ABSTRACT
The balance between the symmetric and asymmetric division of stem cells governs tissue homeostasis, and the deregulation of this balance initiates tumor formation. Although many functions of Numb have been demonstrated in normal stem cells, the role of Numb in cancer stem cells is relatively unclear. We recently demonstrated that in colorectal cancer stem cells, Numb was suppressed by miR-146a-5p, which resulted in the activation of the Wnt signaling pathway and symmetric template DNA division. Here, we demonstrate that the PKH26-labeled subcellular foci are enriched for endosomal markers such as EEA1 and RAB11. In colorectal cancer stem cells, the PKH-26-labeled vesicles are segregated equally at the first mitotic division; in contrast, they are unequally segregated in parental cells or in cancer stem cells undergoing serum-induced differentiation. The PKHBright progeny of colorectal cancer stem cells harbors a stem cell phenotype, whereas the PKHDim progeny behaves as the differentiating cells. The miR-146a-5p-regulated Numb controls the distribution of PKH26 vesicles. Our results suggest a critical role of Numb in controlling the segregation of subcellular vesicles during division of colorectal cancer stem cells.
Introduction and results
The role of Numb in cell fate determination and tumorigenesis
Asymmetric cell division (ACD) is a fundamental process by which progenitor cells generate daughter cells with distinct cell fates, which is essential for maintaining cell diversity and tissue homeostasis.Citation1 The mechanism of ACD involves the asymmetric distribution of intrinsic molecules of progenitor cells, which results in the asymmetric signaling of the generated daughter cells. Among the asymmetrically segregated molecules during ACD, Numb is one of the most important cell fate determinants in both Drosophila and mammalian cells. The segregation of Numb into daughter cells favors a differentiated cell fate in both Drosophila and mammalian cells. In the Drosophila nerve system, the external sensory organs comprising 4 cell types are generated from one single sensory organ progenitor (SOP) cell by a sequence of ACD. The correct cell fate specification of SOP cell requires Numb.Citation2-4 In mouse cortical progenitor cells undergoing active neurogenesis, Numb is largely segregated into neural daughter cells, which suggests that Numb promotes a differentiated cell fate.Citation5 In haematopoietic precursors isolated from TNR mice that contain a GFP transgene to report Notch signaling activity, the expression of Numb is found to be lower in GFP(+)SKL cells (Sca-1/c-Kit positive, low-to-negative lineage markers) and preferentially segregated to the GFP(-) differentiated cells.Citation6 The above findings suggest that Numb tends to segregate into the differentiated cells during the division of progenitor/stem cells.
In mammalian cancer cells, lines of evidence support that Numb functions as a tumor suppressor. Numb suppresses the Notch signaling pathway in cancer cells, which plays a pivotal role in tumorigenesis and cancer progression.Citation7,8 Mechanistically, Numb reduces Notch1 stability by interacting with Itch to promote the polyubiquitination of the membrane-tethered Notch1Citation9 or by inducing Notch1 endosomal sorting for degradation.Citation10 Numb interacts with the p53/MDM2 complex, which leads to the prevention of polyubiquitination of p53 by MDM2 to increase p53 expression.Citation11 The methylation of Numb at lysine 158 and lysine 163 sites by Set8 disrupts the Numb-p53 interaction, which results in an increase of p53 degradation.Citation12 In summary, Numb is correlated with a differentiated cell fate and acts as a tumor suppressor.
The role of the miR-146a-5p-Numb axis in determining the mode of cell division of colorectal cancer stem cells
Cancer stem cells (CSCs) are a small population of heterogeneous tumors that possess traits of normal stem cells and are crucial for tumor initiation and progression.Citation13 However, whether an increase in symmetric cell division is responsible for expanding CSCs and the mechanisms that guide the mode of cellular division in CSCs are unclear. In our previous studies, we demonstrated that colorectal cancer stem cells (CRCSCs) underwent Snail-predominant epithelial-mesenchymal transition (EMT).Citation14 Furthermore, we identified miR-146a-5p as a major microRNA in CRCSCs that targets Numb to regulate the Wnt-β-catenin pathway. In line with the tumor suppressor functions of Numb, the restoration of Numb in miR-146a-5p-dominant CRCSCs attenuated the spheroid formation capacity in vitro and tumorigenicity in vivo.Citation15
Next, we marked the DNA strands with a long-term BrdU pulse based on the immortal strand hypothesis, i.e., the stem cells are prone to retaining the original DNA strand (template strand) to surpassing replication errors and preserve genetic features.Citation16 We found that CRCSCs divided symmetrically under stem cell medium, and the percentage of asymmetric DNA segregation increased upon serum-induced differentiation. Intriguingly, recent results from independent groups showed the important role of another miRNA, miR-34a, in CRCSCs. In CRCSCs, miR-34a has a distinct role from miR-146a-5p: miR-146a-5p induces Wnt-β-catenin signaling in the progeny and tends to segregate into cancer stem cells,Citation15 whereas the distribution of miR-34a into differentiated daughter cells inhibits Notch signaling and promotes the differentiation of CRCSCs.Citation17 Moreover, miR-34a and Snail form a negative feedback loop to regulate EMT.Citation18 Together with our findings, we suggest that in Snail-dominant CRCSCs, the miR-146a-5p-Numb pathway activates the β-catenin/Wnt pathway to maintain cancer stemness for symmetric expansion, whereas miR-34a suppresses Notch to generate daughter cells with a more differentiated phenotype.
