KEYWORDS:
Repetitive DNA constitutes a large part of most vertebrate genomes. However, it is unclear how DNA containing repetitive sequences is organized in the context of chromatin and nuclear structures, how it is duplicated and how it responds to replication stress. Some key chromosome regions like the centromere are particularly enriched in highly repetitive DNA sequences known as alpha, minor and major satellites. Repetitive DNA sequences can easily misalign during DNA synthesis, likely requiring specific mechanisms to ensure accurate DNA replication to avoid events leading to copy number variation and structural rearrangement. The chromatin structure underlying regions containing repetitive sequences such as the centromeres might also alter replication fork progression due to its compact nature. It was unclear so far how replication fidelity and smooth progression of replication forks was ensured on these templates.
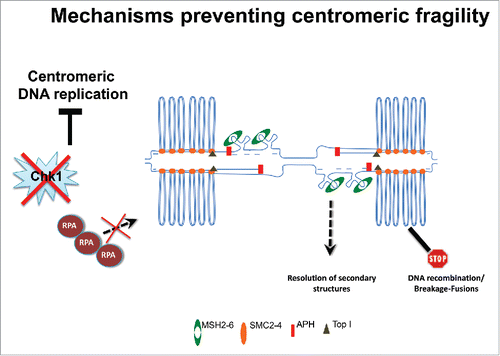
Using Xenopus laevis egg extract Aze et alCitation1 successfully reconstituted the replication of repetitive human centromeric DNA, unveiling unexpected structural and regulatory features. To this end bacterial artificial chromosomes (BACs) bearing centromeric alpha satellite DNA sequences of different human chromosomes were used. Large bacmids were useful for their ability to induce efficient formation of nuclear structures, a pre-requisite for semiconservative DNA replication in these extracts. Formation of nuclei also ensured that DNA and chromatin reached a conformation similar to the one found in intact cell nuclei. Centromeric DNA was efficiently replicated and replication efficiency was comparable to bacmid containing non-repetitive sequences, although the replication kinetic was slightly slower. The high replication efficiency of the centromeric DNA was surprising given the intrinsic complexity of the repetitive DNA, which was expected to create strong impairments to the progression of the replication machinery. However, mass spectrometry analysis of the proteome associated with centromeric DNA revealed the enrichment of several DNA repair and DNA structural proteins, some of which were shown to be required for efficient replication of centromeric templates. Among these factors the MSH2/MSH6 complex, which is involved in mismatch repair,Citation2 the Mre11 and Mus81 complexes, Topoisomerases, CENP proteins and the condensin components SMC2/SMC4 were found. In contrast, other common DNA metabolism proteins were under-represented, such as the single-stranded DNA (ssDNA) binding complex RPA and TopBP1. The enrichment of DNA repair proteins, whose binding was dependent upon DNA replication, indicated that centromeric DNA replication was under some sort of stress. This hypothesis was directly tested by monitoring the activation of the ATR dependent checkpoint pathway, which is activated by replication stress and requires RPA for its activation.Citation3 Surprisingly, ATR activation, detected by phosphorylation of its downstream target CHK1 was suppressed on centromeric templates even in the presence of aphidicolin, which induces polymerase stalling. This was due to reduced accumulation of RPA on centromeric DNA compared to normal templates. On the other hand, aphidicholin stimulated the loading of MSH6, which was required for efficient replication of centromeric templates, suggesting the presence of alternative replication programs requiring the presence of accessory DNA repair proteins for unimpeded fork progression. Strikingly, further analyses performed using Electron Microscopy revealed that centromeric DNA accumulated positively supercoiled DNA loops upon replication. The existence of DNA loops at centromeres had been previously hypothesized and suggested to be responsible for some of the biological and biophysical properties of centromeric chromatin.Citation4 This particular arrangement might be related to the topological structures formed in vitro by condensins in the presence of active