ABSTRACT
Genome maintenance requires coordinated actions of diverse DNA metabolism processes. Scaffolding proteins, such as those containing multiple BRCT domains, can influence these processes by collaborating with numerous partners. The best-studied examples of multi-BRCT scaffolds are the budding yeast Dpb11 and its homologues in other organisms, which regulate DNA replication, repair, and damage checkpoints. Recent studies have shed light on another group of multi-BRCT scaffolds, including Rtt107 in budding yeast and related proteins in other organisms. These proteins also influence several DNA metabolism pathways, though they use strategies unlike those employed by the Dpb11 family of proteins. Yet, at the same time, these 2 classes of multi-BRCT proteins can collaborate under specific situations. This review summarizes recent advances in our understanding of how these multi-BRCT proteins function in distinct manners and how they collaborate, with a focus on Dpb11 and Rtt107.
Introduction
A critical aspect of genome maintenance is faithful DNA replication. It begins with the assembly of replisomes that are comprised of replicative enzymes and their cofactors. This multi-step process relies on a conserved cascade of protein-protein interactions involving more than 30 proteins.Citation1 Once formed, replisomes collaborate with another group of proteins that help to manage replication perturbation. These proteins, referred to as replication accessory factors, include DNA metabolism enzymes, protein modification enzymes, and scaffold proteins. DNA metabolism enzymes, such as nucleases and helicases, help eliminate DNA lesions, protein barriers, and secondary DNA structures that block replisome progression.Citation2,3 Protein modification enzymes conjugate substrates with small modifiers, such as phosphate and acetyl groups, or large modifiers, such as ubiquitin and SUMO, and can generate fast and diverse effects on many substrates in a coordinated manner to facilitate replication. Unlike DNA metabolism and protein modification enzymes, scaffold proteins usually do not possess enzymatic activities; rather they assemble protein complexes needed for sophisticated DNA transactions or signaling events. As such, scaffold proteins are central for the extensive network of interactions found among replisome members and replication accessory factors. Prominent scaffolds important for both replisome assembly and progression include proteins with multiple BRCT domains.
The BRCT (90–100 amino acids) domain was initially found in the tumor suppressor protein BRCA1, and has been subsequently identified in more than 2 dozen proteins in bacteria, archaea, and eukaryotes.Citation4-7 Biochemical and sequence analyses suggest that BRCT domains mostly support protein-protein interactions and that BRCT containing proteins act almost exclusively in genome maintenance, with many involved in replication.Citation6,7 For several of these proteins, 2 BRCT domains are connected by a linker of up to 60 amino acids to form tandem BRCT (or tBRCT). Some tBRCTs bind phospho-peptides wherein both units determine binding specificity.Citation8,9 As such, tBRCTs often act in response to kinase signals by establishing situation-specific protein-protein interactions that facilitate replication and response to DNA damage.
While most BRCT proteins contain only 1 or 2 such domains, 2 classes contain 4 or more.Citation6,7 The first includes the budding yeast Dpb11, the fission yeast Cut5/Rad4, and the vertebrate TopBP1. Members of this class share similar roles in DNA replication and repair as well as DNA damage checkpoint response, with TopBP1 acquiring more extensive functions compared to its yeast counterparts.Citation10 The second class includes the budding yeast Rtt107, the fission yeast Brc1, and vertebrate PTIP. Recent studies have begun to delineate the shared and distinct features of these of BRCT scaffolds, and how they collaborate with Dpb11 and related proteins to execute specific functions. In the sections below, we summarize these features with a focus on the budding yeast Dpb11 and Rtt107.
Dpb11 and its homologs function as molecular bridges and in other capacities
Dpb11 and Cut5 in replication initiation
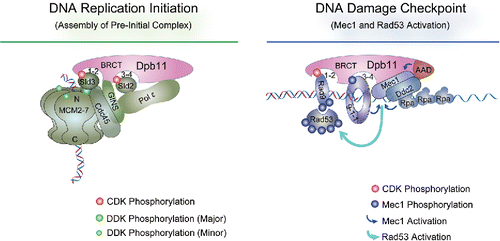
The budding yeast Dpb11 and the fission yeast Cut5 contain 4 BRCT domains arranged as 2 pairs of tBRCT, named BRCT1-2 and BRCT3-4. During replisome assembly, S-CDK mediated phosphorylation of the replication factors Sld3 and Sld2 (or Drc1) enables them to bind to BRCT3-4 and BRCT1-2, respectively ().Citation11-14 In addition, the region between the 2 tBRCTs in Dpb11 interacts with the replicative helicase cofactor GINS ().Citation15 A complex formed by Dpb11, GINS, Sld2, and the replicative polymerase epsilon is detected during S phase.Citation15,16 In the meantime, Sld3 binds to the DNA helicase MCM at replication origins ().Citation17,18 Consequently, Dpb11, in conjunction with Sld2 and Sld3, plays a major role in delivering polymerase epsilon and GINS to the origin-loaded MCM, enabling an important intermediate step during replisome assembly.
Figure 1. The roles of Dpb11 in DNA replication initiation and DNA damage checkpoint. The four BRCT domains of Dpb11 are depicted as 2 tBRCT pairs (BRCT1-2 and 3–4). As described in the text, Dpb11 binds Sld2 and Sld3, both phosphorylated by S-CDK, during DNA replication initiation (left panel).Citation13,14 Dpb11 also interacts with GINS and Pol epsilon. Sld3, in complex with Cdc45, recognizes and interacts with MCM that is phosphorylated by DDK (Dbf4-dependent kinase).Citation16,17 MCM, Cdc45 and GINS form the CMG complex that catalyzes template strand unwinding during replication.Citation129 In DNA damage checkpoint (right panel), Dpb11 binds 9-1-1 that is located at 5′ ss- and ds-DNA junction and phosphorylated by Mec1.Citation24,26 Dpb11 also binds to CDK-phosphorylated Rad9, which can associate with chromatin by interacting with γH2A and H3 with K79 methylation (not depicted here).Citation25,26 Subsequently Mec1 is activated by Ddc2, Ddc1 of the 9-1-1 complex, and the AAD (ATR-activation domain) of Dpb11.Citation29,30,130 Mec1 then phosphorylates and activates Rad53, which is recruited to DNA lesion sites by its association with phosphorylated Rad9.Citation31,32
Dpb11 and Cut5 likely contribute to replication initiation by other means. For example, a recent study has implicated Dpb11 in delivering another MCM cofactor Cdc45 to MCM, as well as in ssDNA binding, which may facilitate the hand-off of GINS from Dpb11 to MCM.Citation19 The ssDNA binding feature appears to be conserved in TopBP1.Citation20 In addition, an association between Cut5 and the replication factor Mcm10 appears to facilitate the latter to be recruited to replication origins.Citation21 Understanding whether these additional interactions and functions are conserved and how they are temporally controlled will further clarify how various functions of Dpb11 and Cut5 are integrated during replication initiation.
Dpb11 and Cut5 in DNA checkpoint response
Upon replication perturbation or stress, Dpb11 and Cut5 engage 2 DNA damage checkpoint adaptor proteins: Rad9 (S. p. Crb2) and the 9-1-1 complex ().Citation22,23,24,25 Both adaptors transmit signals from the apical checkpoint kinase, Mec1 (S. p. Rad3), to a downstream effector kinase, Rad53 (S. p. Cds1).Citation26 Using budding yeast as an example, Mec1 and its partner Ddc2 localize to DNA lesion sites via interactions with RPA coated single strand DNA (ssDNA) ().Citation27 Mec1 can phosphorylate the Ddc1 subunit of the 9-1-1 complex that localizes to the ssDNA/dsDNA junctions at DNA lesions ().Citation26 Phosphorylated Ddc1 then binds to BRCT3-4 of Dpb11, helping to recruit the latter to DNA lesions.Citation24 In the meantime, S-CDK phosphorylation of Rad9 permits its association with BRCT1-2 of Dpb11 ().Citation25 In this manner, Dpb11 serves as a bridge between Rad9 and 9-1-1. Interestingly, both 9-1-1 and Dpb11 possess a region that can directly stimulate Mec1 kinase activity and is termed AAD (ATR activation domain).Citation28-30 One consequence of Mec1 activation is to hyper-phosphorylate Rad9, which then binds to and recruits Rad53 to the vicinity of Mec1 ().Citation31,32 Once there, Rad53 is phosphorylated and activated by Mec1 (), and diffuses to other cellular locations to phosphorylate a large number of targets, enabling a coordinated response to facilitate DNA replication and segregation.Citation26 In this sophisticated signaling transduction process, Dpb11 contributes to Rad53 activation by linking 2 checkpoint adaptor proteins through its tBRCT pairs and by stimulating the Mec1 kinase activity directly through a different region of the proteins.
