ABSTRACT
The first hematopoietic cells are generated very early in ontogeny to support the growth of the embryo and to provide the foundation to the adult hematopoietic system. There is a considerable therapeutic interest in understanding how these first blood cells are generated in order to try to reproduce this process in vitro. This would allow generating blood products, or hematopoietic cell populations from embryonic stem (ES) cells, induced pluripotent stem cells or through directed reprogramming. Recent studies have clearly established that the first hematopoietic cells originate from a hemogenic endothelium (HE) through an endothelial to hematopoietic transition (EHT). The molecular mechanisms underlining this transition remain largely unknown with the exception that the transcription factor RUNX1 is critical for this process. In this Extra Views report, we discuss our recent studies demonstrating that the transcriptional repressors GFI1 and GFI1B have a critical role in the EHT. We established that these RUNX1 transcriptional targets are actively implicated in the downregulation of the endothelial program and the loss of endothelial identity during the formation of the first blood cells. In addition, our results suggest that GFI1 expression provides an ideal novel marker to identify, isolate and study the HE cell population.
The development of the vertebrate hematopoietic system is characterized by 3 successive waves of blood progenitor generation. The first 2 waves of blood cell emergence take place in the extra-embryonic yolk sac and generate mainly primitive erythrocytes by E7.5 and erythroid-myeloid progenitors (EMPs) by E.8.5.Citation1-6 It is only during the third wave of blood development that the first hematopoietic stem cells (HSCs) with multi-lineage and long-term repopulating potential arise in the intra-embryonic aorta-gonad-mesonephros (AGM) region.Citation7-10 Seminal experiments have indicated that blood cells initially emerge from endothelial cells (i.e. from a hemogenic endothelium).Citation11-14 Recent elegant live imaging studies of the AGM regionCitation15-19 or using differentiated embryonic stem (ES) cellsCitation20-23 have provided evidences that endothelial cells directly become blood cells through an endothelial to hematopoiesis transition (EHT). This is consistent with the observations that the first progenitors with HSC activity are detected in the major arteries in the developing embryoCitation24-26 and that the intra-aortic hematopoietic clusters (IAHC) generated through the EHT in the ventral wall of the dorsal aorta (vDA) contain cells with a HSC phenotype.Citation15 Altogether these findings suggest that HSCs directly originate from hemogenic endothelial cells. More recently, hemogenic endothelium (HE) cells have been shown to also give rise to EMPs generated in the yolk sac,Citation3 a process that is recapitulated in vitro in ES cell culture systems.Citation22,27,28 In contrast to AGM derived HE, yolk sac HE expresses the hematopoietic marker c-KIT.Citation3 Even reprogramming of fibroblasts to blood cells was shown to proceed through a HE intermediate.Citation29-31 Together, these recent results highlight the pivotal role of the HE cell population and the EHT process in the de novo generation of blood cells.
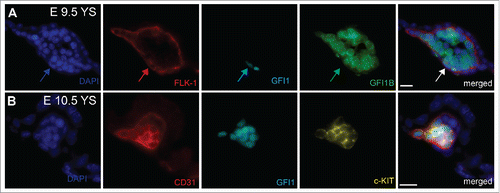
Although HE has now been clearly established as the cellular source of the first blood cells in vivo and in vitro, the molecular and cellular mechanisms orchestrating this intriguing trans-differentiation remain largely unknown. The EHT process is characterized by the loss of endothelial identity concomitant with the acquisition of a round, non-adherent, cellular morphology and the gain of hematopoietic cell surface marker expression. One important clue in understanding this process was provided by the observation that the transcription factor RUNX1 is critical for the generation of definitive blood cells by EHT.Citation17,21,32-35 In the absence of this transcription factor, HE cells do not lose their endothelial identity nor do they acquire a hematopoietic fate. Instead, the cells remain mostly adherent, associated in tight clusters and the few cells that separate from HE clusters rapidly die through apoptosis.Citation21,36 Taking advantage of this critical role of RUNX1 in the EHT process, we identified through genome-wide gene expression studies the transcriptional repressors GFI1 and GFI1B as direct transcriptional targets of RUNX1 during the EHT.Citation37 These 2 homologous nuclear zinc finger proteins share a C-terminal domain containing 6 C2-H2-type zing finger motifs mediating their DNA binding activity and a N-terminal SNAIL/GFI-1 (SNAG) domain required for their repressive activity.Citation38,39 GFI1 and GF1B have already been implicated in the adult hematopoietic system.Citation39,40 Gfi1 is expressed in HSCs, granulocyte-macrophage progenitors, B cells, granulocytes and immature T lymphocytes Citation41-44 whereas Gfi1b is highly expressed in HSCs, erythroid and megakaryocytic cells.Citation45 To investigate the relevance of these 2 proteins in the EHT, we evaluated their ability to rescue this transition in Runx1−/− HE cells. We observed that ectopic expression of Gfi1, or Gfi1b, restored many features of the EHT process. The cells expressing either GFI1 proteins acquired both a round, non-adherent morphology and the expression of early hematopoietic markers, while losing the expression of endothelial genes. However, these newly generated round cells were not able to generate hematopoietic colonies, indicating that the rescue of the EHT was incomplete. To confirm the association of GFI1 and GFI1B with the EHT in vivo, we analyzed in detail their expression in the mouse AGM region.Citation46 We observed that during the EHT process, Gfi1 is specifically expressed within the dorsal aorta in endothelial cells and cells within emerging IAHC, whereas Gfi1b expression was more associated with the fully formed IAHC. Furthermore, transplantation of the E11.5 AGM endothelial cells expressing Gfi1 and/or Gfi1b resulted in long-term repopulation of irradiated recipient mice directly demonstrating that HSC potential at E11.5 resides within the GFI1(s) expressing endothelial cell compartment. These results indicate that the expression of Gfi1 in endothelial cells readily distinguishes HE from normal, non-hemogenic endothelial cells and that GFI1 could be an important effector of RUNX1 function in the EHT process. Interestingly, we also found that in the yolk sac, Gfi1 expression was associated with FLK1+ or CD31+ endothelial cells at sites of EMPs emergence (). In contrast, GFI1B was mostly found in cells negative for endothelial markers. Gfi1 expression in yolk sac endothelium also coincided with the expression of c-KIT, a marker of hemogenic endothelial cells in the yolk sac,Citation3 but not in the AGM where its expression marks subsequent hematopoietic clusters.Citation46 The observation that GFI1 concurs with c-KIT and endothelial markers expression, and therefore potential hemogenic endothelium, suggested that GFI1 could also be critical for the extra-embryonic EHT.