Segregation of subcellular vesicles in colorectal cancer stem cells
The endocytic pathways function in connection with the cell membrane for endocytosis, vesicle recycling and signaling transduction.Citation19 Given that Numb regulates post-endocytic traffickingCitation20 and is preferentially segregated into differentiated daughter cells,Citation6 we hypothesized that the endocytic components may be distributed non-randomly during the cell division of CRCSCs. To prove this assumption, we labeled the colon cancer cell line HT29 cells and HT29-derived CRCSCs (SDCSCs, sphere-derived cancer stem cells) with PKH26 dye to monitor the dynamic distribution of membrane-derived vesicles and their distribution during cell division. The PKH26 dye was initially stained homogeneously and more than 99% of cells were PKH26 dye-positive cells according to flow cytometry analysis (Fig. S1A, upper panel). Immediately upon staining, bright PKH26 fluorescent signals were observed on plasma membranes of HT29 parental cells (, 0 hr). The punctuated fluorescent signals occurred and accumulated along with the decreased fluorescent signal from cell membrane (, 10 min, 1 hr, and 24 hr), which suggested that the PKH26-labeled structures are engulfed from the plasma membrane and become endocytic components.
Figure 1. The symmetrical distribution of PKH26-labeled subcellular compartments occurs in colorectal cancer stem cells. (A) The representative images for PKH26 dye labeling in HT29 cells at the indicated time points. PKH26 dye, red; DNA, blue. Scale bar = 20 μm. (B) The immunofluorescent pictures for showing the association of PKH26-labeled vesicles and endosome markers. PKH26 dye, red; endosome markers (EEA1, RAB11 and RAB7), green; DNA, blue. Scale bar = 5 μm. Arrows indicate the association or colocolization. Insert: magnified vesicle association indicated by yellow arrows. (C) The quantification results of association of PKH26-vesicles and indicated endosome markers in HT29 parental cells. Data represent mean ± SEM ***, p < 0.001 (Student's t test). (D) Representative images of symmetric or asymmetric segregation of PKH26-labeled vesicles in HT29 SDCSCs cultured under stem cell medium or in FBS-induced differentiation, respectively. PKH26 dye, red; DNA, blue. insert: phase pictures for showing paired-cells. (E) The percentage of the asymmetry/symmetry of PKH26-labeled vesicles in parental cells, SDCSCs and serum-differentiated SDCSCs (differentiation) in HT29 and HCT15 cells. n (total counted cells over 2 independent experiments) = 142, 223, 83, 144, 196, and 54 for HT29 parental cells, HT29 SDCSCs, Differentiation (HT29 SDCSCs), HCT15 parental cells, HCT15 SDCSCs, and Differentiation (HCT15 SDCSCs), respectively. PKH-Sym, symmetric segregation of PKH26-labeled vesicles; PKH-Asym, asymmetric segregation of PKH26-labeled vesicles. The p-value is estimated by χ2 test. *, p < 0.05; **, p < 0.01 ***, p < 0.001.