A recent structural study has detailed the interactions between the 2 tBRCTs of Cut5 and corresponding checkpoint adaptor proteins.Citation23 It shows that although each BRCT repeat adopts a classical fold, the relative orientation of the 2 repeats within BRCT1-2 (perpendicular) is distinct from that in BRCT3-4 (anti-parallel), and both are distinct from canonical tBRCTs (parallel). Other unique features include the engagement of both BRCT1 and 2 with phospho-peptide, allowing association with a Crb2 dimer, and the interaction of BRCT4, but not BRCT3, with a phospho-peptide of Rad9. Some of these features appear to be conserved in TopBP1,Citation33,34 suggesting a common strategy by which this class of proteins engage ligands. Currently, it is unclear whether the same strategy is used for their interactions with replication factors, namely Sld2/Drc1 and Sld3, though it is worth noting that like Crb2, Sld3 also forms a dimer when in complex with its partner protein Sld7.Citation35 Further structural and in vivo studies will be needed to provide additional understanding of how different ligands are engaged with this family of scaffold proteins.
TopBP1 in replication initiation and checkpoint response, a comparison to its yeast homologs
Compared with Dpb11 and Cut5, 5 additional BRCT domains are present in TopBP1. Consequently, TopBP1 has a larger interactome, only a subset of which is conserved. For example, the N-terminal BRCTs of TopBP1, as seen for its yeast homologs, interact with the Sld3 homolog, Treslin, in a CDK-dependent manner and this interaction is critical for replication initiation.Citation36,37 On the other hand, while TopBP1 binds to RecQL4, a protein containing a Sld2-like domain, this binding is independent of CDK, unlike in yeasts.Citation38-40 During the DNA damage checkpoint response, TopBP1 deploys both conserved and vertebrate-specific interactions. As in yeasts, 2 sets of BRCT domains in TopBP1 bind to phosphorylated 9-1-1 and 53BP1 (S.c. Rad9) and its AAD directly stimulates ATR kinase activity.Citation28,41-44 Additionally, TopBP1 engages with RPA-coated single-stranded DNA and other checkpoint proteins, such as the MRN complex, BACH1/FANCJ, to be recruited to DNA damage site or to stimulate activity.Citation45-48 These additional interactions fit with the more complex requirements of DNA damage responses in higher eukaryotes, which depend on specific situations, such as tissue types, nutrient availability, and chromatin environment.
Dpb11 and homologs function in recombinational repair
Apart from the roles in replisome assembly and checkpoint response as described above, Dpb11 and TopBP1 also promote early steps of recombinational repair and prevent anaphase ultrafine DNA bridges.Citation49-53 Though detailed mechanisms remain to be elucidated, recent studies suggest that TopBP1 executes these functions by interacting with specific partner proteins. One study found that human TopBP1 binds to Polo-like kinase and stimulates phosphorylation of the recombinase Rad51 at serine 14, a modification important for Rad51 recruitment to chromatin and recombination initiation.Citation53 Others studies have shown that TopBP1 aids the resolution of DNA non-disjunctions by recruiting the scaffold SLX4 and Topoisomerase 2A to chromatin.Citation52,54 Furthermore TopBP1 can promote DNA synthesis at under-replicated regions during mitosis by unknown mechanisms.Citation52 Both functions help prevent the formation of DNA bridges and reduce the transmission of damaged DNA to daughter cells. Whether similar strategies are used by Dpb11 and Cut5 remain to be determined.
In summary, Dpb11 and homologs have conserved functions in DNA replication initiation, DNA checkpoint responses, and recombinational repair. They function not only as scaffolding proteins through multiple BRCT domains and linker regions, but also act as DNA binding factors and co-activators for the ATR kinase. While some of these functions have the same underlying mechanisms among Dpb11 homologs, additional mechanisms have evolved as TopBP1 has acquired additional BRCT domains and sequences. A more detailed summary of TopBP1 and its homologs has been described in 2 recent reviews.Citation10,55 It is clear that further elucidation of the protein-protein and protein-DNA interactions involving the conserved versus species-specific BRCT domains and other regions of the proteins will be important for generating a more integrated picture regarding their functions, and for understanding how they help to coordinate different DNA metabolism processes.
Rtt107-like proteins interact with histones and distinct sets of proteins
The budding yeast Rtt107, the fission yeast Brc1, and vertebrate PTIP all contain 6 BRCT domains (BRCT1-6). Their C-terminal BRCT5-6 forms a tBRCT with residues critical for phospho-peptide binding and is the most conserved BRCT pair among these proteins.Citation56-58 Recent studies demonstrate that the BRCT5-6 of all 3 proteins adopt a similar structure and directly bind to γH2A, a phosphorylated form of H2A generated by Mec1 (hATR) or its paralog Tel1 (hATM).Citation59-63 The interaction between γH2A and BRCT5-6 helps recruit Rtt107 and Brc1 to sites where γH2A is enriched, such as DNA double strand breaks (DSBs) and regions behind replication forks ().Citation59,64-66 A similar role may also apply to PTIP, which has been detected at sites of DNA breaks and is required for complete genome replication.Citation61,67,68
Figure 2. The Rtt107 interactome and functions. As described in the text, Rtt107 uses its BRCT5-6 to interact with γH2A for targeting at DNA breaks and regions behind replication forks. The Rtt107 N-terminal region containing BRCT1-4 interact with 3 different complexes as depicted. The main functions conferred by each of the interactions are indicated. Note that in the middle panel, the Rtt101 complex, but not Rtt107, is involved in histone ubiquitination.
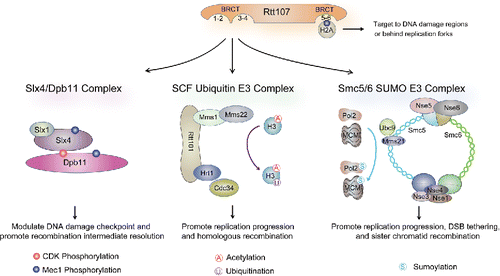
The more N-terminally located BRCT1-4 domains of Rtt107, Brc1, and PTIP appear to be more divergent from each other, though all lack the conserved residues for phospho-peptide binding.Citation5 Thus far, different sets of partners have been found to associate with these domains on PTIP vs. Rtt107, whereas interactors for Brc1 have not been reported. PTIP partners include histone methyltransferases MLL3 and 4, the DNA nuclease Artemis, and the yeast Rad9 homolog 53BP1; these proteins collaborate with PTIP in transcriptional regulation and DSB repair pathway choices.Citation69-73 The N-terminal BRCT domains of Rtt107 interact with a completely different set of proteins, all of which have roles in replication stress. These include the scaffold protein Slx4, a cullin ubiquitin E3 complex composed of Rtt101, Mms1, and Mms22, and an 8-subunit SUMO E3 complex, called Smc5/6 ().Citation56-58,74 Unlike Dpb11, which simultaneously interacts with 2 partners using its 2 tBRCT domains, Rtt107 forms different complexes with its partners.Citation75 Currently it is unclear how Rtt107 achieves such binding exclusivity and what the biological consequences of this mode of interaction are. However recent studies suggest that Rtt107, in collaboration with its partners, serves as a replication accessory protein that influences DNA repair, replisome progression and checkpoint signaling.Citation56,57,75-77 In these capacities, these proteins are critical for genome stability, and studies of their functions have provided important insights into how replication accessory proteins facilitate genome duplication. The remaining sections describe new developments regarding Rtt107 and its partners.