Figure 1. Immunostaining on E9.5 and E10.5 Yolk sacs (A) Arrows indicate the expression of GFI1 in flat FLK-1+ endothelial cells in E9.5 yolk sac. GFI1B is detected in intravascular round cells. (B) Co-expression of GFI1 and c-KIT in CD31+ E10.5 hemogenic endothelial cells. YS = Yolk Sacs. Scale bar = 10μm.
Although these previous findings strongly suggested the importance of GFI1 and GFI1B in the EHT, none of their respective knockout recapitulated the early block in EHT and the embryonic lethality observed at E12.5 in the absence of RUNX1.Citation47,48 GFI1 deficiency is not embryonic lethal and results mainly in deafness, neutropenia and reduction in HSC self-renewal capacity,Citation41,43,44,49,50 whereas Gfi1b knockout leads to embryonic lethality at E14.5 due to a deficiency in erythroid and megakaryocyte development.Citation51 We hypothesized that the lack of an early phenotype might be due to a functional compensation for the loss of one gene by the other. The two GFI1 and GFI1B proteins exhibit very high level of homology in their functional domains and were previously shown to be functionally interchangeable in the adult hematopoietic system.Citation52 In addition, both proteins auto-regulate themselves and cross-regulate each other.Citation53-57 In line with a possible functional compensation, we observed the up-regulation of Gfi1b expression in Gfi1 deficient AGM HE cells Citation46 although Gfi1b is not normally expressed in these HE cells in wild type embryos. To therefore evaluate the functional relevance of GFI1 and GFI1B in EHT, we examined the consequences of deleting both proteins during embryonic development using Gfi1 and Gfi1b GFP knock-in mice. We first observed that deficiency in both proteins resulted in an earlier lethality than either single deficiency, further supporting the hypothesis of a functional compensation between these 2 highly homologous proteins. In the double knockout embryos, strong defects in the EHT were also observed; GFP+ blood cells normally generated from the yolk sac in heterozygous animals were absent from the circulation in the double knockout animals. Furthermore, IAHC were not observed in the AGM. Instead, we found GFP+ cells accumulating in the yolk sac or embedded within the endothelial lining of the dorsal aorta. Interestingly when these yolk sac GFP+ cells from the double knockout embryos were isolated and replated, they readily generated hematopoietic colonies. These results indicate that although the GFP+ cells were not disseminated in the circulation they had already committed to a hematopoietic cell fate. In contrast, the GFP+ endothelial cells present in the dorsal aorta did not generate any hematopoietic colonies following either direct replating or after a maturation step by co-culture on OP9 cells. These findings suggest that blood commitment can take place in absence of both GFI1 and GFI1B in the yolk sac but not in the AGM. Alternatively, committed blood cells are generated in the AGM in the absence of both GFI1 proteins, but these hematopoietic cells, such as HSCs, might be more dependent on the presence of at least one of the GFI1 protein. Supporting this hypothesis, the conditional deletion of Gfi1b in the bone marrow of adult Gfi1 deficient animals, generating double knockout cells, result in complete loss of HSCs indicating that either GFI1 or GFI1B are required to maintain HSC in vivo.Citation50,58
Although these data demonstrated the critical requirement for GFI1 and GFI1B in the EHT, the molecular mechanism associated with their function in this process still remained unknown. GFI1 and GFI1B have been shown to repress transcription in MEL (murine erythroleukemia) cell line, by recruiting the chromatin regulatory CoREST complex.Citation59,60 The CoREST complex includes the histone demethylase LSD1 (KDM1A) and the histone deacetylases, HDAC1 and HDAC2.Citation61 To investigate if this complex was involved in EHT, we examined the consequences of pharmacological LSD1 inactivation on this transition during the in vitro differentiation of ES cells. LSD1 inhibition impaired the emergence of round non-adherent cells in the supernatant of those cultures, affected the acquisition of early hematopoietic markers and perturbed the loss of endothelial markers. A similar phenotype was obtained upon the genetic deletion of Lsd1 in HE cells generated from ESCs carrying a tamoxifen-inducible conditional Lsd1 knockout.Citation62
In order to identify the genome-wide transcriptional changes induced by GFI1 and GFI1B through the recruitment of LSD1 during EHT, we compared global gene expression profiles upon LSD1 inhibition. We found that genes implicated in the development of the cardiovascular system were expressed at higher levels in LSD1-inhibited cells than in control cells. Conversely, transcripts associated with hematological system development/function and cell morphology were found at lower levels. We also mapped GFI1 and GFI1B binding sites in HE cells using the DamID (DNA adenine methyltransferase Identification) strategy. This alternative approach to chromatin immune-precipitation relies on the deposition of “methylation tags” around the binding sites of the Dam-fused transcription factor under investigation by the E.coli DNA adenine methyltransferase (Dam).Citation63,64 We cross-compared the lists of genes bound by GFI1 and/or GFI1B with the list of genes differentially expressed when LSD1 activity is blocked to identify direct transcriptional targets. The resulting list of candidates contained several genes with well-established role in stemness (Lgr5, Lin28A, Sall1), as well as genes involved in cardiovascular development, blood vessels maintenance and remodelling. Interestingly, many of these genes were also previously shown to be bound by RUNX1 during EHT, or to contain RBPJ binding sites, suggesting that RUNX1, NOTCH and GFI1(s) might be participating together in the regulation of these genes.