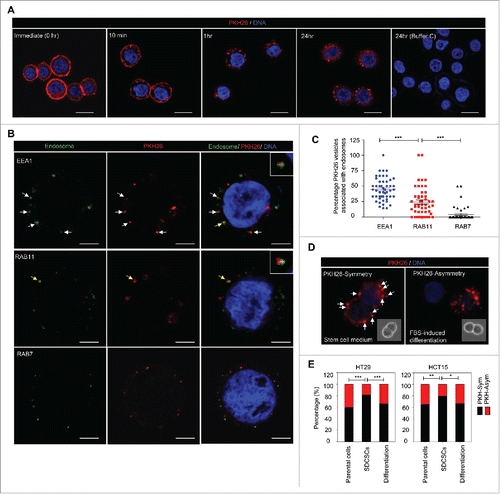
Next, we co-stained several endocytic and organelle markers with PKH26 dye to investigate the major subcellular components for PKH26 vesicles. The results showed that 1 hour after initial dye labeling, the PKH26-labeled structures distributed in the cytoplasm and were positively associated with EEA1 (early endosome marker, the top row) and, to a lesser extent, RAB11 (recycling vesicle marker, the middle row), but not RAB7 (late endosome marker, the bottom row) (). The EEA1- and RAB11-positive endosomes comprised up to 71% of PKH26 vesicles (). However, these PKH26 vesicles did not colocalize with CD81 (exosome marker), calreticulin (endoplasmic reticulum marker), or mitochondria (Fig. S1B). Collectively, these results suggested that the PKH26 vesicles were enriched for endosomal components with newly synthesized membranes engulfed from the plasma membrane.
To investigate the segregation of PKH26 vesicles during cell division in HT29- and HCT15-derived SDCSCs, PKH26-labeled SDCSCs were dissociated to a single cell suspension and cultured under stem cell medium (SCM) or fetal bovine serum (FBS)-containing medium for the induction of differentiation until the next round of cell division. First, we confirmed that labeling with PKH26 dye did not influence the cell viability and proliferation or sphere-forming capacity of HT29 SDCSCs (Fig. S1C-D). By quantifying the integrated fluorescent signal in 2 dividing progenies, we found that the pre-engulfed PKH26 vesicles were segregated symmetrically in both HT29- and HCT15-SDCSCs when cultivated in SCM. However, a non-random distribution of PKH26 vesicles was noted upon serum-induced differentiation, which resembled that in parental cells (). By tracking the cell division through time-lapsed microscopy, we found that the PKH26 vesicles were distributed either equally or unequally in twin cells of HT29 parental cells (Movie S1), which confirmed the existence of asymmetry/symmetry segregation of PKH26 vesicles in cancer cells. Moreover, 81% of the asymmetrically segregated PKH26 vesicles were positive for endosome markers (Fig. S2A-B, 50% for EEA1- and 31% for RAB11-positive endosomes, respectively). This symmetry/asymmetric segregation of the subcellular vesicles coincided with that of DNA segregation observed in our previous study.Citation15
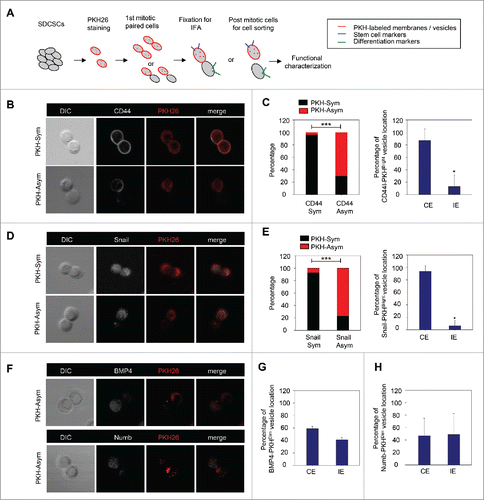
To investigate the cells' fate and to validate the functional divergence in PKHBright/PKHDim progeny generated from the asymmetric cell division of CRCSCs, the mitotic paired cells were enriched with a thymidine-nocodazole sequence for immunofluorescence assay or sequential functional characterization as shown in . CD44 and Snail were selected as markers for CRCSCs because of their abundant expression in CRCSC.Citation15 We found that the pattern of asymmetry/symmetry of PKH26 vesicles was correlated with that of CD44 (), and PKHBright progeny largely co-expressed CD44 (). A similar result was observed in Snail (). However, the PKHDim cells were not co-expressed with the differentiation marker BMP4Citation15 () or Numb ().