Topoismerases.Citation5 Consistent with this model, inhibition of Topoisomerase I activity prevented the formation of these loops. Significantly, interference with this arrangement also restored RPA accumulation and ATR activation under stress. Formation of loops during centromeric DNA replication might restrict RPA binding and ATR checkpoint activation to allow replication to proceed across these repetitive sequences (). This could avoid hyper-loading of RPA at repetitive DNA and continuous activation of the checkpoint at natural stalling sites. It is not clear whether other regions of the genome behave in a similar fashion. However, it is worth mentioning that telomeric repeats also actively suppress checkpoint activation, form positively supercoiled DNA around TRF2 proteinsCitation6 and are organized in t-loops. Localized suppression of the checkpoint may suggest that repetitive sequences are replicated with lower fidelity and that alternative mechanisms are needed to promote genome stability. The loading of DNA repair factors such as the MSH2/MSH6 complex might ensure this function by helping to resolve secondary structures, whose presence might slow down replication fork, avoiding potential misalignments between DNA repeats. The enrichment of condensins at centromeric regions could indicate early condensation processes taking place at centromere in S-phase and then spreading to other regions of the chromosome in mitosis. Condensation can occur in the context of DNA replication in bacterial organisms, where it serves to promote DNA segregation and to prevent misalignment of DNA templates.Citation7 Additional studies in other organisms will show whether these regulatory and structural patterns are conserved in other species. It will be particularly important to understand whether these mechanisms ensure stability of large centromeric DNA repeats, which often undergo aberrant breakage-fusion events in cancer and other diseases in which DNA repair is deficient.
Figure 1. Structural and regulatory features of centromeric DNA. DNA loops form on centromeric chromatin in the presence of condensins and topoisomerase I. Centromeric repeats might form secondary structures on unwound ssDNA at replication forks, attracting MSH2 /6 proteins, which might be required to resolve them. This topological arrangement limits the formation of extensive ssDNA regions and RPA hyper-loading preventing activation of ATR, facilitating centromeric DNA replication.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- Aze A, Sannino V, Soffientini P, Bachi A, Costanzo V. Centromeric DNA replication reconstitution reveals DNA loops and ATR checkpoint suppression. Nat Cell Biol 2016; 18:684-91; PMID:27111843; http://dx.doi.org/10.1038/ncb3344
- Zhao XN, Usdin K. The repeat expansion diseases: The dark side of DNA repair. DNA Repair 2015; 32:96-105; PMID:26002199; http://dx.doi.org/10.1016/j.dnarep.2015.04.019
- Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev 2005; 19:1040-52; PMID:15833913; http://dx.doi.org/10.1101/gad.1301205
- Stephens AD, Haase J, Vicci L, Taylor RM, 2nd, Bloom K. Cohesin, condensin, and the intramolecular centromere loop together generate the mitotic chromatin spring. J Cell Biol 2011; 193:1167-80; PMID:21708976; http://dx.doi.org/10.1083/jcb.201103138
- Hirano T. Condensin-based chromosome organization from bacteria to vertebrates. Cell 2016; 164:847-57; PMID:26919425; http://dx.doi.org/10.1016/j.cell.2016.01.033
- Benarroch-Popivker D, Pisano S, Mendez-Bermudez A, Lototska L, Kaur P, Bauwens S, Djerbi N, Latrick CM, Fraisier V, Pei B, et al. TRF2-mediated control of telomere DNA topology as a mechanism for chromosome-end protection. Mol Cell 2016; 61:274-86; PMID:26774283; http://dx.doi.org/10.1016/j.molcel.2015.12.009
- Wang X, Le TB, Lajoie BR, Dekker J, Laub MT, Rudner DZ. Condensin promotes the juxtaposition of DNA flanking its loading site in Bacillus subtilis. Genes Dev 2015; 29:1661-75; PMID:26253537; http://dx.doi.org/10.1101/gad.265876.115