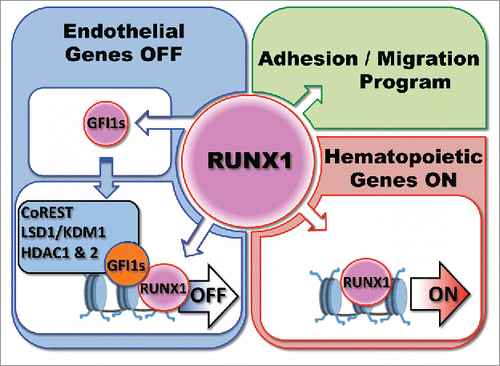
Altogether, these studies suggest a model of regulation of the EHT process where RUNX1 is first expressed in HE and induces the expression of Gfi1 and Gfi1b. These two proteins bind to genes associated with the global maintenance of endothelium identity, and cellular adhesion, and recruit the CoREST complex to epigenetically silence the endothelial program (). This leads to the acquisition of a round non-adherent cellular morphology, allowing the release of newly formed blood cells into the circulation. The precise role of NOTCH signaling in the context of RUNX1 is still unclear but is comprehensively investigated by other laboratories.Citation65-67 It is interesting to note here that GFI1, and GFI1B, have also been respectively shown to be important for the reprogramming of endothelial cells to blood cellsCitation68 or of fibroblasts to hemogenic endothelium.Citation30 We also recurrently found RUNX1 binding sites with GFI1 and GFI1B binding sites,Citation46,69 suggesting a model in which RUNX1 and GFI1(s) interact and cooperate to shut down the endothelial program. Indeed, RUNX1 and GFI1 were shown to be part of common transcriptional complexes in hematopoietic progenitorsCitation70 and we observed that these proteins can immuno-precipitate each other (unpublished data). We therefore propose that RUNX1 first binds to endothelial genes and increases the expression of GFI1(s) repressors that along with LSD1 actively induce the epigenetic silencing of the endothelial program ().
Figure 2. Model of Regulation by RUNX1 and GFI1(s) of the Endothelial to Hematopoietic Transition. The model is discussed in the text.
The observation that the expression of Gfi1, or Gfi1b, in Runx1−/− deficient HE cells induces the loss of endothelial identity but does not confer hematopoietic potential suggest that RUNX1 also activates the expression of a different set of genes required for blood commitment (). One possibility is that the acquisition of a blood cell fate proceeds through a global activation of the expression of hematopoietic genes. Indeed we found in collaboration with the Bonifer laboratory that in the absence of RUNX1, many regulatory genes such as Scl/Tal1, Fli1, Lmo2 and Cebpβ are already expressed and many hematopoietic genes are bound by C/EBPβ, SCL/TAL1 and FLI1.Citation69,71 RUNX1 expression causes a rapid shift in the binding pattern of these transcription factors toward that observed in hematopoietic precursor cells in the absence of overt precursor formation. Moreover, RUNX1 initiates the formation of new transcription factor complexes with a concomitant increase in histone acetylation at a large number of newly formed cis regulatory elements. These data suggest that the acquisition of hematopoietic cell fate might result from a global reorganization of lineage-specific transcription factor assembly controlled by RUNX1 rather than the induced expression of a few critical genes. Nonetheless the exact mechanisms by which RUNX1 promotes hematopoietic cell fate, the stage at which this is initiated, as well as the complete inventory and function of key downstream target genes, remains to be uncovered.
Intriguingly, although we previously found that Gfi1 is among the earliest target genes bound and up-regulated by RUNX1b, the isoform of RUNX1 expressed in HE, the ontology of RUNX1b regulated genes in early HE revealed an upregulation of the expression of genes involved in angiogenesis, cell adhesion and migration.Citation63,72 This suggests that at the initial stage of hematopoietic development, RUNX1b first organizes the formation of HE clusters that is required for the release of blood progenitors. Outside of the hematopoietic context, this endothelial-epithelial RUNX1 transcriptional signature might also reflect the recently uncovered role of RUNX1 in epithelial-based tumor formation and progression. In particular RUNX1 targets that associate with cell migration in HE may represent important regulators of the potential metastatic role of RUNX1 in solid tumors. It is therefore possible that RUNX1 performs 2 seemingly opposite functions in HE, first binding and activating the expression of genes involved in a cell adhesion program followed by the recruitment of GFI1(s) to silence the endothelial identity of HE.
In conclusion, our recent studies provided exciting new insights into the molecular mechanisms driving the EHT and revealed the critical roles of GFI1(s) in this process. These findings extend previous studies that have identified critical transcription factors for the upstream cell fate choice leading to the development of the HE from mesodermal/hemangioblast progenitorsCitation27,73,74 or implicated in the EHT process. These include ETS factors,Citation75-77 in particular ETV2,Citation78-82 and other transcription factors such as SOX7,Citation83-86 SCL/TAL1,Citation21,87-90 HOXA3,Citation91 SOX17,Citation92-94 GATA2 Citation95,96 and FOXF1.Citation97 The next challenge will be to determine how the combination of these transcription factors and the epigenetic machinery together dynamically orchestrate the gene regulatory networks that drive the generation of blood cells.Citation74 Along this line, our finding that LSD1 is critical for the EHT is starting to unravel how this process is regulated at the epigenetic level. In addition, it will be important to identify the components of the cellular niches that trigger and support the generation of the different types of blood cells by EHT in order to reproduce these processes in vitro to produce cell populations appropriate for clinical purposes.Citation98-103 Along this line, we recently demonstrated the relevance of a specific combination of cytokines,Citation104 heparin sulfatesCitation105,106 and graphene oxideCitation107 in supporting the development of blood cells in vitro. Altogether the Gfi1 GFP knockin model represents a powerful tool to address in the future these outstanding questions and study the EHT process in more detail as the expression of Gfi1 in the endothelium can be used to accurately identify and purify hemogenic endothelium and cells undergoing EHT. This should not only allow a better characterization of the molecular program that underpins blood cell emergence but should also lead to the identification of the specific molecular and cellular mechanisms that control the generation of different lineages during the successive waves of blood generation.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
Work in our laboratories is funded by Cancer Research UK, Biotechnology and Biological Sciences Research Council (BBSRC) and Bloodwise.