Figure 2. The PKH26 vesicles co-segregate into daughter stem cells divided from SDCSCs. (A) A schema for illustrating the paired-cell assay. IFA, immunofluorescence analysis. (B) Representative images of paired-cell assay of serum-differentiated HT29 SDCSCs. CD44, white; PKH26 dye, red. PKH-Sym, symmetric segregation of PKH26-labeled vesicles; PKH-Asym, asymmetric segregation of PKH26-labeled vesicles. (C) Left: The percentage of PKH26 vesicles symmetry/asymmetry in serum-differentiated HT29 SDCSCs. CD44 Sym, symmetric CD44 segregation; CD44 Asym, asymmetric CD44 segregation. n (total counted cells over 2 independent experiments) = 66 and 37 for CD44 Sym and CD44 Asym, respectively. ***, p < 0.001 (χ2 test). Right: The percentage of CD44 and PKH26 vesicles co-expression (CE) or inverse expression (IE) in PKHBright daughter cells undergoing asymmetric cell division. Data represent mean ± SD n = 3 independent experiments. *P < 0.05 (Student's t test). (D) Representative images of paired-cell assay of serum-differentiated HT29 SDCSCs. Snail, white; PKH26 dye, red. (E) Left: The percentage of PKH26 vesicles symmetry/asymmetry in serum-differentiated HT29 SDCSCs. Snail Sym, symmetric Snail segregation; Snail Asym, asymmetric Snail segregation. n (total counted cells over 2 independent experiments) = 87 and 38 for Snail Sym and Snail Asym, respectively. ***, p < 0.001 (χ2 test). Right: The percentage of Snail and PKH26 vesicles co-expression (CE) or inverse expression (IE) in PKHBright daughter cells undergoing asymmetric cell division. *, p < 0.05 (Student's t test). (F) Representative images of paired-cell assay of serum-differentiated HT29 SDCSCs. BMP4 and Numb, white; PKH26 dye, red. (G) The percentage of BMP4 and PKH26 vesicles co-expression (CE) or inverse expression (IE) in PKHDim daughter cells undergoing asymmetric cell division. (H) The percentage of Numb and PKH26 vesicles co-expression (CE) or inverse expression (IE) in PKHDim daughter cells undergoing asymmetric cell division.
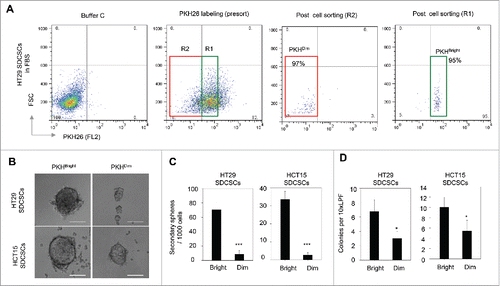
Next, the PKHBrigh and PKHDim cells were sorted from post-mitotic HT29-and HCT15-derived SDCSCs for functional characterization. Only cells with purity greater than 85% were subjected to further analysis (). Compared to the PKHDim cells, the PKHBrigh cells showed an enhanced capability to form secondary spheres () and anchorage-independent colonies (). In summary, the PKHBright progeny derived from PKH26-labeled CRCSCs had the same characteristics as CSCs, whereas the PKHDim progeny exhibited the characteristics of differentiated cancer cells.
Figure 3. The PKHBright daughter stem cells exhibit a higher capability of forming tumorispheres and colonies. (A) Flow cytometry results for showing the gated regions of PKHBright (R1) and PKHDim (R2) of serum-differentiated HT29 SDCSCs before sorting and the purities after sorting. The percentage of cells in the gated region is illustrated in the corresponding panels. (B) Representative images of spheroids formed from HT29 SDCSCs- and HCT15 SDCSCs-derived PKHBright and PKHDim daughter cells. Scale bar = 100μm. (C) Histograms for showing the secondary sphere formation capacities of HT29 SDCSCs- and HCT15 SDCSCs-derived PKHBright and PKHDim daughter cells. Bright, PKHBright cells; Dim, PKHDim cells. Data represent mean ± SD n = 3 independent experiments. ***, p < 0.001 (Student's t test). (D) Histograms for showing the anchorage-independent colony forming capacities of HT29 SDCSCs- and HCT15 SDCSCs-derived PKHBright and PKHDim daughter cells. LPF, low power field. *, p < 0.05 (Student's t test).
Numb directs the asymmetric segregation of PKH26 vesicles
To investigate the impact of Numb on the segregation of PKH26 vesicles during cell division, we labeled the established stable cell linesCitation15 that express full-length Numb or truncated Numb which can not interact with β-catenin. The result showed that the restoration of full-length Numb to the level of parental cells promoted the asymmetric distribution of PKH26 vesicles in HT29- and HCT15-derived SDCSCs. However, the ectopic expression of the β-catenin non-interacting Numb fragment (FLAG-Numb 162-400) did not have such an effect. The co-expression of unphosphorylatable β-catenin restored the symmetric segregation of PKH26 vesicles in Numb-restored SDCSCs (). Second, the asymmetric segregation of PKH vesicles increased in HT29 SDCSCs with the stable knockdown of miR-146a-5p (Fig. S3A). Third, the stable expression of miR-146a-5p in HT29 and HCT15 parental cells engendered the symmetric distribution of PKH26 vesicles, and the restoration of Numb expression in these cells attenuated the miR-146a-5p-induced effect (). Cumulatively, these findings suggested the involvement of Numb in the spatial distribution of PKH26-labeled vesicles of CRCSCs.
