ABSTRACT
For every eukaryotic cell to grow and divide, intricately coordinated action of numerous proteins is required to ensure proper cell-cycle progression. The fission yeast Schizosaccharomyces pombe has been instrumental in elucidating the fundamental principles of cell-cycle control. Mutations in S. pombe ‘cut’ (cell untimely torn) genes cause failed coordination between cell and nuclear division, resulting in catastrophic mitosis. Deletion of cbf11, a fission yeast CSL transcription factor gene, triggers a ‘cut’ phenotype, but the precise role of Cbf11 in promoting mitotic fidelity is not known. We report that Cbf11 directly activates the transcription of the acetyl-coenzyme A carboxylase gene cut6, and the biotin uptake/biosynthesis genes vht1 and bio2, with the former 2 implicated in mitotic fidelity. Cbf11 binds to a canonical, metazoan-like CSL response element (GTGGGAA) in the cut6 promoter. Expression of Cbf11 target genes shows apparent oscillations during the cell cycle using temperature-sensitive cdc25–22 and cdc10-M17 block-release experiments, but not with other synchronization methods. The penetrance of catastrophic mitosis in cbf11 and cut6 mutants is nutrient-dependent. We also show that drastic decrease in biotin availability arrests cell proliferation but does not cause mitotic defects. Taken together, our results raise the possibility that CSL proteins play conserved roles in regulating cell-cycle progression, and they could guide experiments into mitotic CSL functions in mammals.
Introduction
Studies in fission yeast, Schizosaccharomyces pombe, have yielded fundamental insights into the principles of cell-cycle control.Citation1 The S. pombe cell cycle consists of a prolonged period of growth (G2 phase) followed by a rapid sequence of mitosis, a very short G1 phase, and DNA replication (S phase). Under standard laboratory conditions, division septum formation takes place while the 2 daughter nuclei are already in S phase.Citation2,3 In fission yeast, as in other eukaryotes, numerous genes play a role in the coordination of cell and nuclear division. When this coordination is perturbed, a catastrophic mitosis may occur, resulting in the so-called ‘cut’ (for cell untimely torn) phenotype, in which the nucleus, whose membrane does not break down during fission yeast mitosis, is split by the division septum before completion of mitosis.Citation4 ‘Cut’ mutant phenotypes have been described for genes whose products function in various aspects of chromosome biology, such as chromosome condensation or sister chromatid separation.Citation5 Curiously, mutations in some lipid metabolism genes also produce the ‘cut’ phenotype, although little is known why or how mitotic fidelity is compromised in these mutants.Citation6
The gene expression program of the fission yeast cell cycle comprises some 500 genes, the expression of which oscillates periodically with the cell-cycle phase. Many of these periodic genes are regulated at the transcriptional level, and several transcription factors regulating particular subsets of periodic genes have been identified.Citation7-9 These factors include Fkh2, Sep1, Mbx1 and the recently identified RFX protein Sak1, that regulate genes involved in mitosis and cytokinesis,Citation10-14 Ace2 regulating cell separation genes,Citation15 the MBF complex, Nrm1 and Yox1 controlling DNA replication genes,Citation16-18 the histone gene regulator Ams2,Citation19 or Toe1 and Toe2 implicated in the pyrimidine-salvage pathway and division septum formation, respectively.Citation20 Importantly, the transcription factors driving periodic expression of many cell cycle-regulated genes are yet to be identified.
CSL transcription factors regulate development, cell fate determination and cell-cycle progression in Metazoa, mostly via the Notch signaling pathway.Citation21 Fungal CSL proteins also exist,Citation22,23 with Cbf11 and Cbf12, the CSL proteins of fission yeast, also functioning as transcription factors.Citation24 Cbf11 and Cbf12 have been implicated in the control of cell-cycle progression,Citation25,26 and regulate directly and indirectly the expression of ∼80 cell-cycle periodic genes.Citation26 Cbf11 also regulates several lipid metabolism genes.Citation26 Interestingly, cells lacking cbf11 frequently undergo catastrophic mitosis and display the ‘cut’ phenotype.Citation25 Furthermore, the penetrance of the ‘cut’ phenotype in Δcbf11 cells is affected by growth media composition.Citation26 However, the molecular mechanism underlying this severe cell-cycle defect is not known.