References
- Palis J, Robertson S, Kennedy M, Wall C, Keller GM. Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse. Development 1999; 126:5073-84; PMID:10529424
- McGrath KE, Frame JM, Fegan KH, Bowen JR, Conway SJ, Catherman SC, Kingsley PD, Koniski AD, Palis J. Distinct sources of hematopoietic progenitors emerge before HSCs and provide functional blood cells in the mammalian embryo. Cell Reports 2015; 11:1892-904
- Frame JM, Fegan KH, Conway SJ, McGrath KE, Palis J. Definitive hematopoiesis in the yolk sac emerges from wnt-responsive hemogenic endothelium independently of circulation and arterial identity. Stem Cells 2016; 34:431-44; PMID:26418893; http://dx.doi.org/10.1002/stem.2213
- Costa G, Kouskoff V, Lacaud G. Origin of blood cells and HSC production in the embryo. Trends Immunol 2012; 33:215-23; PMID:22365572; http://dx.doi.org/10.1016/j.it.2012.01.012
- Lux CT, Yoshimoto M, McGrath K, Conway SJ, Palis J, Yoder MC. All primitive and definitive hematopoietic progenitor cells emerging before E10 in the mouse embryo are products of the yolk sac. Blood 2008; 111:3435-8; PMID:17932251; http://dx.doi.org/10.1182/blood-2007-08-107086
- Tober J, Koniski A, Mcgrath KE, Vemishetti R, Emerson R, de Mesy-Bentley KKL, Waugh R, Palis J. The megakaryocyte lineage originates from hemangioblast precursors and is an integral component both of primitive and of definitive hematopoiesis. Blood 2007; 109:1433-41; PMID:17062726; http://dx.doi.org/10.1182/blood-2006-06-031898
- Medvinsky AJ, Samoylina NL, Müller AM, Dzierzak EA. An early pre-liver intraembryonic source of CFU-S in the developing mouse. Nature 1993; 364:64-7; PMID:8316298; http://dx.doi.org/10.1038/364064a0
- Medvinsky AJ, Dzierzak EA. Definitive hematopoiesis is autonomously initiated by the AGM region. Cell 1996; 86:897-906; PMID:8808625; http://dx.doi.org/10.1016/S0092-8674(00)80165-8
- Medvinsky AJ, Rybtsov S, Taoudi S. Embryonic origin of the adult hematopoietic system: advances and questions. Development 2011; 138:1017-31; PMID:21343360; http://dx.doi.org/10.1242/dev.040998
- Samokhvalov IM. A long way to stemness. Cell Cycle 2012; 11:2965-6; PMID:22871731; http://dx.doi.org/10.4161/cc.21388
- Nishikawa S, Nishikawa SI, Kawamoto H, Yoshida H, Kizumoto M, Kataoka H, Katsura Y. In vitro generation of lymphohematopoietic cells from endothelial cells purified from murine embryos. Immunity 1998; 8:761-9; PMID:9655490; http://dx.doi.org/10.1016/S1074-7613(00)80581-6
- Jaffredo T, Gautier R, Eichmann A, Dieterlen-Lièvre F. Intraaortic hemopoietic cells are derived from endothelial cells during ontogeny. Development 1998; 125:4575-83; PMID:9778515
- Zovein AC, Hofmann JJ, Lynch M, French WJ, Turlo KA, Yang Y, Becker MS, Zanetta L, Dejana E, Gasson JC, et al. Fate tracing reveals the endothelial origin of hematopoietic stem cells. Cell Stem Cell 2008; 3:625-36; PMID:19041779; http://dx.doi.org/10.1016/j.stem.2008.09.018
- Yokomizo T, Ogawa M, Osato M, Kanno T, Yoshida H, Fujimoto T, Fraser S, Nishikawa S, Okada H, Satake M, et al. Requirement of Runx1/AML1/PEBP2alphaB for the generation of hematopoietic cells from endothelial cells. Genes Cells 2001; 6:13-23; PMID:11168593; http://dx.doi.org/10.1046/j.1365-2443.2001.00393.x
- Boisset JC, van Cappellen W, Andrieu-Soler C, Galjart N, Dzierzak EA, Robin C. In vivo imaging of hematopoietic cells emerging from the mouse aortic endothelium. Nature 2010; 464:116-20; PMID:20154729; http://dx.doi.org/10.1038/nature08764
- Bertrand JY, Chi NC, Santoso B, Teng S, Stainier DY, Traver D. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature 2010; 464:108-11; PMID:20154733; http://dx.doi.org/10.1038/nature08738
- Kissa K, Herbomel P. Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature 2010; 464:112-5; PMID:20154732; http://dx.doi.org/10.1038/nature08761
- Lam EY, Hall CJ, Crosier PS, Crosier KE, Flores MV. Live imaging of Runx1 expression in the dorsal aorta tracks the emergence of blood progenitors from endothelial cells. Blood 2010; 116:909-14; PMID:20453160; http://dx.doi.org/10.1182/blood-2010-01-264382
- Boisset JC, Robin C. Imaging the founder of adult hematopoiesis in the mouse embryo aorta. Cell Cycle 2010; 9:2489-90; PMID:21081839; http://dx.doi.org/10.4161/cc.9.13.12319
- Eilken HM, Nishikawa S, Nishikawa SI, Schroeder T. Continuous single-cell imaging of blood generation from haemogenic endothelium. Nature 2009:457:896-900. PMID:19212410; http://dx.doi.org/ 10.1038/nature07760
- Lancrin C, Sroczynska P, Stephenson C, Allen T, Kouskoff V, Lacaud G. The haemangioblast generates hematopoietic cells through a haemogenic endothelium stage. Nature 2009; 457:892-5; PMID:19182774; http://dx.doi.org/10.1038/nature07679
- Sroczynska P, Lancrin C, Pearson S, Kouskoff V, Lacaud G. In vitro differentiation of mouse embryonic stem cells as a model of early hematopoietic development. Methods Mol Biol 2009; 538:317-34; PMID:19277585; http://dx.doi.org/10.1007/978-1-59745-418-6_16
- Pearson S, Cuvertino S, Fleury M, Lacaud G, Kouskoff V. In vivo repopulating activity emerges at the onset of hematopoietic specification during embryonic stem cell differentiation. Stem Cell Reports 2015; 4:431-44; PMID:25660408; http://dx.doi.org/10.1016/j.stemcr.2015.01.003