Figure 4. Restoration of Numb directs the asymmetry of PKH26 vesicles. (A) Histograms for showing percentages of PKH26 vesicle asymmetry/symmetry in the indicated stable cell lines. PKH-Asym, asymmetric segregation of PKH26 vesicles. n (total counted cells over 2 independent experiments) = 110, 34, 75, 123, 145, 137, 137, and 138 for HT29 SDCSC-Vec, HT29 SDCSCs-FLAG-Numb(162-400), HT29 SDCSCs-Numb(FL) and HT29 SDCSCs-Numb(FL)/β-cat(mut), HCT15 SDCSC-Vec, HCT15 SDCSCs-FLAG-Numb(162-400), HCT15 SDCSCs-Numb(FL) and HCT15 SDCSCs-Numb(FL)/β-cat(mut), respectively. The p-value is estimated by χ2 test. *, p < 0.05; ***, p < 0.001. (B) Histograms for showing percentages of PKH26 vesicle asymmetry/symmetry at indicated stable cell lines. n (total counted cells over 2 independent experiments) = 146, 148, 143, 147, 106, 75, 115 and 119 for HT29-Vec, HT29-146a, HT29-Puro-Vec, HT29-146a/Puro-Numb, HCT15-Vec, HCT15-146a, HCT15-Puro-Vec and HCT15-146a/Puro-Numb, respectively. The p-value is estimated by χ2 test. *, p < 0.05; **, p < 0.01. (C) Upper, RT-qPCR result for showing the expression of miR-146a-5p in HCT15 SDCSC-Vec and HCT15 SDCSC-Numb(FL) stable lines. Lower, RT-qPCR result for showing the expression of miR-146a-5p in PKHBright and PKHDim daughter cells of HCT15 SDCSCs-Numb(FL) cultured in stem-cell medium. PKHBright cells; Dim, PKHDim cells. *, p < 0.05; **, p < 0.01 (Student's t test). (D) A schema for summarizing the impact of Numb in switching division modes of colorectal cancer stem cells.
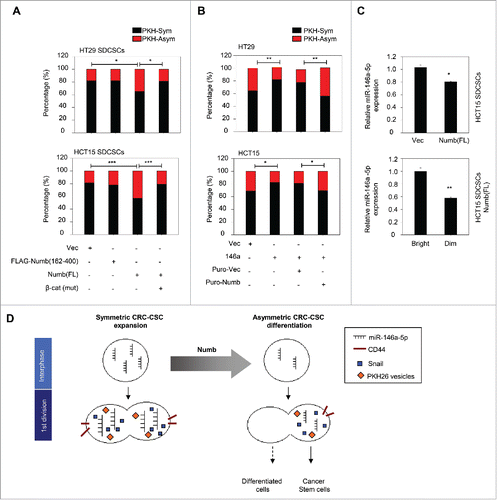
The cortical condensation and segregation of Numb are considered to be critical steps in cell fate determination, and Numb has been shown to be segregated into BrdU-exclusive differentiated progenies at the second mitosis of CRCSCs.Citation15 Nevertheless, we found that at the first round of CRCSC division Numb was not co-expressed in PKHDim cells upon serum-induced differentiation () or Numb restoration (Fig. S3B) in this study, which suggests that the spatial distribution of Numb is not the major mechanism responsible for the segregation of PKH26 vesicles. Because the asymmetric segregation of miR146a-5p to daughter stem cells occurred when its endogenous quantity was reducedCitation15 and the endosome/multivesicular bodies have been implicated in the assembly of the miRNA-induced silencing complex (miRISC),Citation21,22 we speculated that the Numb-regulated distribution of PKH26 vesicles may be attributed to the quantity of miR-146a-5p, and miRISC (miRNA-induced silencing complex) may carry endosomal components along with miR-146a-5p to daughter stem cells. In support of this assumption, we observed that Wnt target genes, including CD44, LGR5, CCND1 and the primary MIR-146A transcript, were downregulated in Numb-restored HCT15 SDCSCs compared with that of vector control cells (Fig. S3C). As expected, the total quantity of miR-146a-5p was decreased in Numb-restored HCT15 SDCSCs (). Intriguingly, in the Numb-restored SDCSCs, miR-146a-5p was unequally distributed and higher in PKHBrigh progenies than the PKHDim progenies at the first round of cell division (), which implicates the co-segregation of miR-146a-5p and PKH26 vesicles to daughter stem cells. However, the mechanism of miRNA-dictated endosomal asymmetry is still unclear and needs further investigation. To summarize our findings in the miR-146a-5p/Numb-regulated segregation of PKH26-labeled vesicles, we found that the re-expression of Numb firstly downregulates the level of miR-146a-5p in parental CRCSCs before mitosis. A lower level of miR-146a-5p in CRCSCs is prone to distribute unequally to the daughter stem cells (PKHBrigh progeny) at the first mitosis, whereas a higher level of miR-146a-5p in CRCSCs causes the equal distribution in progenies to generate 2 daughter stem cells. The hypothetical model is illustrated in .