In this study, we identify Cbf11 target genes contributing to the ‘cut’ phenotype of Δcbf11 cells, and describe the dynamics of their regulation using different cell synchronization methods. We show that Cbf11 is an activator of genes required for proper coordination of cell and nuclear division to prevent catastrophic mitosis.
Results
Mutations in cbf11 or cut6 both cause catastrophic mitosis
Our previous analyses revealed that deletion of cbf11 (Δcbf11) causes impaired coordination of cell and nuclear division, resulting in catastrophic mitosis and the ‘cut’ phenotype (Fig. S1, and refs. Citation25, 26). The ‘cut’ phenotype was also described in cells bearing a temperature-sensitive mutation in cut6 (cut6–621), an essential gene encoding an acetyl-CoA/biotin carboxylase, when grown at 36°C. The precise mechanism of how Cut6 promotes mitotic fidelity is not known.Citation6 The cut6–621 cells display the ‘cut’ phenotype even at the semi-restrictive temperature of 30°C (Fig. S1). Notably, we identified cut6 as one of the putative Cbf11 target genes,Citation26 among which cut6 was actually the only gene associated with the ‘cut’ phenotype.Citation27 This finding raises the possibility that the catastrophic mitosis observed in Δcbf11 cells is mediated by cut6.
Cbf11 binding to DNA oscillates after cdc25–22 block-release
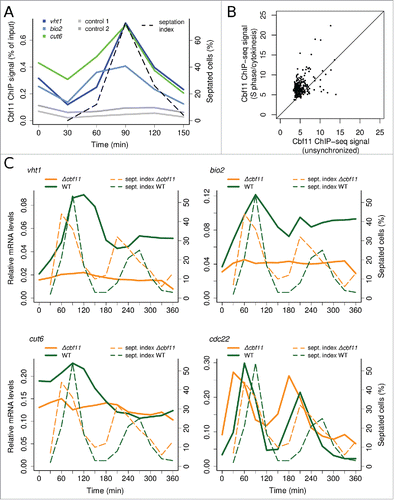
One study identified the cut6 gene as being periodically expressed during the cell cycle with maximum expression in S phase,Citation8 which in fission yeast coincides with cytokinesis (septation).Citation2 Our previous genome-wide chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) identified a region in the cut6 promoter bound by Cbf11. Within that region we found a short sequence element recognized specifically by Cbf11 in vitro.Citation26 We carried out ChIP from cells synchronized using the cdc25–22 temperature-sensitive allele and confirmed that Cbf11 bound to the cut6 promoter in vivo and, notably, maximum binding occurred in S phase/cytokinesis ( and Fig. S2A).
Figure 1. Cbf11 regulates gene expression in an apparently cell-cycle phase-specific manner after cdc25–22 block-release. (A) Timecourse ChIP analysis of Cbf11 binding to cut6, vht1 and bio2 promoters and control unbound loci in cells synchronized in late G2 by cdc25–22 block-release. S phase coincides with division septum formation (see ‘septation index’). Results representative of 2 independent experiments are shown. (B) Comparison of normalized Cbf11 ChIP-seq signal (area under ChIP-seq peak) from unsynchronizedCitation26 and S-phase/cytokinesis cells synchronized by cdc25–22 block-release. Pooled data from 2 independent experiments are shown. (C) Timecourse RT-qPCR analysis of cut6, vht1 and bio2 mRNA levels in wild-type and Δcbf11 cells synchronized by cdc25–22 block-release. The cdc22 gene is a positive control, peaking at the G1/S boundary.Citation32 Results representative of 2 independent experiments are shown.