The Rtt107 and Slx4 association and its functions in conjunction with Dpb11
Slx4 and the associated Slx1 protein interact with Rtt107 constitutively, and this interaction appears to be required for Mec1-mediated phosphorylation of Slx4 and Rtt107 under replication stress and DNA damage situations ().Citation57,78 Interestingly, upon Mec1-and S-CDK-mediated phosphorylation of Slx4, Slx4 and Rtt107 become associated with Dpb11 ().Citation74 The latter modification, particularly at Serine 486 of Slx4, contributes to its interaction with Dpb11.Citation77,79 These studies also suggest that phosphorylated Slx4 likely directly binds to Dpb11, while Rtt107 stabilizes this interaction. The Slx4-Dpb11 interaction is conserved as phosphorylated human SLX4 also associates with TopBP1.Citation79
The complex composed of Rtt107, Slx4, and Dpb11 can diminish Dpb11 interaction with Rad9, thus reducing Rad53 checkpoint activation, a process termed checkpoint dampening ().Citation77,80,81 An underlying mechanism may be that Slx4 and Rad9 compete for binding to BRCT1-2 of Dpb11, though Slx4 can also associate with BRCT3-4 of Dpb11.Citation77,79 It appears that BRCT1-2 and BRCT3-4 of Dpb11 can engage Slx4 and the 9-1-1 complex.Citation77,79,80 As such, Dpb11 can bridge Slx4 and 9-1-1 (). Future biochemical studies dissecting the precise manner by which Rtt107, Slx4, Dpb11, and 9-1-1 associate will provide in-depth understanding on how their association dampens Rad9 functions. As another way to reduce Rad9 function, Rtt107 also disfavors the Rad9-γH2A association, an interaction that helps recruit Rad9 to damaged chromatin ().Citation80,82 It is possible that the affinity of the tBRCT of Rad9 toward γH2A is less than that of Rtt107, a hypothesis worthy of additional testing. Despite this potential difference, BRCT5-6 of Rtt107 and tBRCT of Rad9 are interchangeable and the domain swapped constructs can partially support the functions of either protein.Citation83
Figure 3. The Rtt107-Slx4 interaction and its functions. Three kinases promote specific interactions as depicted. Several functions of the Rtt107-Slx4-Dpb11 complex are indicated. See text for details.
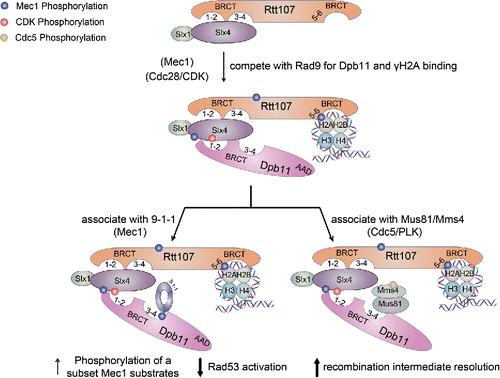
Taking into account the newly found role of Dpb11 in checkpoint dampening and its classical role in checkpoint activation, how might Dpb11 juggle these 2 functions? This is related to a more general issue of how cells transit from a “pro-checkpoint” state, established upon initial exposure to stress, to an “anti-checkpoint” state following alleviation of stress. One clue has emerged from studies showing that Dpb11, Slx4 and Rtt107 can uncouple Mec1 activity from that of Rad53.Citation77,84,65 The Slx4-Rtt107-Dpb11 complex can actually increase Mec1-mediated phosphorylation of Rtt107, H2A and Dpb11, presumably due to the Mec1-activation function of Dpb11 ().Citation65 Such selective increase of Mec1 activity in principle can reduce Rad53 functions, because Rtt107 phosphorylation disfavors Rad53 activation (see above). As such, Slx4-Rtt107-Dpb11 plays a dual role in tuning down Rad53 activity: through dampening Rad9 and through selective activation of Mec1. It is likely that the formation of Dpb11-Slx4-Rtt107 complex marks an important turning point for Rad53 inactivation. Interestingly, Mec1 and Rad53 uncoupling also occurs in normal S phase, where Mec1, but not Rad53, is highly active.Citation85 It will be interesting to explore whether this uncoupling also requires the complex containing Slx4-Rtt107-Dpb11.
A second role of the Slx4-Dpb11 interaction is to facilitate the removal of recombination intermediates (RIs).Citation79 A recent study found that the Slx4-Dpb11-Rtt107 complex interacts with the RI resolvase Mus81-Mms4 only in G2/M phase of the cell cycle ().Citation79 Unlike mammalian SLX4, which binds directly to Mus81-Mms4 homologs,Citation86 yeast Slx4 interacts with this nuclease through Dpb11 ().Citation79 In addition, Mms4 phosphorylation by the polo-like kinase Cdc5 is required for Mms4-Dpb11 association, although whether this is through a direct interaction is not yet known.Citation79 It remains to be ascertained how Slx4-Dpb11 influences Mus81-Mms4 functions. The physical interaction between Slx4-Dpb11 and Mus81-Mms4 suggests a direct effect, perhaps through regulation of enzymatic activities or RI association. In addition, since Cdc5-mediated Mms4 phosphorylation is disfavored by Rad53, suppression of Rad53 functions by Slx4-Dpb11 may in turn boost Mms4 phosphorylation.Citation79,87-89 These two models are not mutually exclusive and further in vivo and in vitro experiments should provide more insight into how the association of Slx4 and Dpb11 can influence RI resolution.
Roles of Rtt107 when partnered with Rtt101 cullin ubiquitin ligase
Rtt107 also interacts with the SCF ubiquitin ligase composed of the cullin subunit Rtt101 (mammalian Cul4) and the substrate adaptor subunits Mms1 and Mms22 ().Citation56,90-92 Within this ubiquitin ligase (referred to as the Rtt101 E3 complex), Mms1 serves as a bridge between Rtt101 and Mms22 ().Citation91,92 Mms22 interacts with Rtt107 directly and this interaction requires the region of Rtt107 encompassing its N-terminal BRCTs.Citation75,91,92 Several lines of evidence suggest that the Rtt101 complex facilitates histone H3 modification and nucleosome assembly. During replication, newly synthesized histone H3, in complex with H4 and the histone chaperone Asf1, is acetylated at lysine 56 (H3K56ac) by Rtt109.Citation93,94 The modified H3 is then ubiquitinated by the Rtt101 complex at 3 lysines, weakening its association with Asf1 and allowing its transfer to downstream histone chaperones for deposition on DNA.Citation95,96 As H3 lysine 56 is located at the DNA entry site of the nucleosome, H3K56ac favors nucleosome removal, a step thought to enable recombinational repair behind replication forks.Citation97 Indeed, removing Rtt109 or the Rtt101 complex impairs recombinational repair and replication, and sensitizes cells to replication stress.Citation93,94,98-101 Once replication is completed, histone deacetylases Hst3 and Hst4 remove the acetyl group from H3K56ac to establish a more stable nucleosome state.Citation102-104 Cells lacking Hst3 and Hst4 or containing the acetylation mimetic H3 mutation (H3K56Q) exhibit defects such as persistent checkpoint and sensitivity to higher temperature and genotoxins.Citation102,103,105,106 The fact that H3K56 acetylation and deacetylation are both important suggests that the 2 states must be carefully regulated. Indeed, the removal of Rtt109 or the Rtt101 complex suppresses several hst3Δ hst4Δ defects.Citation93,94,107,108
The role of Rtt107 in the above process is not completely understood. Like Rtt109 and the Rtt101 complex, Rtt107 loss also suppresses hst3Δ hst4Δ defects; however, Rtt107 does not affect H3 ubiquitination.Citation96,107 It is therefore thought that Rtt107 acts downstream of Rtt109 and Rtt101. The close relationship among these factors is also evidenced in their chromatin association. Stable association of Rtt107 with chromatin requires Rtt109 and Rtt101; conversely, Rtt101 chromatin association requires Rtt107 and Rtt109.Citation90 Considering that the E3 has multiple substrates, Rtt107 may collaborate with it in several functions.Citation91,109-111 Our recent findings suggest that when cells are under replication stress, Rtt107 and the Rtt101 complex favor the synthesis of large replicons or regions far from fired origins that occurs in the late stages of S phase.Citation75 This conclusion is consistent with a Xenopus study that implicates PTIP (or Swift) in later stages of replication.Citation67 Another study implicates the Rtt101 complex in regulating recombinational repair during replication stress.Citation112 This study and our recent work found that removal of Mrc1, which suppresses late origin firing and recombination, rescues replication stress sensitivity of mutants of Rtt107 and/or the Rtt101 complex. The suppression could be due to the combined effects of increasing recombination and reducing large replicons.Citation75,112 It is likely that Rtt107, in association with the Rtt101 E3 complex, promotes replisome progression and recombinational repair at stalled replication forks, ultimately contributing to replication recovery and large replicon synthesis. Future studies will be required to test these models and examine the molecular mechanisms by which H3 modification and chromatin assembly influence these processes.
