwang DS. Knockdown of human MCM10 activates G2 checkpoint pathway. Biochem Biophys Res Commun 2008; 365:490-5; PMID:17997981; http://dx.doi.org/10.1016/j.bbrc.2007.11.004.
- Paulsen RD, Soni DV, Wollman R, Hahn AT, Yee MC, Guan A, Hesley JA, Miller SC, Cromwell EF, Solow-Cordero DE, et al. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol Cell 2009; 35:228-39; PMID:19647519; http://dx.doi.org/10.1016/j.molcel.2009.06.021.
- Alver RC, Zhang T, Josephrajan A, Fultz BL, Hendrix CJ, Das-Bradoo S, Bielinsky AK. The N-terminus of Mcm10 is important for interaction with the 9-1-1 clamp and in resistance to DNA damage. Nucleic Acids Res 2014; 42:8389-404; PMID:24972833; http://dx.doi.org/10.1093/nar/gku479.
- Wawrousek KE, Fortini BK, Polaczek P, Chen L, Liu Q, Dunphy WG, Campbell JL. Xenopus DNA2 is a helicase/nuclease that is found in complexes with replication proteins And-1/Ctf4 and Mcm10 and DSB response proteins Nbs1 and ATM. Cell Cycle 2010; 9:1156-66; PMID:20237432; http://dx.doi.org/10.4161/cc.9.6.11049.
- Blow JJ, Watson JV. Nuclei act as independent and integrated units of replication in a Xenopus cell-free DNA replication system. EMBO J 1987; 6:1997-2002; PMID:3653079.
- Newport J. Nuclear reconstitution in vitro: stages of assembly around protein-free DNA. Cell 1987; 48:205-17; PMID:3026635; http://dx.doi.org/10.1016/0092-8674(87)90424-7.
- Blow JJ, Sleeman AM. Replication of purified DNA in Xenopus egg extract is dependent on nuclear assembly. J Cell Sci 1990; 95:383-91; PMID:2384521.
- Finlay DR, Newmeyer DD, Price TM, Forbes DJ. Inhibition of in vitro nuclear transport by a lectin that binds to nuclear pores. J Cell Biol 1987; 104:189-200; PMID:3805121; http://dx.doi.org/10.1083/jcb.104.2.189.
- Cox LS. DNA replication in cell-free extracts from Xenopus eggs is prevented by disrupting nuclear envelope function. J Cell Sci 1992; 101:43-53; PMID:1569128.
- Poh WT, Chadha GS, Gillespie PJ, Kaldis P, Blow JJ. Xenopus Cdc7 executes its essential function early in S phase and is counteracted by checkpoint-regulated protein phosphatase 1. Open Biol 2014; 4:130138; PMID:24403013; http://dx.doi.org/10.1098/rsob.130138.
- Woodward AM, Gohler T, Luciani MG, Oehlmann M, Ge X, Gartner A, Jackson DA, Blow JJ. Excess Mcm2-7 license dormant origins of replication that can be used under conditions of replicative stress. J Cell Biol 2006; 173:673-83; PMID:16754955; http://dx.doi.org/10.1083/jcb.200602108.
- Jares P, Blow JJ. Xenopus Cdc7 function is dependent on licensing but not on XORC, XCdc6, or CDK activity and is required for XCdc45 loading. Genes Dev 2000; 14:1528-40; PMID:10859170.
- Walter JC. Evidence for sequential action of cdc7 and cdk2 protein kinases during initiation of DNA replication in Xenopus egg extracts. J Biol Chem 2000; 275:39773-8; PMID:11005825; http://dx.doi.org/10.1074/jbc.M008107200.
- Du W, Josephrajan A, Adhikary S, Bowles T, Bielinsky AK, Eichman BF. Mcm10 self-association is mediated by an N-terminal coiled-coil domain. PLoS One 2013; 8:e70518; PMID:23894664; http://dx.doi.org/10.1371/journal.pone.0070518.
- Okorokov AL, Waugh A, Hodgkinson J, Murthy A, Hong HK, Leo E, Sherman MB, Stoeber K, Orlova EV, Williams GH. Hexameric ring structure of human MCM10 DNA replication factor. EMBO Rep 2007; 8:925-30; PMID:17823614; http://dx.doi.org/10.1038/sj.embor.7401064.
- Matsuno K, Kumano M, Kubota Y, Hashimoto Y, Takisawa H. The N-terminal noncatalytic region of Xenopus RecQ4 is required for chromatin binding of DNA polymerase α in the initiation of DNA replication. Mol Cell Biol 2006; 26:4843-52; PMID:16782873; http://dx.doi.org/10.1128/MCB.02267-05.
- Sangrithi MN, Bernal JA, Madine M, Philpott A, Lee J, Dunphy WG, Venkitaraman AR. Initiation of DNA replication requires the RECQL4 protein mutated in Rothmund-Thomson syndrome. Cell 2005; 121:887-98; PMID:15960976; http://dx.doi.org/10.1016/j.cell.2005.05.015.
- Sanuki Y, Kubota Y, Kanemaki MT, Takahashi TS, Mimura S, Takisawa H. RecQ4 promotes the conversion of the pre-initiation complex at a site-specific origin for DNA unwinding in Xenopus egg extracts. Cell Cycle 2015; 14:1010-23; PMID:25602506; http://dx.doi.org/10.1080/15384101.2015.1007003.
