First postulated by Theodor Boveri in 1902, abnormalities in chromatin structure and function are well-known hallmarks of cancer. Indeed, aberrant nuclear structure is the key feature that distinguishes a cancer cell from a normal cell histologically. The mechanisms that govern nuclear structure, however, are only beginning to emerge. The recent paper by Pierantoni et al. from Italy begins to shed light on this process by providing further evidence for high mobility group A1 (HMGA1) proteins in regulating genes involved in spindle assembly checkpoint (SAC).Citation1
Regulation of SAC genes by HMGA1 is of great interest because HMGA1 proteins are among the most abundant, non-histone chromatin binding proteins in the nucleus of cancer cells.Citation2-4 The HMGA1 gene is highly expressed in embryonic and adult stem cells, and most aggressive cancers studied to date, with low or undetectable levels in differentiated cells.Citation2-6 The HMGA1a/HMGA1b protein isoforms contain AT-hook DNA binding domains that mediate binding to AT-rich regions in the minor groove of chromosomal DNA.Citation2 They function as architectural transcription factors by bending chromatin to facilitate recruitment of other transcriptional regulators to promoters and enhancers throughout the genome. Prior studies identified HMGA1 as a master regulator in poorly differentiated, refractory tumors and pluripotent stem cells, where it drives a de-differentiated, stem-like state by inducing developmental transcriptional networks.Citation2-6
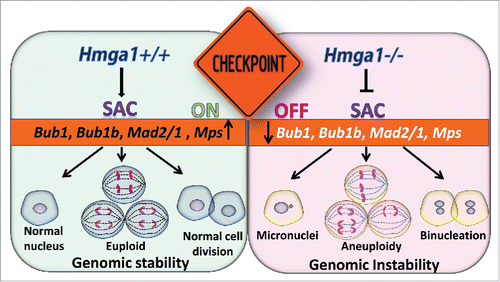
The recent study by Dr. Pierantoni and colleagues showed that Hmga1 deficiency in mouse embryonic fibroblasts (MEFs) leads to repression in SAC genes, including Bub1, Bub1b (BubR1), Mad2 (Mad2l1), and Ttk (Mps).Citation1 The first evidence that HMGA1 regulates SAC genes came from a study of lymphoid tumors in a transgenic mice overexpressing Hmga1a.Citation4 Bub1b was induced in the Hmga1-driven lymphoid tumors compared to lymphoid cells from wildtype mice. This pathway was validated in human Jurkat T-cell leukemic blasts where Bub1b was repressed following HMGA1 knock-down. SAC is a fundamental signaling mechanism of higher organisms that helps to ensure equal distribution of each genome with cell division. This checkpoint delays mitosis until all chromosomes form stable, bipolar attachments to spindle microtubules. The core components of SAC are highly conserved, including several kinases [budding uninhibited by benzimidazoles (Bub1), budding uninhibited by benzimidazoles related 1 (BubR1 or Bub1b), monopolar spindle 1 (Mps or Ttk), aurora B]. They cooperate with other proteins (Mad1, Mad2, Bub3, Cdc20) forming a kinetochore, or proteinaceous framework, that assembles onto the centromeric region of chromosomes, functioning as a platform and signaling hub to orchestrate chromosomal attachments, SAC activity, and cell cycle progression from metaphase to anaphase.
The recent study by Dr. Pierantoni builds on prior work in which this group reported that HMGA1 overexpression in colorectal cancer correlates with overexpression of SAC genes, which could contribute to chromosomal instability (CIN).Citation6 In the former study, they propose a model whereby HMGA1 promotes CIN by inducing SAC genes. In the more recent work, they find MEFs deficient in Hmga1 exhibit CIN and repression in SAC genes. There is a precedence for these seemingly contradictory findings; mice overexpressing Bub1b develop chromosomal missegregation, aneuploidy and spontaneous tumors, while hypomorphic mice with Bub1b deficiency also develop chromosomal missegregation, aneuploidy, and increased tumorigenesis with aging. The Hmga1 deficient MEFs exhibited an increase in micronuclei, binucleation, and aneuploidy, similar to abnormalities observed in cancer cells overexpressing HMGA1.