The roles of Rtt107 in collaboration with Smc5/6 SUMO ligase
Among the 3 partners associated with the N-terminal BRCTs of Rtt107, the roles of the Smc5/6 complex are the least understood and the most perplexing. The core of the complex includes the Smc5 and Smc6 proteins that can adopt 3-partite configurations common to SMC proteins ().Citation113-115 The Smc5-6 proteins use their globular head and hinge domains and long coiled coil arm to interact with 6 Non-SMC elements, Nse1-6: the Nse1-3-4 subcomplex interacts with the Smc5/6 head regions, the Nse5-6 complex with the Smc5/6 hinge regions, and Nse2 (or Mms21) binds to the arm of Smc5.Citation114-117 Besides the ATPase activity of the Smc5/6 head regions, thought to regulate the loading of Smc5/6 onto chromatin,Citation113,118,119 the SUMO E3 ligase activity of the Mms21 subunit is also conserved.Citation115,120,121
Rtt107 appears to interact with the Smc5/6 complex through the Nse6 subunit based on yeast 2-hybrid analysis.Citation122 As mutants of the Smc5/6 complex do not show increased Rad53 levels or defects associated with reduced H3K56ac levels, its association with Rtt107 unlikely affects checkpoint dampening or the histone modification functions assigned to Slx4 and the Rtt101 complex, respectively.Citation75 This conclusion concurs with the physical separation of the Rtt107-Smc5/6 complex from that of Rtt107-Slx4 and Rtt107-Rtt101.Citation75 One function of the Rtt107 and Smc5/6 association appears to DSB targeting ().Citation64,76,122 Their inter-dependence in DSB localization implicates both protein entities in recombinational repair and DSB tethering to the nuclear envelope Citation56,123 ().
The Rtt107 and Smc5/6 interaction also contributes to replication under replication stress (). They both affect the sumoylation of DNA polymerase epsilon and MCM helicase subunits ().Citation75 As Rtt107 interacts with these substrates, it may facilitate Smc5/6-mediated transfer of SUMO to these proteins.Citation75 Whether and how the observed sumoylation events affect replication remains to be addressed, though several lines of evidence suggest this may be related to the replication of large replicons. For example, replication stress sensitivity of smc5/6 mutants is suppressed by reducing the size of large replicons achieved by firing hundreds of repressed origins.Citation75 We envision that Rtt107 partners with both Smc5/6 and Rtt101 complexes to promote replication progression using SUMO and ubiquitin modifications, respectively. Testing this model will likely shed light on the roles of Rtt107 and the 2 E3s in replication.
Conclusions and perspectives
Studies of multi-BRCT scaffolds and their partners have elucidated the various strategies used by cells to duplicate the genome. In the above sections, we have mainly summarized recent findings detailing the roles of Dpb11 and Rtt107, and some of their interactors. While both contribute to DNA replication, Dpb11 plays a critical role in initiation and Rtt107 promotes progression in collaboration with its ubiquitin and SUMO E3 partners. On the other hand, Dpb11 and Rtt107 cooperate to prevent checkpoint over-activation. Finally, both Dpb11 and Rtt107 regulate DNA repair, but through different mechanisms. While these findings highlight their importance in genome maintenance and the complexity of their actions, many mechanistic questions remain to be addressed as exemplified above. Answering these questions will provide insight into how Dpb11 and Rtt107 execute diverse, coordinated and antagonistic functions in genome maintenance.
As described earlier, a good degree of functional conservation has been observed for Dpb11 and homologs. In comparison, there appears to be fewer similarities between Rtt107 and its homologs, though some common features have been noted. In particular, BRCT5-6 targets these proteins to γH2A at DNA lesions. In addition, Brc1 acts in the same pathway as Mms22Citation124 and its overexpression suppresses the DNA damage sensitivity of smc6 mutants.Citation125 Furthermore, like Rtt107, Brc1 and PTIP are implicated in replication fork stability or replication stress responses, and Brc1 influences nuclease pathways.Citation66-68,126,127 Despite these similarities, differences have been noted among Rtt107, Brc1 and PTIP. For example, unlike Rtt107, Brc1 does not have a checkpoint-dampening function and the genetic interaction profiles of Brc1 and Rtt107 mutants are quite different.Citation66,128 Moreover, unlike the positive influence of Rtt107 in homologous recombination, PTIP disfavors this repair pathway through its interaction with 53BP1 and the Artemis nucleases.Citation71,73 These diverse functions may have evolved to suit the specific needs of each particular organism. Future studies in multiple organisms will better elucidate the similarities and differences among these multi-function proteins, and their roles in genome maintenance and human diseases.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Nalini Dhingra for comments on the manuscript.
Funding
Work in the Zhao lab is supported by the US National Institutes of Health grant GM080670.
References
- Tanaka S, Araki H. Helicase activation and establishment of replication forks at chromosomal origins of replication. Cold Spring Harb Symp Quant Biol 2013; 5:a010371; http://dx.doi.org/10.1101/cshperspect.a010371
- Chang DJ, Cimprich KA. DNA damage tolerance: when it's OK to make mistakes. Nat Chem Biol 2009; 5:82-90; PMID:19148176; http://dx.doi.org/10.1038/nchembio.139
- Yeeles JTP, Poli J, Marians KJ, Pasero P. Rescuing stalled or damaged replication forks. Cold Spring Harb Symp Quant Biol 2013; 5:a012815-a; PMID:23637285
- Koonin EV, Altschul SF, Bork P. BRCA1 protein products … Functional motifs. Nat Genet 1996; 13:266-8; PMID:8673121; http://dx.doi.org/10.1038/ng0796-266
- Bork P, Hofmann K, Bucher P, Neuwald AF, Altschul SF, Koonin EV. A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J 1997; 11:68-76; PMID:9034168
- Gerloff DL, Woods NT, Farago AA, Monteiro ANA. BRCT domains: A little more than kin, and less than kind. FEBS Lett 2012; 586:2711-6; PMID:22584059; http://dx.doi.org/10.1016/j.febslet.2012.05.005
- Leung CCY, Glover JNM. BRCT domains: easy as one, two, three. Cell Cycle 2011; 10:2461-70; PMID:21734457; http://dx.doi.org/10.4161/cc.10.15.16312
- Yu X, Chini C, He M, Mer G, Chen J. The BRCT domain is a phospho-protein binding domain. Science 2003; 302:639-42; PMID:14576433; http://dx.doi.org/10.1126/science.1088753
- Manke IA. BRCT Repeats as phosphopeptide-binding modules involved in protein targeting. Science 2003; 302:636-9; PMID:14576432; http://dx.doi.org/10.1126/science.1088877
- Wardlaw CP, Carr AM, Oliver AW. TopBP1: A BRCT-scaffold protein functioning in multiple cellular pathways. DNA Repair 2014; 22:165-74; PMID:25087188; http://dx.doi.org/10.1016/j.dnarep.2014.06.004