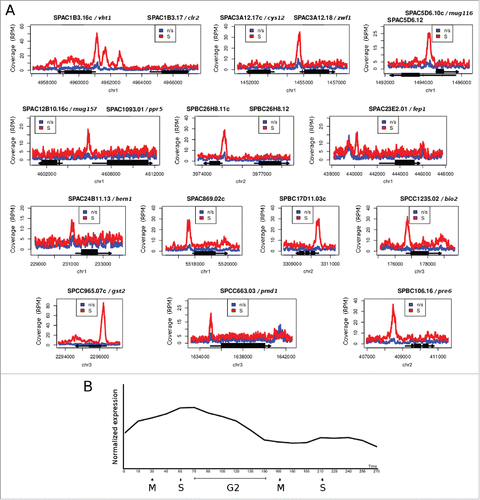
Previous ChIP-seq experiments in unsynchronized cell cultures (which contain mostly G2 cells) revealed rather low binding of Cbf11 to DNA.Citation26 It is therefore possible that increased DNA binding in S phase/cytokinesis is a more general property of Cbf11. To test this hypothesis we performed ChIP-seq from cells progressing synchronously through S phase/cytokinesis after cdc25–22 block-release. Indeed, we found higher Cbf11 occupancy at most of the 284 previously identified Cbf11 target loci in S phase/cytokinesis compared to unsynchronized cells, even though the degree of change in Cbf11 occupancy was locus-specific (). We also identified novel loci bound by Cbf11, which showed little Cbf11 binding in unsynchronized cells but markedly increased Cbf11 occupancy in S phase/cytokinesis (). Interestingly, genes adjacent to these novel Cbf11-bound loci typically showed increased expression in S phase (, expression data taken from ref. 7).
Figure 2. Genes with markedly increased Cbf11 binding to their promoters during S phase/cytokinesis following cdc25–22 block-release. (A) Pooled data from 2 independent ChIP-seq experiments are shown. ‘n/s’ - not synchronized, ‘S’ - S phase/cytokinesis. (B) Average expression levels of genes from (A) during 2 cell cycles, post cdc25–22 block-release. Data were taken from ref. 7. Expression peaks around S/G2.
Two of these novel putative Cbf11 target genes are linked to biotin metabolism: vht1 and bio2 encode a biotin (vitamin H) transporterCitation28 and a biotin synthase,Citation29 respectively. Moreover, vht1 is expressed periodically during the cell cycle, with maximum expression in early G2 phase.Citation30,31 Since the Cut6 enzyme requires biotin for its function, we selected these 2 genes for further analysis. The promoter of vht1 contains 2 Cbf11 response elements (GTGGGAA),Citation24,25 both corresponding to peaks of Cbf11 ChIP-seq signal; a variant GTGAGTA motif is located in the center of the Cbf11 ChIP-seq peak in the bio2 promoter (data not shown). As expected, maximum Cbf11 binding to vht1 and bio2 promoters was detected in S phase/cytokinesis by cell-cycle timecourse ChIP ( and Fig. S2A). We conclude that the DNA binding activity of Cbf11 is modulated after cdc25–22 block-release synchronization and peaks at S phase/cytokinesis.
Cbf11 is required for oscillatory expression of cut6, vht1 and bio2 after cdc25–22 block-release
Next we wanted to determine how Cbf11 affects expression of its target genes and how this regulation is related to cell-cycle progression. We carried out a longer timecourse experiment, spanning 2 cell cycles synchronized by cdc25–22 block-release for wild-type and Δcbf11 cells, and measured cut6, vht1 and bio2 mRNA levels using reverse transcription followed by quantitative PCR (RT-qPCR). The expression of cut6, vht1 and bio2 in wild-type cells oscillated, with peaks at S phase/cytokinesis or early G2 phase, although the expression peak was considerably less pronounced in the second synchronous cell-cycle ( and Fig. S2B). Notably, the mRNA levels of these genes were decreased in Δcbf11 cells compared to wild type and any oscillations of expression were lost. The cdc22 ribonucleotide reductase large subunit gene, a well-studied periodic gene with peak expression at the G1/S boundary,Citation32 showed periodic expression irrespective of Cbf11 presence. Thus, Cbf11 functions as an activator of expression of the cut6, vht1 and bio2 genes, likely via directly regulating their transcription. Furthermore, this regulation shows oscillatory dynamics following cdc25–22 block-release synchronization.