Conclusion and future directions
During the division of CRCSCs, intracellular components distribute equally into progenies to orchestrate stem cell properties to maintain the number of stem cells. An increase in Numb suppresses the expression of miR-146a-5p, which results in the asymmetric division of CRCSCs and reduces the stem cell number. To our knowledge, we are the first to observe the asymmetric/symmetric distribution of subcellular vesicles in CRCSCs and to demonstrate the involvement of Numb in the asymmetric distribution of subcellular vesicles of CSCs.
It has been noted that endocytosis and the endosomal pathway transduces signals from the plasma membrane.Citation23,24 Upon ligand binding, the phosphorylated TGFβ receptor (TGFβR) is internalized to endosomes. The FYVE domain protein endofin binds to the type I TGFβ receptor and potentiates SMAD2-SMAD4 complex formation in endosomes.Citation25 EEA1, a Rab5 effector protein, tethers to early endosomes via its zinc-binding FYVE finger and phosphatidylinositol-3-phosphate (PI3P) on early endosomes.Citation26 The EEA1-positive endosomes are suggested to function as signaling scaffolds for angiotensin II-induced AKT activation.Citation27 Emerging evidence suggests the involvement of the endosomal component in stem cell division. RAB11-positive recycling endosomes regulates the activities of the Notch ligand Delta, and RAB11 asymmetry determines cell fate specification in Drosophila SOP cells.Citation28 Here, we indicated that Numb might regulate the segregation of endosomal components indirectly through miR-146a-5p. Previous reports have shown that the miRISC component Ago2 is associated with EEA1- and RAB11-positive endosomes by interacting with PICK1 in neuronal dendrites,Citation29 and its phosphorylation on S387 controls its localization to endosomes as well as sorting to exosomes in colon cancer cells.Citation30 We suggest that the argonaute protein might provide a protein scaffold between the miRNA machinery and endosomal pathway. miR-146a-5p is the most abundant miRNA identified in CRCSCs, and asymmetrical miR-146a-5p segregation may contribute to endosomal asymmetry via miRISC components, such as argonaute proteins. However, the impact of the symmetric/asymmetric segregation of endosomal components and the interplay between endosomes and miRNAs during cancer stem cell division deserves further investigation.
Abbreviations
| ACD | = | Asymmetric cell division |
| PKH-Asym | = | Asymmetric segregation of PKH26-labeled vesicles |
| Buffer C | = | Buffer diluent C |
| CSCs | = | Cancer stem cells |
| CRCSCs | = | Colorectal cancer stem cells |
| EMT | = | Epithelial-mesenchymal transition |
| FBS | = | Fetal bovine serum |
| miRISC | = | miRNA-induced silencing complex |
| MVBs | = | Multivescular bodies |
| PI3P | = | Phosphatidylinositol-3-phosphate |
| SDCSCs | = | sphere-derived cancer stem cells |
| SOP | = | Sensory organ progenitor |
| SKL cells | = | Sca-1/c-Kit positive, low-to-negative lineage markers |
| SCD | = | Symmetric cell division |
| SCM | = | Stem cell medium |
| PKH-Sym | = | Symmetric segregation of PKH26-labeled vesicles |
| TGFβR | = | TGFβ receptor |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
KCCY_A_1218101_supplement.zip
Download Zip (2.9 MB)Acknowledgment
The authors would like to dedicate this paper to the memory of Prof. Hsei-Wei Wang (Institute of Microbiology and Immunology, National Yang-Ming University), who passed away during the period of this research. Prof. Wang originated the bioinformatics mining of the targets of miR-146a-5p and found Numb as a target of miR-146a-5p. This paper could not be completed without his long-lasting devotion to bioinformatics and colon cancer research. We thank Hsin-Yi Lan (Institute of Clinical Medicine of National Yang-Ming University, Taiwan) for providing technical supports in paired-cell assay.