- Fukuura M, Nagao K, Obuse C, Takahashi TS, Nakagawa T, Masukata H. CDK promotes interactions of Sld3 and Drc1 with Cut5 for initiation of DNA replication in fission yeast. Mol Cell Biol 2011; 22:2620-33; http://dx.doi.org/10.1091/mbc.E10-12-0995
- Noguchi E, Shanahan P, Noguchi C, Russell P. CDK phosphorylation of Drc1 regulates DNA replication in fission yeast. Curr Biol 2002; 12:599-605; PMID:11937031; http://dx.doi.org/10.1016/S0960-9822(02)00739-X
- Tak Y-S, Tanaka Y, Endo S, Kamimura Y, Araki H. A CDK-catalysed regulatory phosphorylation for formation of the DNA replication complex Sld2-Dpb11. EMBO J 2006; 25:1987-96; PMID:16619031; http://dx.doi.org/10.1038/sj.emboj.7601075
- Zegerman P, Diffley JFX. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature 2007; 445:281-5; PMID:17167417; http://dx.doi.org/10.1038/nature05432
- Tanaka S, Komeda Y, Umemori T, Kubota Y, Takisawa H, Araki H. Efficient initiation of DNA replication in eukaryotes requires Dpb11/TopBP1-GINS interaction. Mol Cell Biol 2013; 33:2614-22; PMID:23629628; http://dx.doi.org/10.1128/MCB.00431-13
- Muramatsu S, Hirai K, Tak Y-S, Kamimura Y, Araki H. CDK-dependent complex formation between replication proteins Dpb11, Sld2, Pol (epsilon}, and GINS in budding yeast. Genes Dev 2010; 24:602-12; PMID:20231317; http://dx.doi.org/10.1101/gad.1883410
- Deegan TD, Yeeles JT, Diffley JF. Phosphopeptide binding by Sld3 links Dbf4-dependent kinase to MCM replicative helicase activation. EMBO J 2016; 35:961-73; PMID:26912723; http://dx.doi.org/10.15252/embj.201593552
- Bruck I, Kaplan DL. The Dbf4-Cdc7 kinase promotes Mcm2-7 ring opening to allow for single-stranded DNA extrusion and helicase assembly. J Biol Chem 2015; 290:1210-21; PMID:25471369; http://dx.doi.org/10.1074/jbc.M114.608232
- Dhingra N, Bruck I, Smith S, Ning B, Kaplan DL. Dpb11 protein helps control assembly of the Cdc45.Mcm2-7.GINS replication fork helicase. J Biol Chem 2015; 290:7586-601; PMID:25659432; http://dx.doi.org/10.1074/jbc.M115.640383
- Choi JH, Lindsey-Boltz LA, Sancar A. Cooperative activation of the ATR checkpoint kinase by TopBP1 and damaged DNA. Nucleic Acids Res 2009; 37:1501-9; PMID:19139065; http://dx.doi.org/10.1093/nar/gkn1075
- Taylor M, Moore K, Murray J, Aves SJ, Price C. Mcm10 interacts with Rad4/Cut5(TopBP1) and its association with origins of DNA replication is dependent on Rad4/Cut5(TopBP1). DNA Repair (Amst) 2011; 10:1154-63; PMID:21945095; http://dx.doi.org/10.1016/j.dnarep.2011.09.001
- Saka Y, Esashi F, Matsusaka T, Mochida S, Yanagida M. Damage and replication checkpoint control in fission yeast is ensured by interactions of Crb2, a protein with BRCT motif, with Cut5 and Chk1. Genes Dev 1997; 11:3387-400; PMID:9407031; http://dx.doi.org/10.1101/gad.11.24.3387
- Qu M, Rappas M, Wardlaw CP, Garcia V, Ren JY, Day M, Carr AM, Oliver AW, Du LL, Pearl LH. Phosphorylation-dependent assembly and coordination of the DNA damage checkpoint apparatus by Rad4(TopBP1). Mol Cell 2013; 51:723-36; PMID:24074952; http://dx.doi.org/10.1016/j.molcel.2013.08.030
- Puddu F, Granata M, Di Nola L, Balestrini A, Piergiovanni G, Lazzaro F, Giannattasio M, Plevani P, Muzi-Falconi M. Phosphorylation of the budding yeast 9-1-1 complex is required for Dpb11 function in the full activation of the UV-induced DNA damage checkpoint. Mol Cell Biol 2008; 28:4782-93; PMID:18541674; http://dx.doi.org/10.1128/MCB.00330-08
- Pfander B, Diffley JFX. Dpb11 coordinates Mec1 kinase activation with cell cycle-regulated Rad9 recruitment. EMBO J 2011; 30:4897-907; PMID:21946560; http://dx.doi.org/10.1038/emboj.2011.345
- Hustedt N, Gasser S, Shimada K. Replication checkpoint: tuning and coordination of replication forks in S phase. Genes 2013; 4:388-434; PMID:24705211; http://dx.doi.org/10.3390/genes4030388
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 2003; 300:1542-8; PMID:12791985; http://dx.doi.org/10.1126/science.1083430
- Mordes DA, Glick GG, Zhao R, Cortez D. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev 2008; 22:1478-89; PMID:18519640; http://dx.doi.org/10.1101/gad.1666208
- Navadgi-Patil VM, Burgers PM. Yeast DNA replication protein Dpb11 activates the Mec1/ATR checkpoint kinase. J Biol Chem 2008; 283:35853-9; PMID:18922789; http://dx.doi.org/10.1074/jbc.M807435200
- Navadgi-Patil VM, Burgers PM. The unstructured C-terminal tail of the 9-1-1 clamp subunit Ddc1 activates Mec1/ATR via two distinct mechanisms. Mol Cell 2009; 36:743-53; PMID:20005839; http://dx.doi.org/10.1016/j.molcel.2009.10.014
- Gilbert CS, Green CM, Lowndes NF. Budding yeast Rad9 is an ATP-dependent Rad53 activating machine. Mol Cell 2001; 8:129-36; PMID:11511366; http://dx.doi.org/10.1016/S1097-2765(01)00267-2
- Sweeney FD, Yang F, Chi A, Shabanowitz J, Hunt DF, Durocher D. Saccharomyces cerevisiae Rad9 acts as a Mec1 adaptor to allow Rad53 activation. Curr Biol 2005; 15:1364-75; PMID:16085488; http://dx.doi.org/10.1016/j.cub.2005.06.063
- Huo YG, Bai L, Xu M, Jiang T. Crystal structure of the N-terminal region of human Topoisomerase IIbeta binding protein 1. Biochem Biophys Res Commun 2010; 401:401-5; PMID:20858457; http://dx.doi.org/10.1016/j.bbrc.2010.09.066
- Rappas M, Oliver AW, Pearl LH. Structure and function of the Rad9-binding region of the DNA-damage checkpoint adaptor TopBP1. Nucleic Acids Res 2011; 39:313-24; PMID:20724438; http://dx.doi.org/10.1093/nar/gkq743
- Itou H, Shirakihara Y, Araki H. The quaternary structure of the eukaryotic DNA replication proteins Sld7 and Sld3. Acta Crystallogr D Biol Crystallogr 2015; 71:1649-56; PMID:26249346; http://dx.doi.org/10.1107/S1399004715010457
- Kumagai A, Shevchenko A, Shevchenko A, Dunphy WG. Treslin collaborates with TopBP1 in triggering the initiation of DNA replication. Cell 2010; 140:349-59; PMID:20116089; http://dx.doi.org/10.1016/j.cell.2009.12.049
- Boos D, Sanchez-Pulido L, Rappas M, Pearl LH, Oliver AW, Ponting CP, Diffley JF. Regulation of DNA replication through Sld3-Dpb11 interaction is conserved from yeast to humans. Curr Biol 2011; 21:1152-7; PMID:21700459; http://dx.doi.org/10.1016/j.cub.2011.05.057
- Sangrithi MN, Bernal JA, Madine M, Philpott A, Lee J, Dunphy WG, Venkitaraman AR. Initiation of DNA replication requires the RECQL4 protein mutated in Rothmund-Thomson syndrome. Cell 2005; 121:887-98; PMID:15960976; http://dx.doi.org/10.1016/j.cell.2005.05.015
- Matsuno K, Kumano M, Kubota Y, Hashimoto Y, Takisawa H. The N-terminal noncatalytic region of Xenopus RecQ4 is required for chromatin binding of DNA polymerase alpha in the initiation of DNA replication. Mol Cell Biol 2006; 26:4843-52; PMID:16782873; http://dx.doi.org/10.1128/MCB.02267-05
- Ohlenschlager O, Kuhnert A, Schneider A, Haumann S, Bellstedt P, Keller H, Saluz HP, Hortschansky P, Hanel F, Grosse F, et al. The N-terminus of the human RecQL4 helicase is a homeodomain-like DNA interaction motif. Nucleic Acids Res 2012; 40:8309-24; PMID:22730300; http://dx.doi.org/10.1093/nar/gks591