- de Bruijn MFTR, Speck NA, Peeters MC, Dzierzak EA. Definitive hematopoietic stem cells first develop within the major arterial regions of the mouse embryo. EMBO J 2000; 19:2465-74; PMID:10835345; http://dx.doi.org/10.1093/emboj/19.11.2465
- Gordon-Keylock S, Sobiesiak M, Rybtsov S, Moore K, Medvinsky AJ. Mouse extra-embryonic arterial vessels harbor precursors capable of maturing into definitive HSCs. Blood 2013; 122(14):2338-45; PMID:23863896; http://dx.doi.org/10.1182/blood-2012-12-470971
- Taoudi S, Medvinsky AJ. Functional identification of the hematopoietic stem cell niche in the ventral domain of the embryonic dorsal aorta. Proc Natl Acad Sci USA 2007; 104:9399-403; PMID:17517650; http://dx.doi.org/10.1073/pnas.0700984104
- Lancrin C, Sroczynska P, Serrano AG, Gandillet A, Ferreras C, Kouskoff V, Lacaud G. Blood cell generation from the hemangioblast. J Mol Med 2010; 88:167-72; PMID:19856139; http://dx.doi.org/10.1007/s00109-009-0554-0
- Keller GM. Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes Dev 2005; 19:1129-55; PMID:15905405; http://dx.doi.org/10.1101/gad.1303605
- Batta K, Florkowska M, Kouskoff V, Lacaud G. Direct reprogramming of murine fibroblasts to hematopoietic progenitor cells. CellReports 2014; 9:1871-84
- Pereira CF, Chang B, Qiu J, Niu X, Papatsenko D, Hendry CE, Clark NR, Nomura-Kitabayashi A, Kovacic JC, Ma'ayan A, et al. Induction of a hemogenic program in mouse fibroblasts. Cell Stem Cell 2013; 13:205-18; PMID:23770078; http://dx.doi.org/10.1016/j.stem.2013.05.024
- Pereira CF, Lemischka IR, Moore KA. “There will be blood” from fibroblasts. Cell Cycle 2014; 13:0-1; http://dx.doi.org/10.4161/cc.27507
- Chen MJ, Yokomizo T, Zeigler BM, Dzierzak EA, Speck NA. Runx1 is required for the endothelial to hematopoietic cell transition but not thereafter. Nature 2009; 457:887-91; PMID:19129762; http://dx.doi.org/10.1038/nature07619
- North T, Gu TL, Stacy T, Wang Q, Howard L, Binder M, Marín-Padilla M, Speck NA. Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Development 1999; 126:2563-75; PMID:10226014
- Lacaud G, Gore L, Kennedy M, Kouskoff V, Kingsley PD, Hogan C, Carlsson L, Speck NA, Palis J, Keller GM. Runx1 is essential for hematopoietic commitment at the hemangioblast stage of development in vitro. Blood 2002; 100:458-66; PMID:12091336; http://dx.doi.org/10.1182/blood-2001-12-0321
- Ichikawa M, Asai T, Chiba S, Kurokawa M, Ogawa S. Runx1/AML-1 ranks as a master regulator of adult hematopoiesis. Cell Cycle 2004; 3:722-4; PMID:15213471; http://dx.doi.org/10.4161/cc.3.6.951
- Kissa K, Herbomel P. Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature 2010; 464:112-5; PMID:20154732; http://dx.doi.org/10.1038/nature08761
- Lancrin C, Mazan M, Mazan M, Stefanska M, Patel R, Patel R, Lichtinger M, Costa G, Vargel O, Vargel O, et al. GFI1 and GFI1B control the loss of endothelial identity of hemogenic endothelium during hematopoietic commitment. Blood 2012; 120:314-22; PMID:22668850; http://dx.doi.org/10.1182/blood-2011-10-386094
- Phelan J, Shroyer N, Cook T, Gebelein B, Grimes H. Gfi1-cells and circuits: unraveling transcriptional networks of development and disease. Curr Opin Hematol 2010; 17:300; PMID:20571393; http://dx.doi.org/10.1097/MOH.0b013e32833a06f8
- van der Meer L, Jansen J, van der Reijden B. Gfi1 and Gfi1b: key regulators of hematopoiesis. Leukemia 2010; 24(11):1834-43; PMID:20861919; http://dx.doi.org/10.1038/leu.2010.195
- Duan Z, Horwitz M. Gfi-1 oncoproteins in hematopoiesis. Hematology 2003; 8:339-44; PMID:14530176; http://dx.doi.org/10.1080/10245330310001612116
- Karsunky H, Zeng H, Schmidt T, Zevnik B, Kluge R, Schmid KW, Dührsen U, Möröy T. Inflammatory reactions and severe neutropenia in mice lacking the transcriptional repressor Gfi1. Nat Genet 2002; 30:295-300; PMID:11810106; http://dx.doi.org/10.1038/ng831
- Hock H, Hamblen MJ, Rooke HM, Traver D, Bronson RT, Cameron S, Orkin S. Intrinsic requirement for zinc finger transcription factor Gfi-1 in neutrophil differentiation. Immunity 2003; 18:109-20; PMID:12530980; http://dx.doi.org/10.1016/S1074-7613(02)00501-0
- Hock H, Hamblen MJ, Rooke HM, Schindler JW, Saleque S, Fujiwara Y, Orkin S. Gfi-1 restricts proliferation and preserves functional integrity of hematopoietic stem cells. Nature 2004; 431:1002-7; PMID:15457180; http://dx.doi.org/10.1038/nature02994
- Zeng H, Yücel R, Kosan C, Klein-Hitpass L, Möröy T. Transcription factor Gfi1 regulates self-renewal and engraftment of hematopoietic stem cells. EMBO J 2004; 23:4116-25; PMID:15385956; http://dx.doi.org/10.1038/sj.emboj.7600419
- Vassen L, Okayama T, Möröy T. Gfi1b:green fluorescent protein knock-in mice reveal a dynamic expression pattern of Gfi1b during hematopoiesis that is largely complementary to Gfi1. Blood 2007; 109:2356-64; PMID:17095621; http://dx.doi.org/10.1182/blood-2006-06-030031
- Thambyrajah R, Mazan M, Mazan M, Patel R, Patel R, Moignard V, Stefanska M, Marinopoulou E, Marinopoulou E, Li Y, et al. GFI1 proteins orchestrate the emergence of hematopoietic stem cells through recruitment of LSD1. Nat Cell Biol 2016; 18:21-32; PMID:26619147; http://dx.doi.org/10.1038/ncb3276
- Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 1996; 84:321-30; PMID:8565077; http://dx.doi.org/10.1016/S0092-8674(00)80986-1
- Wang Q, Stacy T, Miller JD, Lewis AF, Gu TL, Huang X, Bushweller JH, Bories JC, Alt FW, Ryan G, et al. The CBFbeta subunit is essential for CBFalpha2 (AML1) function in vivo. Cell 1996; 87:697-708; PMID:8929538; http://dx.doi.org/10.1016/S0092-8674(00)81389-6
- Kazanjian A. Growth factor Independence-1 is expressed in primary human neuroendocrine lung carcinomas and mediates the differentiation of murine pulmonary neuroendocrine cells. Cancer Res 2004; 64:6874-82; PMID:15466176; http://dx.doi.org/10.1158/0008-5472.CAN-04-0633
- Khandanpour C, Kosan C, Gaudreau M-C, Dührsen U, Hébert J, Zeng H, Möröy T. Growth factor independence 1 protects hematopoietic stem cells against apoptosis but also prevents the development of a myeloproliferative-like disease. Stem Cells 2011; 29:376-85; PMID:21732494; http://dx.doi.org/10.1002/stem.575
- Saleque S, Orkin S, Cameron S. The zinc-finger proto-oncogene Gfi-1b is essential for development of the erythroid and megakaryocytic lineages. Genes Dev 2002; 16:301-6; PMID:11825872; http://dx.doi.org/10.1101/gad.959102
- Fiolka K, Hertzano R, Vassen L, Zeng H, Hermesh O, Avraham KB, Dührsen U, Möröy T. Gfi1 and Gfi1b act equivalently in haematopoiesis, but have distinct, non-overlapping functions in inner ear development. EMBO Rep 2006; 7:326-33; PMID:16397623; http://dx.doi.org/10.1038/sj.embor.7400618
- Vassen L, Fiolka K, Mahlmann S, Möröy T. Direct transcriptional repression of the genes encoding the zinc-finger proteins Gfi1b and Gfi1 by Gfi1b. Nucleic Acids Res 2005; 33:987-98; PMID:15718298; http://dx.doi.org/10.1093/nar/gki243
- Doan LL, Porter SD, Duan Z, Flubacher MM, Montoya D, Tsichlis PN, Horwitz M, Gilks CB, Grimes HL. Targeted transcriptional repression of Gfi1 by GFI1 and GFI1B in lymphoid cells. Nucleic Acids Res 2004; 32:2508-19; PMID:15131254; http://dx.doi.org/10.1093/nar/gkh570
- Yücel R, Kosan C, Heyd F, Möröy T. Gfi1:green fluorescent protein knock-in mutant reveals differential expression and autoregulation of the growth factor independence 1 (Gfi1) gene during lymphocyte development. J Biol Chem 2004; 279:40906-17; PMID:15252036; http://dx.doi.org/10.1074/jbc.M400808200
- Anguita E, Villegas A, Iborra F, Hernandez A. GFI1B controls its own expression binding to multiple sites. Haematologica 2010; 95:36; PMID:19773260; http://dx.doi.org/10.3324/haematol.2009.012351
- Huang D-Y, Kuo Y-Y, Chang Z-F. GATA-1 mediates auto-regulation of Gfi-1B transcription in K562 cells. Nucleic Acids Res 2005; 33:5331-42; PMID:16177182; http://dx.doi.org/10.1093/nar/gki838
- Khandanpour C, Sharif-Askari E, Vassen L, Vassen L. Evidence that Growth factor independence 1b (Gfi1b) regulates dormancy and peripheral blood mobilization of hematopoietic stem cells. Blood 2010; 116(24):5149-61; PMID:20826720; http://dx.doi.org/10.1182/blood-2010-04-280305
- Saleque S, Orkin S, Kim J, Rooke HM. Epigenetic regulation of hematopoietic differentiation by Gfi-1 and Gfi-1b is mediated by the cofactors CoREST and LSD1. Molecular Cell 2007; 27:562-72; PMID:17707228; http://dx.doi.org/10.1016/j.molcel.2007.06.039
- Chowdhury AH, Ramroop JR, Upadhyay G, Sengupta A, Andrzejczyk A, Saleque S. Differential Transcriptional Regulation of meis1 by Gfi1b and Its Co-Factors LSD1 and CoREST. PLoS One 2013; 8:e53666; PMID:23308270; http://dx.doi.org/10.1371/journal.pone.0053666
- Kelly RD, Cowley SM. The physiological roles of histone deacetylase (HDAC) 1 and 2: complex co-stars with multiple leading parts. Biochem Soc Trans 2013; 41:741-9; PMID:23697933; http://dx.doi.org/10.1042/BST20130010
- Foster CT, Dovey OM, Lezina L, Luo JL, Gant TW, Barlev N, Bradley A, Cowley SM. Lysine-specific demethylase 1 regulates the embryonic transcriptome and CoREST stability. Mol Cell Biol 2010; 30:4851-63; PMID:20713442; http://dx.doi.org/10.1128/MCB.00521-10
- Lie-A-Ling M, Marinopoulou E, Li Y, Patel R, Stefanska M, Bonifer C, Miller C, Kouskoff V, Lacaud G. RUNX1 positively regulates a cell adhesion and migration program in murine hemogenic endothelium prior to blood emergence. Blood 2014; 124(11):e11-20; PMID:25082880; http://dx.doi.org/10.1182/blood-2014-04-572958
- van Steensel B, Henikoff S. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat Biotech 2000; 18:424-8; ; http://dx.doi.org/10.1038/74487
- Bigas A, Espinosa L. Hematopoietic stem cells: to be or Notch to be. Blood 2012; PMID:22308291
- Kanz D, Konantz M, Alghisi E, North TE, Lengerke C. Endothelial-to-hematopoietic transition: Notch-ing vessels into blood. Annals N York Acad Sci 2016; 1370(1):97-108; PMID:27015586; http://dx.doi.org/10.1111/nyas.13030
- Gering M, Patient R. Notch signalling and hematopoietic stem cell formation during embryogenesis. J Cell Physiol 2010; 222:11-6; PMID:19725072; http://dx.doi.org/10.1002/jcp.21905