Funding
This work is supported by Taipei Medical University (TMU104-AE1-B11 to W-L.H), Ministry of Science and Technology (104-2321-B-010-005, 104-0210-01-09-02, and 103-2633-H-010-001 to M-H.Y; 105-2320-B-038-009-MY2 to W-L.H), a grant from Ministry of Education, Aim for the Top University Plan (to M-H.Y), and a grant from Ministry of Health and Welfare, Center of Excellence for Cancer Research (MOHW105-TDU-B-211-134003 to M-H.Y.).
Reference
- Jan YN, Jan LY. Asymmetric cell division in the Drosophila nervous system. Nat Rev Neurosci 2001; 2:772-9; PMID:11715054; http://dx.doi.org/10.1038/35097516
- Uemura T, Shepherd S, Ackerman L, Jan LY, Jan YN. numb, a gene required in determination of cell fate during sensory organ formation in Drosophila embryos. Cell 1989; 58:349-60; PMID:2752427; http://dx.doi.org/10.1016/0092-8674(89)90849-0
- Rhyu MS, Jan LY, Jan YN. Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell 1994; 76:477-91; PMID:8313469; http://dx.doi.org/10.1016/0092-8674(94)90112-0
- Berdnik D, Torok T, Gonzalez-Gaitan M, Knoblich JA. The endocytic protein alpha-Adaptin is required for numb-mediated asymmetric cell division in Drosophila. Dev Cell 2002; 3:221-31; PMID:12194853; http://dx.doi.org/10.1016/S1534-5807(02)00215-0
- Shen Q, Zhong W, Jan YN, Temple S. Asymmetric Numb distribution is critical for asymmetric cell division of mouse cerebral cortical stem cells and neuroblasts. Development 2002; 129:4843-53; PMID:12361975
- Wu M, Kwon HY, Rattis F, Blum J, Zhao C, Ashkenazi R, Jackson TL, Gaiano N, Oliver T, Reya T. Imaging hematopoietic precursor division in real time. Cell Stem Cell 2007; 1:541-54; PMID:18345353; http://dx.doi.org/10.1016/j.stem.2007.08.009
- Louvi A, Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat Rev Neurosci 2006; 7:93-102; PMID:16429119; http://dx.doi.org/10.1038/nrn1847
- Rizzo P, Osipo C, Foreman K, Golde T, Osborne B, Miele L. Rational targeting of Notch signaling in cancer. Oncogene 2008; 27:5124-31; PMID:18758481; http://dx.doi.org/10.1038/onc.2008.226
- McGill MA, McGlade CJ. Mammalian numb proteins promote Notch1 receptor ubiquitination and degradation of the Notch1 intracellular domain. J Biol Chem 2003; 278:23196-203; PMID:12682059; http://dx.doi.org/10.1074/jbc.M302827200
- McGill MA, Dho SE, Weinmaster G, McGlade CJ. Numb regulates post-endocytic trafficking and degradation of Notch1. J Biol Chem 2009; 284:26427-38; PMID:19567869; http://dx.doi.org/10.1074/jbc.M109.014845
- Colaluca IN, Tosoni D, Nuciforo P, Senic-Matuglia F, Galimberti V, Viale G, Pece S, Di Fiore PP. NUMB controls p53 tumour suppressor activity. Nature 2008; 451:76-80; PMID:18172499; http://dx.doi.org/10.1038/nature06412
- Dhami GK, Liu H, Galka M, Voss C, Wei R, Muranko K, Kaneko T, Cregan SP, Li L, Li SS. Dynamic methylation of Numb by Set8 regulates its binding to p53 and apoptosis. Mol Cell 2013; 50:565-76; PMID:23706821; http://dx.doi.org/10.1016/j.molcel.2013.04.028
- Visvader JE, Lindeman GJ. Cancer stem cells: current status and evolving complexities. Cell Stem Cell 2012; 10:717-28; PMID:22704512; http://dx.doi.org/10.1016/j.stem.2012.05.007
- Hwang WL, Yang MH, Tsai ML, Lan HY, Su SH, Chang SC, Teng HW, Yang SH, Lan YT, Chiou SH, et al. SNAIL regulates interleukin-8 expression, stem cell-like activity, and tumorigenicity of human colorectal carcinoma cells. Gastroenterology 2011; 141:279-91, 91 e1-5; PMID:21640118