- Greer DA, Besley BD, Kennedy KB, Davey S. hRad9 rapidly binds DNA containing double-strand breaks and is required for damage-dependent topoisomerase II beta binding protein 1 focus formation. Cancer Res 2003; 63:4829-35; PMID:12941802
- Delacroix S, Wagner JM, Kobayashi M, Yamamoto K, Karnitz LM. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev 2007; 21:1472-7; PMID:17575048; http://dx.doi.org/10.1101/gad.1547007
- Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell 2006; 124:943-55; PMID:16530042; http://dx.doi.org/10.1016/j.cell.2005.12.041
- Cescutti R, Negrini S, Kohzaki M, Halazonetis TD. TopBP1 functions with 53BP1 in the G1 DNA damage checkpoint. EMBO J 2010; 29:3723-32; PMID:20871591; http://dx.doi.org/10.1038/emboj.2010.238
- Acevedo J, Yan S, Michael WM. Direct Binding to Replication Protein A (RPA)-coated Single-stranded DNA Allows Recruitment of the ATR Activator TopBP1 to Sites of DNA Damage. J Biol Chem 2016; 291:13124-31; PMID:27129245; http://dx.doi.org/10.1074/jbc.M116.729194
- Liu S, Shiotani B, Lahiri M, Marechal A, Tse A, Leung CC, Glover JN, Yang XH, Zou L. ATR autophosphorylation as a molecular switch for checkpoint activation. Mol Cell 2011; 43:192-202; PMID:21777809; http://dx.doi.org/10.1016/j.molcel.2011.06.019
- Leung CC, Gong Z, Chen J, Glover JN. Molecular basis of BACH1/FANCJ recognition by TopBP1 in DNA replication checkpoint control. J Biol Chem 2011; 286:4292-301; PMID:21127055; http://dx.doi.org/10.1074/jbc.M110.189555
- Duursma AM, Driscoll R, Elias JE, Cimprich KA. A Role for the MRN Complex in ATR Activation via TOPBP1 Recruitment. Mol Cell 2013; 50:116-22; PMID:23582259; http://dx.doi.org/10.1016/j.molcel.2013.03.006
- Germann SM, Schramke V, Pedersen RT, Gallina I, Eckert-Boulet N, Oestergaard VH, Lisby M. TopBP1/Dpb11 binds DNA anaphase bridges to prevent genome instability. J Cell Biol 2013; 204:45-59; PMID:24379413; http://dx.doi.org/10.1083/jcb.201305157
- Ogiwara H, Ui A, Onoda F, Tada S, Enomoto T, Seki M. Dpb11, the budding yeast homolog of TopBP1, functions with the checkpoint clamp in recombination repair. Nucleic Acids Res 2006; 34:3389-98; PMID:16840526; http://dx.doi.org/10.1093/nar/gkl411
- Germann SM, Oestergaard VH, Haas C, Salis P, Motegi A, Lisby M. Dpb11/TopBP1 plays distinct roles in DNA replication, checkpoint response and homologous recombination. DNA Repair 2011; 10:210-24; PMID:21130053; http://dx.doi.org/10.1016/j.dnarep.2010.11.001
- Pedersen RT, Kruse T, Nilsson J, Oestergaard VH, Lisby M. TopBP1 is required at mitosis to reduce transmission of DNA damage to G1 daughter cells. J Cell Biol 2015; 210:565-82; PMID:26283799; http://dx.doi.org/10.1083/jcb.201502107
- Moudry P, Watanabe K, Wolanin KM, Bartkova J, Wassing IE, Watanabe S, Strauss R, Troelsgaard Pedersen R, Oestergaard VH, Lisby M, et al. TOPBP1 regulates RAD51 phosphorylation and chromatin loading and determines PARP inhibitor sensitivity. J Cell Biol 2016; 212:281-8; PMID:26811421; http://dx.doi.org/10.1083/jcb.201507042
- Broderick R, Nieminuszczy J, Blackford AN, Winczura A, Niedzwiedz W. TOPBP1 recruits TOP2A to ultra-fine anaphase bridges to aid in their resolution. Nat Comm 2015; 6:6572; http://dx.doi.org/10.1038/ncomms7572
- Liu Y, Smolka MB. TOPBP1 takes RADical command in recombinational DNA repair. J Cell Biol 2016; 212:263-6; PMID:26811424; http://dx.doi.org/10.1083/jcb.201601028
- Chin J, Bashkirov V, Heyer W, Romesberg F. Esc4/Rtt107 and the control of recombination during replication. DNA Repair 2006; 5:618-28; PMID:16569515; http://dx.doi.org/10.1016/j.dnarep.2006.02.005
- Roberts T, Kobor M, Bastin-Shanower S, Ii M, Horte S, Gin J, Emili A, Rine J, Brill S, Brown G. Slx4 regulates DNA damage checkpoint-dependent phosphorylation of the BRCT domain protein Rtt107/Esc4. Mol Biol Cell 2006; 17:539-48; PMID:16267268; http://dx.doi.org/10.1091/mbc.E05-08-0785
- Zappulla DC, Maharaj ASR, Connelly JJ, Jockusch RA, Sternglanz R. Rtt107/Esc4 binds silent chromatin and DNA repair proteins using different BRCT motifs. BMC Mol Biol 2006; 7:40; PMID:17094803; http://dx.doi.org/10.1186/1471-2199-7-40
- Williams JS, Williams RS, Dovey CL, Guenther G, Tainer JA, Russell P. γH2A binds Brc1 to maintain genome integrity during S-phase. EMBO J 2010; 29:1136-48; PMID:20094029; http://dx.doi.org/10.1038/emboj.2009.413
- Li X, Liu K, Li F, Wang J, Huang H, Wu J, Shi Y. Structure of the C-terminal tandem BRCT repeats of Rtt107 reveals a critical role in the interaction with H2A during DNA damage repair. J Biol Chem 2012; 287:9137-46; PMID:22262834; http://dx.doi.org/10.1074/jbc.M111.311860
- Manke IA, Lowery DM, Nguyen A, Yaffe MB. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science 2003; 302:636-9; PMID:14576432; http://dx.doi.org/10.1126/science.1088877
- Yan W, Shao Z, Li F, Niu L, Shi Y, Teng M, Li X. Structural basis of γH2AX recognition by human PTIP BRCT5-BRCT6 domains in the DNA damage response pathway. FEBS Lett 2011; 585:3874-9; PMID:22064073; http://dx.doi.org/10.1016/j.febslet.2011.10.045
- Downs JA, Lowndes NF, Jackson SP. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature 2000; 408:1001-4; PMID:11140636; http://dx.doi.org/10.1038/35050000
- Leung GP, Brown JAR, Glover JNM, Kobor MS. Rtt107 BRCT domains act as a targeting module in the DNA damage response. DNA Repair 2016; 37:22-32; PMID:26641499; http://dx.doi.org/10.1016/j.dnarep.2015.10.007
- Balint A, Kim T, Gallo D, Cussiol JR, Bastos de Oliveira FM, Yimit A, Ou J, Nakato R, Gurevich A, Shirahige K, et al. Assembly of Slx4 signaling complexes behind DNA replication forks. EMBO J 2015; 34:2182-97; PMID:26113155; http://dx.doi.org/10.15252/embj.201591190
- Mejía-Ramírez E, Limbo O, Langerak P, Russell P. Critical function of γH2A in S-phase. PLoS Genet 2015; 11:e1005517; http://dx.doi.org/10.1371/journal.pgen.1005517
- Gohler T, Munoz I, Rouse J, Blow J. PTIP/Swift is required for efficient PCNA ubiquitination in response to DNA damage. DNA Repair 2008; 7:775-87; PMID:18353733; http://dx.doi.org/10.1016/j.dnarep.2008.02.001
- Munoz IM, Jowsey PA, Toth R, Rouse J. Phospho-epitope binding by the BRCT domains of hPTIP controls multiple aspects of the cellular response to DNA damage. Nucleic Acids Res 2007; 35:5312-22; PMID:17690115; http://dx.doi.org/10.1093/nar/gkm493
- Patel SR, Kim D, Levitan I, Dressler GR. The BRCT-domain containing protein PTIP links PAX2 to a histone H3, lysine 4 methyltransferase complex. Dev Cell 2007; 13:580-92; PMID:17925232; http://dx.doi.org/10.1016/j.devcel.2007.09.004
- Jowsey P, Doherty A, Rouse J. Human PTIP facilitates ATM-mediated activation of p53 and promotes cellular resistance to ionizing radiation. J Biol Chem 2004; 279:55562-9; PMID:15456759; http://dx.doi.org/10.1074/jbc.M411021200
- Wang J, Aroumougame A, Lobrich M, Li Y, Chen D, Chen J, Gong Z. PTIP associates with Artemis to dictate DNA repair pathway choice. Genes Dev 2014; 28:2693-8; PMID:25512557; http://dx.doi.org/10.1101/gad.252478.114
