The cohesin complex is a multi-protein ring-like structure responsible for holding together newly replicated sister chromatids (sister chromatid cohesion (SCC)) both in mitotic and in meiotic cells.Citation1 In addition, the cohesion complex constitutes a platform for the formation of a chromosome axis, or axial element (AE), along which replicated meiotic DNA is organized. During meiotic prophase, via a process termed chromosome synapsis, the AEs of each homolog become progressively tethered by the synaptonemal complex (SC). Completion of DNA recombination occurs in the context of synapsed homologues, and is essential for the formation of meiotic crossovers which ensure not only genomic variability but also correct chromosome segregation at the onset of anaphase I.Citation2 In the absence of synapsis or upon illegitimate synapsis between sister chromatids (inter-sister synapsis), meiotic progression and animal fertility are highly compromised.
Several meiosis-specific subunits of the cohesion complex have been identified, including SMC1β, α-kleisins REC8 and RAD21L and stromal antigen protein SA3/STAG3. We have previously shownCitation3 that akin to Rec8 KO mice,Citation4,5 mice with reduced expression of Stag3 and subsequent reduction of REC8 levels at chromosome axes, display inter-sister synapsis, suggesting the existence of a cohesin-based regulatory mechanism that favors SC formation between homologous chromosomes. To better understand the contribution of REC8 to SCC and SC formation in germ cells, we employed super-resolution microscopy in order to visualize individual sister chromatid axes (sister-AEs), and with this method we found that reduced levels of REC8 in Stag3 mutant meiotic cells, resulted in sister-AEs that displayed local regions of axis separation (“axial openings”), allowing SC formation at these sites.Citation6 These regions were decorated by RAD21L and RAD21 (an α-kleisin expressed in both mitotic and meiotic cells), but flanked solely by REC8, strongly suggesting that REC8 localization defines the borders of local axis separation, and thus maintains the close association between sister-AEs. In addition, we show that inter-axis distances between the sister-AEs of wild-type sex chromosomes were significantly narrower than at axial openings in Stag3 mutant cells. Accordingly, sex chromosomes displayed a high density of REC8 foci and rarely inter-sister synapsis.
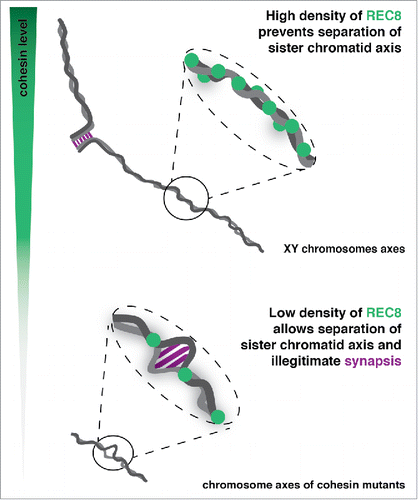
In order to gain insight into how REC8 prevents inter-sister synapsis, we performed a quantitative analysis of REC8 distribution along wild-type sex chromosomes.Citation6 REC8 did not show preference for binding at specific regions along X chromosomes, being detected randomly along the sister-AEs. Interestingly, nearly all REC8 foci were separated by distances smaller than 15% of a chromosome axis length, and this particular deposition of REC8 along sister-AEs was not found to rely on an active mechanism, such as interference. Instead, a high density of randomly placed REC8 foci along a given chromosome axis is sufficient to ensure that most foci are distributed along short intervals, as shown by an overlap of the median inter-REC8 distance obtained along sister-AEs of X chromosomes with the computationally generated distribution of randomly distributed foci. We further tested this result by analyzing the distribution of REC8 along autosomes. As expected by the absence of inter-sister synapsis in autosomes, nearly all inter-REC8 distances were smaller than 15% of a chromosome axis length. In contrast, the remaining REC8 foci detected in Stag3 mutant cells were separated by a significantly larger median distance of chromosome axis length (28%), thus allowing the occurrence of axial openings. In summary, we propose a model where the deposition of a high density of randomly distributed REC8 cohesin complexes at intervals of less than 15% of the chromosome axis length, is sufficient to maintain the close association between sister-AEs and to prevent illegitimate inter-sister synapsis ().
In human oocytes an age-related increase in aneuploidy (chromosome missegregation) has been shown to negatively correlate with the levels of meiosis-specific cohesins,Citation7 prompting the suggestion of the existence of a cohesin threshold level required to maintain meiotic SCC and thus counteract physical tensions during segregation. One could speculate that in human oocytes, during the long period of developmental arrest at the dictyate stage of meiosis I, the occurrence of local separation of sister-AEs along the chromosome arms or close to centromeres due to an increased spacing between REC8 complexes, in combination with the physical tensions applied to bivalent chromosomes, could lead to premature sister chromatid segregation at anaphase I onset.
Figure 1. (Top panel) A high density of REC8 (green) present along sister chromatid axes (gray) in wild-type cells ensures their close association, preventing illegitimate inter-sister synapsis. (Lower panel) In cohesin mutants the absence of a high density of REC8 leads to appearance of axial openings, and illegitimate inter-sister synapsis formation at these sites.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- Mehta GD, Rizvi SM, Ghosh SK. Cohesin: a guardian of genome integrity. Biochim Biophys Acta 2012; 1823:1324-42; PMID:22677545; http://dx.doi.org/10.1016/j.bbamcr.2012.05.027
- Zickler D, Kleckner N. Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harbor Perspectives Biol 2015; 7:a016626; http://dx.doi.org/10.1101/cshperspect.a016626
- Fukuda T, Fukuda N, Agostinho A, Hernández-Hernández A, Kouznetsova A, Höög C. STAG3-mediated stabilization of REC8 cohesin complexes promotes chromosome synapsis during meiosis. EMBO J 2014; 33:1243-55; PMID:24797475; http://dx.doi.org/10.1002/embj.201387329
- Bannister LA, Reinholdt LG, Munroe RJ, Schimenti JC. Positional cloning and characterization of mouse mei8, a disrupted allelle of the meiotic cohesin Rec8. Genesis 2004; 40:184-94; PMID:15515002; http://dx.doi.org/10.1002/gene.20085
- Xu H, Beasley MD, Warren WD, van der Horst GT, McKay MJ. Absence of mouse REC8 cohesin promotes synapsis of sister chromatids in meiosis. Dev Cell 2005; 8:949-61; PMID:15935783; http://dx.doi.org/10.1016/j.devcel.2005.03.018
- Agostinho A, Manneberg O, van Schendel R,Hernández-Hernández A, Kouznetsova A, Blom H, Brismar H, Höög C. High density of REC8 constrains sister chromatid axes and prevents illegitimate synaptonemal complex formation. EMBO Rep 2016; 17(6):901-13; PMID:27170622; http://dx.doi.org/10.15252/embr.201642030
- Tsutsumi M, Fujiwara R, Nishizawa H, Ito M, Kogo H, Inagaki H, Ohye T, Kato T, Fujii T, Kurahashi H. Age-related decrease of meiotic cohesins in human oocytes. PLoS One 2014; 9:e96710; PMID:24806359; http://dx.doi.org/10.1371/journal.pone.0096710