The apparently periodic expression of Cbf11 target genes might be synchronization artifact
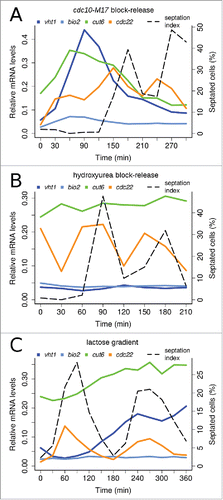
Each cell synchronization method has its limitations and may introduce artifacts.Citation33 Notably, unlike for the cdc22 control G1/S-phase gene, the amplitude of oscillatory expression of Cbf11 target genes in the second synchronous cell cycle following cdc25–22 block-release was markedly diminished ( and Fig. S2B). Therefore, we asked whether these oscillations of Cbf11 targets reflect truly cell-cycle periodic expression or rather artifacts caused by the synchronization procedure which involves a heat shock. We employed diverse additional cell synchronization methods: cdc10-M17 block-release, which uses a temperature-sensitive MBF complex mutant to synchronize cells in G1; hydroxyurea block-release, which synchronizes cells in S phase by reversibly blocking dNTP synthesis; and lactose gradient centrifugation, which selects a subpopulation of small early-G2 cells.Citation2,33 We used these techniques to perform RT-qPCR timecourse analyses of cut6, vht1, bio2 and cdc22 expression during at least 2 synchronous cell cycles in wild-type cells ( and Fig S3). Cbf11 target genes showed oscillatory expression following cdc10-M17 block-release that was similar to the cdc25–22 timecourse pattern ( and Fig. S3A; note that for cdc10-M17 synchronization the very first S phase after release is not accompanied by septation). No clear oscillatory expression was observed for Cbf11 targets after hydroxyurea block-release ( and Fig. S3B). The expression of bio2 appeared constant in lactose gradient-synchronized cells, while the expression of cut6 and vht1 markedly increased only during the second synchronous cell cycle ( and Fig. S3C). Thus, each synchronization method produced a distinct pattern of Cbf11 target gene expression. By contrast, the expression of the cdc22 control was clearly periodic in all cases. In summary, Cbf11 targets show oscillatory expression following some but not all synchronization methods tested, and the apparent cell-cycle periodicity of their regulation by Cbf11 might be an artifact.
Figure 3. Expression patterns of Cbf11 target genes depend on synchronization method. Timecourse RT-qPCR analysis of cut6, vht1 and bio2 mRNA levels in wild-type cells synchronized by cdc10-M17 block-release (A), hydroxyurea block-release (B), and lactose gradient centrifugation (C). S phase coincides with division septum formation (see ‘septation index’); note that for the synchronization method used in panel (A) the very first S phase is never accompanied by septation. The cdc22 gene is a positive control, peaking at the G1/S boundary.Citation32 Note that cdc22 is induced by hydroxyurea treatment even at time 0 in (B).Citation7 Results representative of 3 independent experiments are shown.
Cbf11 is a direct regulator of cut6 expression
Next, we tested whether the putative Cbf11 response element we identified in the cut6 promoter (GTGGGAA)Citation26 could mediate regulation of gene expression by Cbf11. We inserted a 30 bp sequence flanking this Cbf11 response element upstream of a minimal promoter in the pREPORT vector.Citation34 This construct did indeed drive β-galactosidase expression in a Cbf11-dependent manner. No reporter activation was observed when the core 6 nucleotides of the CSL response element were mutated into a XhoI site (GTGGGA → cTcGag) (ref. 34 and data not shown).
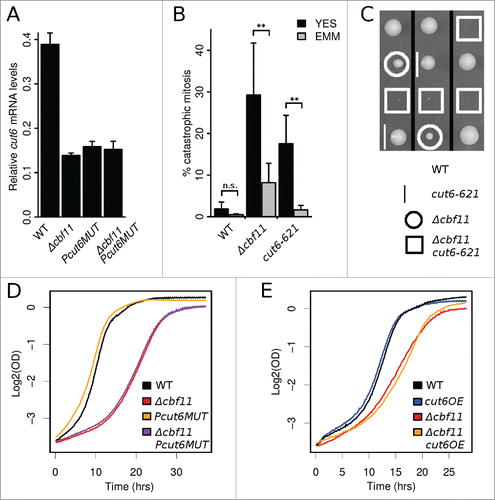
We then introduced the same Cbf11 response element mutation into the native cut6 promoter. The levels of cut6 mRNA were downregulated in the resulting Pcut6MUT strain, to the same extent as in Δcbf11 cells (). No further reduction in cut6 mRNA occurred when the 2 mutations were combined, confirming that the identified Cbf11 response element is both necessary and sufficient for Cbf11 to activate cut6 expression (). Taken together, these findings indicate that Cbf11 is a direct transcriptional activator of cut6.