- Cho Y-W, Hong T, Hong S, Guo H, Yu H, Kim D, Guszczynski T, Dressler GR, Copeland TD, Kalkum M, et al. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J Biol Chem 2007; 282:20395-406; PMID:17500065; http://dx.doi.org/10.1074/jbc.M701574200
- CallEn E, Di Virgilio M, Kruhlak MJ, Nieto-Soler M, Wong N, Chen H-T, Faryabi RB, Polato F, Santos M, Starnes LM, et al. 53BP1 mediates productive and mutagenic DNA repair through distinct phosphoprotein interactions. Cell 2013; 153:1266-80; PMID:23727112; http://dx.doi.org/10.1016/j.cell.2013.05.023
- Ohouo PY, Bastos de Oliveira FM, Almeida BS, Smolka MB. DNA damage signaling recruits the Rtt107-Slx4 scaffolds via Dpb11 to mediate replication stress response. Mol Cell 2010; 39:300-6; PMID:20670896; http://dx.doi.org/10.1016/j.molcel.2010.06.019
- Hang LE, Peng J, Tan W, Szakal B, Menolfi D, Sheng Z, Lobachev K, Branzei D, Feng W, Zhao X. Rtt107 is a multi-functional scaffold supporting replication progression with partner SUMO and ubiquitin ligases. Mol Cell 2015; 60:268-79; PMID:26439300; http://dx.doi.org/10.1016/j.molcel.2015.08.023
- Ullal P, Vilella-Mitjana F, Jarmuz A, Aragon L. Rtt107 phosphorylation promotes localisation to DNA double-stranded breaks (DSBs) and recombinational repair between sister chromatids. PLoS One 2011; 6:e20152; PMID:21647453; http://dx.doi.org/10.1371/journal.pone.0020152
- Ohouo PY, de Oliveira FMB, Liu Y, Ma CJ, Smolka MB. DNA-repair scaffolds dampen checkpoint signalling by counteracting the adaptor Rad9. Nature 2012; 493:120-4; PMID:23160493; http://dx.doi.org/10.1038/nature11658
- Rouse J. Esc4p, a new target of Mec1p (ATR), promotes resumption of DNA synthesis after DNA damage. EMBO J 2004; 23:1188-97; PMID:14988729; http://dx.doi.org/10.1038/sj.emboj.7600129
- Gritenaite D, Princz LN, Szakal B, Bantele SCS, Wendeler L, Schilbach S, Habermann BH, Matos J, Lisby M, Branzei D, et al. A cell cycle-regulated Slx4-Dpb11 complex promotes the resolution of DNA repair intermediates linked to stalled replication. Genes Dev 2014; 28:1604-19; PMID:25030699; http://dx.doi.org/10.1101/gad.240515.114
- Cussiol JR, Jablonowski CM, Yimit A, Brown GW, Smolka MB. Dampening DNA damage checkpoint signalling via coordinated BRCT domain interactions. EMBO J 2015; 34:1704-17; PMID:25896509; http://dx.doi.org/10.15252/embj.201490834
- Dibitetto D, Ferrari M, Rawal CC, Balint A, Kim T, Zhang Z, Smolka MB, Brown GW, Marini F, Pellicioli A. Slx4 and Rtt107 control checkpoint signalling and DNA resection at double-strand breaks. Nucleic Acids Res 2015; 44:669-82; PMID:26490958; http://dx.doi.org/10.1093/nar/gkv1080
- Hammet A, Magill C, Heierhorst J, Jackson SP. Rad9 BRCT domain interaction with phosphorylated H2AX regulates the G1 checkpoint in budding yeast. EMBO Rep 2007; 8:851-7; PMID:17721446; http://dx.doi.org/10.1038/sj.embor.7401036
- Leung GP, Brown JA, Glover JN, Kobor MS. Rtt107 BRCT domains act as a targeting module in the DNA damage response. DNA Repair (Amst) 2016; 37:22-32; PMID:26641499; http://dx.doi.org/10.1016/j.dnarep.2015.10.007
- Cussiol JR, Dibitetto D, Pellicioli A, Smolka MB. Slx4 scaffolding in homologous recombination and checkpoint control: lessons from yeast. Chromosoma 2016; PMID:27165041
- de Oliveira FMB, Kim D, Cussiol JR, Das J, Jeong MC, Doerfler L, Schmidt KH, Yu H, Smolka MB. Phosphoproteomics reveals distinct modes of Mec1/ATR signaling during DNA replication. Mol Cell 2015; 57:1124-32; PMID:25752575; http://dx.doi.org/10.1016/j.molcel.2015.01.043
- Wyatt HDM, Sarbajna S, Matos J, West SC. Coordinated actions of SLX1-SLX4 and MUS81-EME1 for Holliday junction resolution in human cells. Mol Cell 2013; 52:234-47; PMID:24076221; http://dx.doi.org/10.1016/j.molcel.2013.08.035
- Szakal B, Branzei D. Premature Cdk1/Cdc5/Mus81 pathway activation induces aberrant replication and deleterious crossover. EMBO J 2013; 32:1155-67; PMID:23531881; http://dx.doi.org/10.1038/emboj.2013.67
- Gallo-Fernandez M, Saugar I, Ortiz-Bazan MA, Vazquez MV, Tercero JA. Cell cycle-dependent regulation of the nuclease activity of Mus81-Eme1/Mms4. Nucleic Acids Res 2012; 40:8325-35; PMID:22730299; http://dx.doi.org/10.1093/nar/gks599
- Matos J, Blanco MG, West SC. Cell-cycle kinases coordinate the resolution of recombination intermediates with chromosome segregation. Cell Rep 2013; 4:76-86; PMID:23810555; http://dx.doi.org/10.1016/j.celrep.2013.05.039
- Roberts T, Zaidi I, Vaisica J, Peter M, Brown G. Regulation of Rtt107 recruitment to stalled DNA replication forks by the cullin Rtt101 and the Rtt109 acetyltransferase. Mol Biol Cell 2008; 19:171-80; PMID:17978089; http://dx.doi.org/10.1091/mbc.E07-09-0961
- Zaidi IW, Rabut G, Poveda A, Scheel H, Malmstrom J, Ulrich H, Hofmann K, Pasero P, Peter M, Luke B. Rtt101 and Mms1 in budding yeast form a CUL4(DDB1)-like ubiquitin ligase that promotes replication through damaged DNA. EMBO Rep 2008; 9:1034-40; PMID:18704118; http://dx.doi.org/10.1038/embor.2008.155
- Mimura S, Yamaguchi T, Ishii S, Noro E, Katsura T, Obuse C, Kamura T. Cul8/Rtt101 forms a variety of protein complexes that regulate DNA damage response and transcriptional silencing. J Biol Chem 2010; 285:9858-67; PMID:20139071; http://dx.doi.org/10.1074/jbc.M109.082107
- Driscoll R, Hudson A, Jackson SP. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science 2007; 315:649-52; PMID:17272722; http://dx.doi.org/10.1126/science.1135862
- Han J, Zhou H, Horazdovsky B, Zhang K, Xu R-M, Zhang Z. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science 2007; 315:653-5; PMID:17272723; http://dx.doi.org/10.1126/science.1133234
- Li Q, Zhou H, Wurtele H, Davies B, Horazdovsky B, Verreault A, Zhang Z. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell 2008; 134:244-55; PMID:18662540; http://dx.doi.org/10.1016/j.cell.2008.06.018
- Han J, Zhang H, Zhang H, Wang Z, Zhou H, Zhang Z. A Cul4 E3 ubiquitin ligase regulates histone hand-off during nucleosome assembly. Cell 2013; 155:817-29; PMID:24209620; http://dx.doi.org/10.1016/j.cell.2013.10.014
- Neumann H, Hancock SM, Buning R, Routh A, Chapman L, Somers J, Owen-Hughes T, van Noort J, Rhodes D, Chin JW. A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol Cell 2009; 36:153-63; PMID:19818718; http://dx.doi.org/10.1016/j.molcel.2009.07.027
- Duro E, Vaisica JA, Brown GW, Rouse J. Budding yeast Mms22 and Mms1 regulate homologous recombination induced by replisome blockage. DNA Repair 2008; 7:811-8; PMID:18321796; http://dx.doi.org/10.1016/j.dnarep.2008.01.007
- Endo H, Kawashima S, Sato L, Lai MS, Enomoto T, Seki M, Horikoshi M. Chromatin dynamics mediated by histone modifiers and histone chaperones in postreplicative recombination. Genes Cells 2010; 15:945-58; PMID:20718939; http://dx.doi.org/10.1111/j.1365-2443.2010.01435.x
- Clemente-Ruiz M, González-Prieto R, Prado F. Histone H3K56 acetylation, CAF1, and Rtt106 coordinate nucleosome assembly and stability of advancing replication forks. PLoS Genet 2011; 7:e1002376; PMID:22102830; http://dx.doi.org/10.1371/journal.pgen.1002376