- Sandler VM, Sandler VM, Sandler VM, Lis R, Liu Y, Kedem A, James D, Elemento O, Butler JM, Scandura JM, et al. Reprogramming human endothelial cells to hematopoietic cells requires vascular induction. Nature 2014; 511:312-8; PMID:25030167; http://dx.doi.org/10.1038/nature13547
- Lichtinger M, Ingram R, Hannah R, Muller D, Clarke D, Assi SA, Lie-A-Ling M, Noailles L, Vijayabaskar MS, Wu M, et al. RUNX1 reshapes the epigenetic landscape at the onset of haematopoiesis. EMBO J 2012; 31(22):4318-33: 1-16; PMID:23064151; http://dx.doi.org/10.1038/emboj.2012.275
- Wilson NK, Foster SD, Wang X, Knezevic K, Schütte J, Kaimakis P, Chilarska PM, Kinston S, Ouwehand WH, Dzierzak EA, et al. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell 2010; 7:532-44; PMID:20887958; http://dx.doi.org/10.1016/j.stem.2010.07.016
- Hoogenkamp M, Lichtinger M, Krysinska H, Lancrin C, Clarke D, Williamson A, Mazzarella L, Ingram R, Jorgensen H, Fisher AG, et al. Early chromatin unfolding by RUNX1: a molecular explanation for differential requirements during specification versus maintenance of the hematopoietic gene expression program. Blood 2009; 114:299-309; PMID:19339695; http://dx.doi.org/10.1182/blood-2008-11-191890
- Swiers G, Baumann C, O'Rourke J, O'Rourke J, Giannoulatou E, Taylor S, Joshi A, Moignard V, Pina C, Bee T, et al. Early dynamic fate changes in haemogenic endothelium characterized at the single-cell level. Nature Communications 2013; 4:2924; PMID:24326267; http://dx.doi.org/10.1038/ncomms3924
- Stefanska M, Costa G, Lie-A-Ling M, Kouskoff V, Lacaud G. Smooth muscle cells largely develop independently of functional hemogenic endothelium. Stem Cell Res 2014; 12:222-32; PMID:24270161; http://dx.doi.org/10.1016/j.scr.2013.10.009
- Goode DK, Obier N, Vijayabaskar MS, Lie-A-Ling M, Lilly AJ, Hannah R, Lichtinger M, Batta K, Florkowska M, Patel R, et al. Dynamic gene regulatory networks drive hematopoietic specification and differentiation. Dev Cell 2016; 36:572-87; PMID:26923725; http://dx.doi.org/10.1016/j.devcel.2016.01.024
- Ciau-Uitz A, Wang L, Patient R, Liu F. ETS transcription factors in hematopoietic stem cell development. Blood Cells Mol Dis 2013; 51:248-55; PMID:23927967; http://dx.doi.org/10.1016/j.bcmd.2013.07.010
- Liu F, Walmsley M, Rodaway A, Patient R. Fli1 acts at the top of the transcriptional network driving blood and endothelial development. Curr Biol 2008; 18:1234-40; PMID:18718762; http://dx.doi.org/10.1016/j.cub.2008.07.048
- Liu F, Patient R. Genome-wide analysis of the zebrafish ETS family identifies three genes required for hemangioblast differentiation or angiogenesis. Circulation Res 2008; 103:1147-54; PMID:18832752; http://dx.doi.org/10.1161/CIRCRESAHA.108.179713
- Wareing S, Mazan A, Pearson S, Göttgens B, Lacaud G, Kouskoff V. The Flk1-Cre-mediated deletion of ETV2 defines its narrow temporal requirement during embryonic hematopoietic development. Stem Cells 2012; 30:1521-31; PMID:22570122; http://dx.doi.org/10.1002/stem.1115
- Kataoka H, Hayashi M, Nakagawa R, Tanaka Y, Izumi N, Nishikawa S, Jakt ML, Tarui H, Nishikawa S. Etv2/ER71 induces vascular mesoderm from Flk1+PDGFRα+ primitive mesoderm. Blood 2011; 118:6975-86; PMID:21911838; http://dx.doi.org/10.1182/blood-2011-05-352658
- Lee D, Park C, Lee H, Lugus JJ, Kim SH, Arentson E, Chung YS, Gomez G, Kyba M, Lin S, et al. ER71 acts downstream of BMP, Notch, and Wnt signaling in blood and vessel progenitor specification. Cell Stem Cell 2008; 2:497-507; PMID:18462699; http://dx.doi.org/10.1016/j.stem.2008.03.008
- Liu F, Kang I, Park C, Chang LW, Wang W, Lee D, Lim DS, Vittet D, Nerbonne JM, Choi K. ER71 specifies Flk-1+ hemangiogenic mesoderm by inhibiting cardiac mesoderm and Wnt signaling. Blood 2012; 119(14):3295-305; PMID:22343916; http://dx.doi.org/10.1182/blood-2012-01-403766
- Wareing S, Eliades A, Lacaud G, Kouskoff V. ETV2 expression marks blood and endothelium precursors, including hemogenic endothelium, at the onset of blood development. Dev Dyn 2012; 241:1454-64; PMID:22733530; http://dx.doi.org/10.1002/dvdy.23825
- Costa G, Mazan A, Gandillet A, Pearson S, Lacaud G, Kouskoff V. SOX7 regulates the expression of VE-cadherin in the haemogenic endothelium at the onset of hematopoietic development. Development 2012; 139:1587-98; PMID:22492353; http://dx.doi.org/10.1242/dev.071282
- Moignard V, Woodhouse S, Haghverdi L, Lilly AJ, Tanaka Y, Wilkinson AC, Buettner F, Macaulay IC, Jawaid W, Diamanti E, et al. Decoding the regulatory network of early blood development from single-cell gene expression measurements. Nat Biotech 2015; 33:269-76; http://dx.doi.org/10.1038/nbt.3154
- Serrano AG, Gandillet A, Pearson S, Lacaud G, Kouskoff V. Contrasting effects of Sox17- and Sox18-sustained expression at the onset of blood specification. Blood 2010; 115:3895-8; PMID:20228271; http://dx.doi.org/10.1182/blood-2009-10-247395
- Behrens AN, Zierold C, Shi X, Ren Y, Koyano-Nakagawa N, Garry DJ, Martin CM. Sox7 is regulated by ETV2 during cardiovascular development. Stem Cells Dev 2014; 23:2004-13; PMID:24762086; http://dx.doi.org/10.1089/scd.2013.0525