Figure 4. Cbf11 is a direct activator of cut6 expression. (A) RT-qPCR analysis of cut6 expression. Mean values ± SD from 3 independent experiments are shown. (B) Differential occurrence of catastrophic mitosis in cells grown in YES and EMM. Mean values ± SD from 6 independent experiments are shown. Significance was determined by one-sided paired t-test (**p ≤ 0.01; ‘n.s.’ p > 0.05). (C) Deletion of cbf11 shows a severe synthetic sick genetic interaction with cut6–621 at permissive temperature. Segregants from 3 representative meioses are shown. (D) The Δcbf11 strain, but not the Pcut6MUT promoter mutant strain, showed reduced growth compared to wild type. Results representative of ≥3 independent experiments are shown. (D) Effect of cut6 overexpression on wild-type and Δcbf11 cell proliferation. Results representative of 3 independent experiments are shown.
cbf11 interacts genetically with cut6
The penetrance of the ‘cut’ phenotype is markedly lower when Δcbf11 cells are grown in the EMM minimal medium compared to the rich YES medium.Citation26 Notably, this is also true for the cut6–621 mutant grown at semi-restrictive temperature (), supporting possible implication of cut6 in the catastrophic mitosis occurring when Cbf11 is missing. While a similar media-dependent trend was observed even for wild-type cells, it was much less pronounced and not statistically significant (p > 0.05; one-sided paired t-test).
The cut6–621 mutation conferred a mild growth defect even at permissive temperature (data not shown). The Δcbf11 cut6–621 double mutant displayed a severe synthetic-sick phenotype at permissive temperature (). The hypomorphic mutation in cut6 combined with the absence of the Cbf11 activator might thus reduce the overall Cut6 activity to a level no longer tolerated by the cell.
The Δcbf11 cell culture showed strong growth retardation compared to wild type (). Since cell-cycle duration in synchronized Δcbf11 cells was very similar to wild type (), this population-level growth defect was likely attributable to high cell mortality in Δcbf11 cultures, possibly due to catastrophic mitosis. Unexpectedly, the Pcut6MUT strain, featuring Δcbf11-like decreased cut6 expression () did not show impaired growth (), or higher occurrence of catastrophic mitosis compared to wild type (). Decreased cut6 mRNA levels alone are thus not sufficient to cause the ‘cut’ phenotype of Δcbf11 cells.
Figure 5. Cbf11-regulated genes are needed to prevent catastrophic mitosis. (A) Cells were fixed, stained with DAPI and the occurrence of catastrophic mitosis was determined. Mean values ± SD from ≥3 independent experiments are shown. Significance was determined by one-sided unpaired t-test (*p ≤ 0.05; **p ≤ 0.01). (B) Western blots of whole cell extracts were probed with streptavidin-HRP to visualize biotinylated proteins, and with anti-PSTAIR antibody as a loading control. The position of Cut6 (2 bands) and the expected position of pyruvate carboxylase Pyr1 are indicated. Results representative of 3 independent experiments are shown. (C) Densitometric quantification of Cut6 ECL signals from (B). Mean values ± SD of 3 independent repeats are shown. Significance was determined by one-sided paired t-test (*p ≤ 0.05). (D) Model of gene expression regulation by the Cbf11 transcription factor. Cbf11 binds to target promoters and activates transcription of genes required to prevent catastrophic mitosis. Speculative inputs influencing Cbf11 activity are indicated on the left and discussed in the main text.