While this work provides further evidence for yet another critical pathway regulated by HMGA1, it leaves some important questions unanswered. Rescue experiments with SAC genes are needed to determine whether these phenotypes are dependent upon one or more SAC genes. It is also not yet known whether Hmga1 directly induces SAC gene expression in this setting. Intriguing, knock-out of Bub1b in mice results in embryonic lethality,Citation7 whereas this group reported a mouse with Hmga1 deficiency and normal viability, suggesting that Bub1b is also regulated by factors other than Hmga1. Indeed, normal viability in Hmga1 deficient mice is surprising given the extensive CIN observed in these MEFs. Confirmatory studies in other models are needed to validate these findings and uncover mechanisms that underlie regulation of SAC genes by Hmga1. Nonetheless, this work adds to the growing body of literature underscoring the importance of HMGA1 in regulating cell cycle progression and chromosomal stability, 2 pathways required for normal development that become disrupted in carcinogenesis.
Figure 1. Hmga1 helps to maintain the spindle assembly checkpoint (SAC). Left: Hmga1 maintains expression of SAC genes to help ensure genomic stability and faithful replication of the genome with each cell division. Right: In contrast, cells with Hmga1 deficiency down-regulate SAC genes, leading to genomic instability, aneuploidy and nuclear atypia.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- Pierantoni GM, Conte A, Rinaldo C, Tornincasa M, Gerlini R, Valente D, Izzo A, Fusco A. Hmga1 null mouse embryonic fibroblasts display downregulation of spindle assembly checkpoint gene expression associated to nuclear and karyotypic abnormalities. Cell Cycle 2016; 15:812-8; PMID:26889953; http://dx.doi.org/10.1080/15384101.2016.1146835
- Resar LMS. The high mobility group A1 gene: transforming inflammatory signals into cancer? Cancer Res 2010; 70:436-439; PMID:20068164; http://dx.doi.org/10.1158/0008-5472.CAN-09-1212
- Shah SN, Cope L, Poh W, Belton A, Roy S, Talbot CC, Sukumar S, Huso DL, Resar LMS. HMGA1: A master regulator of tumor progression in triple-negative breast cancer cells. PLoS One 2013; 8:e63419; PMID:23658826; http://dx.doi.org/10.1371/journal.pone.0063419
- Schuldenfrei A, Belton A, Kowalski J, Talbot CC, Di Cello F, Poh W, Tsai H-L, Shah SN, Huso TH, et al. HMGA1 drives stem cell, inflammatory pathway, and cell cycle progression genes during lymphoid tumorigenesis. BMC Genomics 2011; 12:549; PMID:22053823; http://dx.doi.org/10.1186/1471-2164-12-549
- Shah SN, Kerr C, Cope L, Zambidis E, Liu C, Hillion J, Belton A, Huso DL, Resar LMS. HMGA1 reprograms somatic cells into pluripotent stem cells by inducing stem cell transcriptional networks. PLoS One 2012; 7:e48533; PMID:23166588; http://dx.doi.org/10.1371/journal.pone.0048533
- Pierantoni GM, Conte A, Rinaldo C, Tornincasa M, Gerlini R, Federico A, Valente D, Medico E, Fusco A. Deregulation of HMGA1 expression induces chromosome instability through regulation of spindle assembly checkpoint genes. Oncotarget 2015; 6:17342-17353; PMID:26009897; http://dx.doi.org/10.18632/oncotarget.3944
- Kapanidou M, Lee S, Bolanos-Garcia VM. BubR1 kinase: protection against aneuploidy and premature aging. Trends Mol Med 2015; 21:364-372.; PMID:25964054; http://dx.doi.org/10.1016/j.molmed.2015.04.003