- Hwang WL, Jiang JK, Yang SH, Huang TS, Lan HY, Teng HW, Yang CY, Tsai YP, Lin CH, Wang HW, et al. MicroRNA-146a directs the symmetric division of Snail-dominant colorectal cancer stem cells. Nat Cell Biol 2014; 16:268-80; PMID:24561623; http://dx.doi.org/10.1038/ncb2910
- Rando TA. The immortal strand hypothesis: segregation and reconstruction. Cell 2007; 129:1239-43; PMID:17604710; http://dx.doi.org/10.1016/j.cell.2007.06.019
- Bu P, Chen KY, Chen JH, Wang L, Walters J, Shin YJ, Goerger JP, Sun J, Witherspoon M, Rakhilin N, et al. A microRNA miR-34a-regulated bimodal switch targets Notch in colon cancer stem cells. Cell Stem Cell 2013; 12:602-15; PMID:23642368; http://dx.doi.org/10.1016/j.stem.2013.03.002
- Siemens H, Jackstadt R, Hunten S, Kaller M, Menssen A, Gotz U, Hermeking H. miR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions. Cell Cycle 2011; 10:4256-71; PMID:22134354; http://dx.doi.org/10.4161/cc.10.24.18552
- Platta HW, Stenmark H. Endocytosis and signaling. Curr Opin Cell Biol 2011; 23:393-403; PMID:21474295; http://dx.doi.org/10.1016/j.ceb.2011.03.008
- Santolini E, Puri C, Salcini AE, Gagliani MC, Pelicci PG, Tacchetti C, Di Fiore PP. Numb is an endocytic protein. J Cell Biol 2000; 151:1345-52; PMID:11121447; http://dx.doi.org/10.1083/jcb.151.6.1345
- Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol 2009; 11:1143-9; PMID:19684575; http://dx.doi.org/10.1038/ncb1929
- Lee YS, Pressman S, Andress AP, Kim K, White JL, Cassidy JJ, Li X, Lubell K, Lim DH, Cho IS, et al. Silencing by small RNAs is linked to endosomal trafficking. Nat Cell Biol 2009; 11:1150-6; PMID:19684574; http://dx.doi.org/10.1038/ncb1930
- Sorkin A, von Zastrow M. Endocytosis and signalling: intertwining molecular networks. Nat Rev Mol Cell Biol 2009; 10:609-22; PMID:19696798; http://dx.doi.org/10.1038/nrm2748
- Dobrowolski R, De Robertis EM. Endocytic control of growth factor signalling: multivesicular bodies as signalling organelles. Nat Rev Mol Cell Biol 2012; 13:53-60
- Chen YG, Wang Z, Ma J, Zhang L, Lu Z. Endofin, a FYVE domain protein, interacts with Smad4 and facilitates transforming growth factor-beta signaling. J Biol Chem 2007; 282:9688-95; PMID:17272273; http://dx.doi.org/10.1074/jbc.M611704200
- Mu FT, Callaghan JM, Steele-Mortimer O, Stenmark H, Parton RG, Campbell PL, McCluskey J, Yeo JP, Tock EP, Toh BH. EEA1, an early endosome-associated protein. EEA1 is a conserved alpha-helical peripheral membrane protein flanked by cysteine “fingers” and contains a calmodulin-binding IQ motif. J Biol Chem 1995; 270:13503-11; PMID:7768953; http://dx.doi.org/10.1074/jbc.270.22.13503
- Nazarewicz RR, Salazar G, Patrushev N, San Martin A, Hilenski L, Xiong S, Alexander RW. Early endosomal antigen 1 (EEA1) is an obligate scaffold for angiotensin II-induced, PKC-alpha-dependent Akt activation in endosomes. J Biol Chem 2011; 286:2886-95; PMID:21097843; http://dx.doi.org/10.1074/jbc.M110.141499
- Emery G, Hutterer A, Berdnik D, Mayer B, Wirtz-Peitz F, Gaitan MG, Knoblich JA. Asymmetric Rab 11 endosomes regulate delta recycling and specify cell fate in the Drosophila nervous system. Cell 2005; 122:763-73; PMID:16137758; http://dx.doi.org/10.1016/j.cell.2005.08.017
- Antoniou A, Baptista M, Carney N, Hanley JG. PICK1 links Argonaute 2 to endosomes in neuronal dendrites and regulates miRNA activity. EMBO Rep 2014; 15:548-56; PMID:24723684; http://dx.doi.org/10.1002/embr.201337631
- McKenzie AJ, Hoshino D, Hong NH, Cha DJ, Franklin JL, Coffey RJ, Patton JG, Weaver AM. KRAS-MEK Signaling Controls Ago2 Sorting into Exosomes. Cell Rep 2016; 15:978-87; PMID:27117408; http://dx.doi.org/10.1016/j.celrep.2016.03.085