- Patterson LJ, Gering M, Eckfeldt CE, Green AR, Verfaillie CM, Ekker SC, Patient R. The transcription factors Scl and Lmo2 act together during development of the hemangioblast in zebrafish. Blood 2007; 109:2389-98; PMID:17090656; http://dx.doi.org/10.1182/blood-2006-02-003087
- Patterson LJ, Gering M, Patient R. Scl is required for dorsal aorta as well as blood formation in zebrafish embryos. Blood 2005; 105:3502-11; PMID:15644413; http://dx.doi.org/10.1182/blood-2004-09-3547
- Gering M. Lmo2 and Scl/Tal1 convert non-axial mesoderm into haemangioblasts which differentiate into endothelial cells in the absence of Gata1. Development 2003; 130:6187-99; PMID:14602685; http://dx.doi.org/10.1242/dev.00875
- van Handel B, Montel-Hagen A, Sasidharan R, Nakano H, Ferrari R, Boogerd CJ, Schredelseker J, Wang Y, Hunter S, Org T, et al. Scl represses cardiomyogenesis in prospective hemogenic endothelium and endocardium. Cell 2012; 150:590-605; PMID:22863011; http://dx.doi.org/10.1016/j.cell.2012.06.026
- Iacovino M, Chong D, Szatmari I, Hartweck L, Rux D, Caprioli A, Cleaver O, Kyba M. HoxA3 is an apical regulator of haemogenic endothelium. Nat Cell Biol 2010; 13:72-8; PMID:21170035; http://dx.doi.org/10.1038/ncb2137
- Nakajima-Takagi Y, Osawa M, Oshima M, Takagi H, Miyagi S, Endoh M, Endo TA, Takayama N, Eto K, Toyoda T, et al. Role of SOX17 in hematopoietic development from human embryonic stem cells. Blood 2013; 121:447-58. PMID:23169777; http://dx.doi.org/10.1182/blood-2012-05-431403
- Clarke RL, Yzaguirre AD, Yashiro-Ohtani Y, Bondue A, Blanpain C, Pear WS, Speck NA, Keller GM. The expression of Sox17 identifies and regulates haemogenic endothelium. Nat Cell Biol 2013; 15:1-10; PMID:23263367; http://dx.doi.org/10.1038/ncb2724
- Nobuhisa I, Osawa M, Uemura M, Kishikawa Y, Anani M, Harada K, Takagi H, Saito K, Kanai-Azuma M, Kanai Y, et al. Sox17-mediated maintenance of fetal intra-aortic hematopoietic cell clusters. Mol Cell Biol 2014; 34:1976-90; PMID:24662049; http://dx.doi.org/10.1128/MCB.01485-13
- Ling KW, Ottersbach K, van Hamburg JP, Oziemlak A, Tsai FY, Orkin S, Ploemacher R, Hendriks RW, Dzierzak EA. GATA-2 plays two functionally distinct roles during the ontogeny of hematopoietic stem cells. J Exp Med 2004; 200:871-82; PMID:15466621; http://dx.doi.org/10.1084/jem.20031556
- de Pater E, Kaimakis P, Vink CS, Yokomizo T, Yamada-Inagawa T, van der Linden R, Kartalaei PS, Camper SA, Speck NA, Dzierzak EA. Gata2 is required for HSC generation and survival. J Exp Med 2013; 464:116
- Fleury M, Eliades A, Carlsson P, Lacaud G, Kouskoff V. FOXF1 inhibits hematopoietic lineage commitment during early mesoderm specification. Development 2015; 142:3307-20; PMID:26293303; http://dx.doi.org/10.1242/dev.124685
- Sugimura R. Bioengineering Hematopoietic Stem Cell Niche toward Regenerative Medicine. Adv Drug Deliv Rev 2016; 99:212–20. PMID:26527127; http://dx.doi.org/10.1016/j.addr.2015.10.010
- Charbord P, Pouget C, Binder H, Dumont F, Stik G, Levy P, Allain F, Marchal C, Richter J, Uzan B, et al. A systems biology approach for defining the molecular framework of the hematopoietic stem cell niche. Cell Stem Cell 2014; 15:376-91; PMID:25042701; http://dx.doi.org/10.1016/j.stem.2014.06.005
- Wagers AJ. The stem cell niche in regenerative medicine. Cell Stem Cell 2012; 10:362-9; PMID:22482502; http://dx.doi.org/10.1016/j.stem.2012.02.018
- Dzierzak EA, Speck NA. Of lineage and legacy: the development of mammalian hematopoietic stem cells. Nat Immunol 2008; 9:129-36; PMID:18204427; http://dx.doi.org/10.1038/ni1560
- Robin C, Ottersbach K, Durand C, Durand C, Peeters M, Vanes L, Tybulewicz V, Dzierzak EA. An unexpected role for IL-3 in the embryonic development of hematopoietic stem cells. Dev Cell 2006; 11:171-80; PMID:16890157; http://dx.doi.org/10.1016/j.devcel.2006.07.002
- Fitch SR, Kimber GM, Wilson N, Parker A, Mirshekar-Syahkal B, Göttgens B, Medvinsky AJ, Dzierzak EA, Ottersbach K. Signaling from the Sympathetic Nervous System Regulates Hematopoietic Stem Cell Emergence during Embryogenesis. Stem Cell 2012; 11:554-66
- Pearson S, Sroczynska P, Lacaud G, Kouskoff V. The stepwise specification of embryonic stem cells to hematopoietic fate is driven by sequential exposure to Bmp4, activin A, bFGF and VEGF. Development 2008; 135:1525-35; PMID:18339678; http://dx.doi.org/10.1242/dev.011767
- Baldwin RJ, ten Dam GB, van Kuppevelt THxs, Lacaud G, Gallagher JT, Kouskoff V, Merry CL. A developmentally regulated heparan sulfate epitope defines a subpopulation with increased blood potential during mesodermal differentiation. Stem Cells 2008; 26:3108-18; PMID:18787209; http://dx.doi.org/10.1634/stemcells.2008-0311
- Holley RJ, Pickford CE, Rushton G, Lacaud G, Gallagher JT, Kouskoff V, Merry CL. Influencing hematopoietic differentiation of mouse embryonic stem cells using soluble heparin and heparan sulfate saccharides. J Biol Chem 2011; 286:6241-52; PMID:21148566; http://dx.doi.org/10.1074/jbc.M110.178483
- Garcia-Alegria E, Iluit M, Stefanska M, Silva C, Heeg S, Kimber SJ, Kouskoff V, Lacaud G, Vijayaraghavan A, Batta K. Graphene oxide promotes embryonic stem cell differentiation to hematopoietic lineage. Sci Rep 2016; 6:25917-12; PMID:27197878; http://dx.doi.org/10.1038/srep25917