- Strausfeld UP, Howell M, Rempel R, Maller JL, Hunt T, Blow JJ. Cip1 blocks the initiation of DNA replication in Xenopus extracts by inhibition of cyclin-dependent kinases. Curr Biol 1994; 4:876-83; PMID:7850420; http://dx.doi.org/10.1016/S0960-9822(00)00196-2.
- Luciani MG, Oehlmann M, Blow JJ. Characterization of a novel ATR-dependent, Chk1-independent, intra-S-phase checkpoint that suppresses initiation of replication in Xenopus. J Cell Sci 2004; 117:6019-30; PMID:15536124; http://dx.doi.org/10.1242/jcs.01400.
- Benatti P, Belluti S, Miotto B, Neusiedler J, Dolfini D, Drac M, Basile V, Schwob E, Mantovani R, Blow JJ, et al. Direct non transcriptional role of NF-Y in DNA replication. Biochim Biophys Acta 2016; 1863:673-85; PMID:26732297; http://dx.doi.org/10.1016/j.bbamcr.2015.12.019.
- low JJ, Laskey RA. Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell 1986; 47:577-87; PMID:3779837; http://dx.doi.org/10.1016/0092-8674(86)90622-7.
- Miotto B, Chibi M, Xie P, Koundrioukoff S, Moolman-Smook H, Pugh D, Debatisse M, He F, Zhang L, Defossez PA. The RBBP6/ZBTB38/MCM10 axis regulates DNA replication and common fragile site stability. Cell Rep 2014; 7:575-87; PMID:24726359; http://dx.doi.org/10.1016/j.celrep.2014.03.030.
- Vo N, Taga A, Inaba Y, Yoshida H, Cotterill S, Yamaguchi M. Drosophila Mcm10 is required for DNA replication and differentiation in the compound eye. PLoS One 2014; 9:e93450; PMID:24686397; http://dx.doi.org/10.1371/journal.pone.0093450.
- Yeeles JT, Deegan TD, Janska A, Early A, Diffley JF. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature 2015; 519:431-5; PMID:25739503; http://dx.doi.org/10.1038/nature14285.
- Im JS, Park SY, Cho WH, Bae SH, Hurwitz J, Lee JK. RecQL4 is required for the association of Mcm10 and Ctf4 with replication origins in human cells. Cell Cycle 2015; 14:1001-9; PMID:25602958; http://dx.doi.org/10.1080/15384101.2015.1007001.
- Sonneville R, Craig G, Labib K, Gartner A, Blow JJ. Both Chromosome Decondensation and Condensation Are Dependent on DNA Replication in C. elegans Embryos. Cell Rep 2015; 12:405-17; PMID:26166571; http://dx.doi.org/10.1016/j.celrep.2015.06.046.
- Reubens MC, Biller MD, Bedsole SE, Hopkins LT, Ables ET, Christensen TW. Mcm10 is required for oogenesis and early embryogenesis in Drosophila. Mech Dev 2015; 138:291-9; PMID:26369283; http://dx.doi.org/10.1016/j.mod.2015.09.002.
- Pacek M, Tutter AV, Kubota Y, Takisawa H, Walter JC. Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol Cell 2006; 21:581-7; PMID:16483939; http://dx.doi.org/10.1016/j.molcel.2006.01.030.
- Blow JJ, Ge XQ. A model for DNA replication showing how dormant origins safeguard against replication fork failure. EMBO Rep 2009; 10:406-12; PMID:19218919; http://dx.doi.org/10.1038/embor.2009.5.
- Becker JR, Nguyen HD, Wang X, Bielinsky AK. Mcm10 deficiency causes defective-replisome-induced mutagenesis and a dependency on error-free postreplicative repair. Cell Cycle 2014; 13:1737-48; PMID:24674891; http://dx.doi.org/10.4161/cc.28652.
- Gillespie PJ, Gambus A, Blow JJ. Preparation and use of Xenopus egg extracts to study DNA replication and chromatin associated proteins. Methods 2012; 57:203-13; PMID:22521908; http://dx.doi.org/10.1016/j.ymeth.2012.03.029.
- Gambus A, Khoudoli GA, Jones RC, Blow JJ. MCM2-7 form double hexamers at licensed origins in Xenopus egg extract. J Biol Chem 2011; 286:11855-64; PMID:21282109; http://dx.doi.org/10.1074/jbc.M110.199521.
- Ferenbach A, Li A, Brito-Martins M, Blow JJ. Functional domains of the Xenopus replication licensing factor Cdt1. Nucleic Acids Res 2005; 33:316-24; PMID:15653632; http://dx.doi.org/10.1093/nar/gki176.
- Prokhorova TA, Blow JJ. Sequential MCM/P1 subcomplex assembly is required to form a heterohexamer with replication licensing activity. J Biol Chem 2000; 275:2491-8; PMID:10644704; http://dx.doi.org/10.1074/jbc.275.4.2491.
- Francon P, Lemaitre JM, Dreyer C, Maiorano D, Cuvier O, Mechali M. A hypophosphorylated form of RPA34 is a specific component of pre-replication centers. J Cell Sci 2004; 117:4909-20; PMID:15456845; http://dx.doi.org/10.1242/jcs.01361.
- Hirano T, Kobayashi R, Hirano M. Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila Barren protein. Cell 1997; 89:511-21; PMID:9160743; http://dx.doi.org/10.1016/S0092-8674(00)80233-0.