- Muñoz-Galván S, Jimeno S, Rothstein R, Aguilera A. Histone H3K56 acetylation, Rad52, and non-DNA repair factors control double-strand break repair choice with the sister chromatid. PLoS Genet 2013; 9:e1003237; PMID:Can't; http://dx.doi.org/10.1371/journal.pgen.1003237
- Celic I, Masumoto H, Griffith WP, Meluh P, Cotter RJ, Boeke JD, Verreault A. The sirtuins Hst3 and Hst4p preserve genome integrity by controlling histone H3 lysine 56 deacetylation. Curr Biol 2006; 16:1280-9; PMID:16815704; http://dx.doi.org/10.1016/j.cub.2006.06.023
- Maas NL, Miller KM, DeFazio LG, Toczyski DP. Cell cycle and checkpoint regulation of histone H3 K56 acetylation by Hst3 and Hst4. Mol Cell 2006; 23:109-19; PMID:16818235; http://dx.doi.org/10.1016/j.molcel.2006.06.006
- Kaplan T, Liu CL, Erkmann JA, Holik J, Grunstein M, Kaufman PD, Friedman N, Rando OJ. Cell cycle- and chaperone-mediated regulation of H3K56ac incorporation in yeast. PLoS Genet 2008; 4:e1000270; PMID:19023413; http://dx.doi.org/10.1371/journal.pgen.1000270
- Celic I, Verreault A, Boeke JD. Histone H3 K56 hyperacetylation perturbs replisomes and causes DNA damage. Genetics 2008; 179:1769-84; PMID:18579506; http://dx.doi.org/10.1534/genetics.108.088914
- Simoneau A, Delgoshaie N, Celic I, Dai J, Abshiru N, Costantino S, Thibault P, Boeke JD, Verreault A, Wurtele H. Interplay between histone H3 lysine 56 deacetylation and chromatin modifiers in response to DNA damage. Genetics 2015; 200:185-205; PMID:25786853; http://dx.doi.org/10.1534/genetics.115.175919
- Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 2007; 446:806-10; PMID:17314980; http://dx.doi.org/10.1038/nature05649
- Che J, Smith S, Kim YJ, Shim EY, Myung K, Lee SE. Hyper-acetylation of histone H3K56 limits break-induced replication by inhibiting extensive repair synthesis. PLoS Genet 2015; 11:e1004990; PMID:25705897; http://dx.doi.org/10.1371/journal.pgen.1004990
- Scholes DT, Banerjee M, Bowen B, Curcio MJ. Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics 2001; 159:1449-65; PMID:11779788
- Luke B, Versini G, Jaquenoud M, Zaidi IW, Kurz T, Pintard L, Pasero P, Peter M. The cullin Rtt101p promotes replication fork progression through damaged DNA and natural pause sites. Curr Biol 2006; 16:786-92; PMID:16631586; http://dx.doi.org/10.1016/j.cub.2006.02.071
- Han J, Li Q, McCullough L, Kettelkamp C, Formosa T, Zhang Z. Ubiquitylation of FACT by the cullin-E3 ligase Rtt101 connects FACT to DNA replication. Genes Dev 2010; 24:1485-90; PMID:20634314; http://dx.doi.org/10.1101/gad.1887310
- Buser R, Kellner V, Melnik A, Wilson-Zbinden C, Schellhaas R, Kastner L, Piwko W, Dees M, Picotti P, Maric M, et al. The replisome-coupled E3 ubiquitin ligase Rtt101-Mms22 counteracts Mrc1 function to tolerate genotoxic stress. PLoS Genet 2016; 12:e1005843; PMID:26849847; http://dx.doi.org/10.1371/journal.pgen.1005843
- Taylor E, Moghraby J, Lees J, Smit B, Moens P, Lehmann A. Characterization of a novel human SMC heterodimer homologous to the Schizosaccharomyces pombe Rad18/Spr18 complex. Mol Biol Cell 2001; 12:1583-94; PMID:11408570; http://dx.doi.org/10.1091/mbc.12.6.1583
- Sergeant J, Taylor E, Palecek J, Andrews E, Sweeney S, Shinagawa H, Watts F, Lehmann A. Composition and architecture of the Schizosaccharomyces pombe Rad18 (Smc5-6) complex. Mol Cell Biol 2005; 25:172-84; PMID:15601840; http://dx.doi.org/10.1128/MCB.25.1.172-184.2005
- Zhao X, Blobel G. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc Natl Acad Sci USA 2005; 102:4777-82; PMID:15738391; http://dx.doi.org/10.1073/pnas.0500537102
- Duan X, Sarangi P, Liu X, Rangi GK, Zhao X, Ye H. Structural and functional insights into the roles of the Mms21 subunit of the Smc5/6 complex. Mol Cell 2009; 35:657-68; PMID:19748359; http://dx.doi.org/10.1016/j.molcel.2009.06.032
- Duan X, Yang Y, Chen YH, Arenz J, Rangi GK, Zhao X, Ye H. Architecture of the Smc5/6 complex of Saccharomyces cerevisiae reveals a unique interaction between the Nse5-6 subcomplex and the hinge regions of Smc5 and Smc6. J Biol Chem 2009; 284:8507-15; PMID:19141609; http://dx.doi.org/10.1074/jbc.M809139200
- Lehmann A, Walicka M, Griffiths D, Murray J, Watts F, McCready S, Carr A. The Rad18 gene of Schizosaccharomyces pombe defines a new subgroup of the SMC superfamily involved in DNA repair. Mol Cell Biol 1995; 15:7067-80; PMID:8524274; http://dx.doi.org/10.1128/MCB.15.12.7067
- Murayama Y, Uhlmann F. DNA entry into and exit out of the cohesin ring by an interlocking gate mechanism. Cell 2015; 163:1628-40; PMID:26687354; http://dx.doi.org/10.1016/j.cell.2015.11.030
- Potts PR, Yu H. Human MMS21/NSE2 is a SUMO ligase required for DNA repair. Mol Cell Biol 2005; 25:7021-32; PMID:16055714; http://dx.doi.org/10.1128/MCB.25.16.7021-7032.2005
- Andrews EA, Palecek J, Sergeant J, Taylor E, Lehmann AR, Watts FZ. Nse2, a component of the Smc5-6 complex, is a SUMO ligase required for the response to DNA damage. Mol Cell Biol 2005; 25:185-96; PMID:15601841; http://dx.doi.org/10.1128/MCB.25.1.185-196.2005
- Leung GP, Lee L, Schmidt TI, Shirahige K, Kobor MS. Rtt107 is required for recruitment of the Smc5/6 complex to DNA double strand breaks. J Biol Chem 2011; 286:26250-7; PMID:21642432; http://dx.doi.org/10.1074/jbc.M111.235200
- Horigome C, Bustard DE, Marcomini I, Delgoshaie N, Tsai-Pflugfelder M, Cobb JA, Gasser SM. PolySUMOylation by Siz2 and Mms21 triggers relocation of DNA breaks to nuclear pores through the Slx5/Slx8 STUbL. Genes Dev 2016; 30:931-45; PMID:27056668; http://dx.doi.org/10.1101/gad.277665.116
- Dovey CL, Russell P. Mms22 preserves genomic integrity during DNA replication in Schizosaccharomyces pombe. Genetics 2007; 177:47-61; PMID:17660542; http://dx.doi.org/10.1534/genetics.107.077255
- Sheedy DM, Dimitrova D, Rankin JK, Bass KL, Lee KM, Tapia-Alveal C, Harvey SH, Murray JM, O'Connell MJ. Brc1-mediated DNA repair and damage tolerance. Genetics 2005; 171:457-68; PMID:15972456; http://dx.doi.org/10.1534/genetics.105.044966
- Lee KM, Nizza S, Hayes T, Bass KL, Irmisch A, Murray JM, O'Connell MJ. Brc1-mediated rescue of Smc5/6 deficiency: requirement for multiple nucleases and a novel Rad18 function. Genetics 2007; 175:1585-95; PMID:17277362; http://dx.doi.org/10.1534/genetics.106.067801
- Lee SY, Russell P. Brc1 links replication stress response and centromere function. Cell Cycle 2013; 12:1665-71; PMID:23656778; http://dx.doi.org/10.4161/cc.24900
- Sanchez A, Roguev A, Krogan NJ, Russell P. Genetic interaction landscape reveals critical requirements for Schizosaccharomyces pombe Brc1 in DNA damage response mutants. G3 2015; 5:953-62; PMID:25795664; http://dx.doi.org/full_text
- Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci U S A 2006; 103:10236-41; PMID:16798881; http://dx.doi.org/10.1073/pnas.0602400103
- Bandhu A, Kang J, Fukunaga K, Goto G, Sugimoto K. Ddc2 mediates Mec1 activation through a Ddc1- or Dpb11-Independent mechanism. PLoS Genet 2014; 10:e1004136; PMID:24586187; http://dx.doi.org/10.1371/journal.pgen.1004136