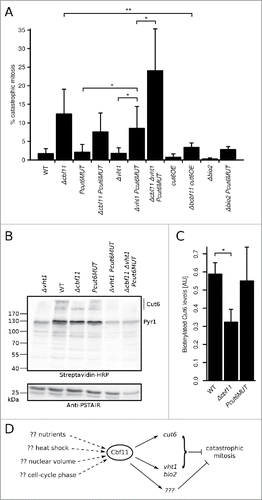
We also tested whether cut6 overexpression could rescue some of the defects observed in Δcbf11 cells. We replaced the native cut6 promoter with a strong constitutive adh1 promoter in both wild-type and Δcbf11 backgrounds; the resulting cut6OE strains showed a ∼5 fold increase in cut6 mRNA levels (data not shown). While cut6 overexpression had little effect in a wild-type background, the cut6OE allele conferred a modest growth improvement (mean doubling time ± SD, n = 3: Δcbf11 227 ± 5 min, Δcbf11 cut6OE 208 ± 14 min; note the differences in growth curve slope in ) and significantly decreased the frequency of mitotic defects in a Δcbf11 background (; p = 0.01; one-sided unpaired t-test). These results further demonstrate an important functional link between Cbf11 and Cut6.
Multiple Cbf11-regulated genes are required to prevent catastrophic mitosis
The Cut6 enzyme is presumably synthesized as an inactive precursor and depends upon the addition of a biotin cofactor for activity.Citation35 In fission yeast, vht1 and bio2 are the only known biotin uptake and biosynthesis genes, and both are regulated by Cbf11 (). It is thus plausible that the cellular biotin pool is decreased in the Δcbf11 mutant, which might affect the functionality of Cut6. To test this hypothesis, we analyzed the cellular content of biotinylated proteins in wild-type, Δcbf11 and Pcut6MUT strains (, C). Notably, biotinylated, and thus likely active, Cut6 levels were significantly decreased in Δcbf11 cells (p = 0.023; one-sided unpaired t-test; compared to wild type), but not in Pcut6MUT cells.
Adding biotin into the medium or overexpressing the vht1 biotin transporter gene did not rescue the mitotic defects of Δcbf11 cells (data not shown). Curiously, when the Pcut6MUT mutation was combined with the deletion of vht1, the double mutant but not the single mutant strains showed increased occurrence of catastrophic mitosis (p = 0.011 and p = 0.014 for double mutant compared to Δvht1 and Pcut6MUT, respectively; one-sided unpaired t-test), which was further exacerbated by deletion of cbf11 (; p = 0.031; one-sided unpaired t-test). However, the dramatic decrease in total biotinylated protein levels (including Cut6) in Δvht1 cells () was not sufficient for increased occurrence of catastrophic mitosis (). Deletion of bio2, either alone or in combination with the Pcut6MUT allele, did not trigger any increase in the frequency of catastrophic mitosis (). Together, these data indicate that the Cbf11 targets cut6 and vht1 are important to prevent catastrophic mitosis, but their decreased expression in Δcbf11 cells can only partially explain the observed mitotic defects. We therefore conclude that other Cbf11-regulated genes are also involved in the coordination of cytokinesis with mitosis.
Complete biotin removal does not lead to catastrophic mitosis
To further examine the impact of biotin availability on mitotic fidelity we performed a biotin depletion experiment. Wild-type cells were grown in the rich YES medium and then shifted to either standard EMM (1 mg/l biotin) or biotin-free EMM. Cultures were then diluted every 5 hours and samples were taken for analysis. Over the course of 20 hours, cells in the biotin-free medium gradually ceased proliferation (Fig. S4A), became small (Fig. S4B, C), and arrested with DNA content corresponding either to G1 or G2 (Fig. S4D). Importantly, we did not observe increased occurrence of catastrophic mitosis at any timepoint in the absence of biotin (Fig. S4C and data not shown), suggesting that a drastic drop in biotin availability leads to cell-cycle arrest rather than mitotic defects.
Discussion
Fission yeast CSL proteins are important for multiple aspects of cell-cycle progression and contribute to the regulation of periodic genes.Citation20,25,26 In the present study, we show that Cbf11 directly regulates the transcription of several genes required to prevent catastrophic mitosis ().
Notably, the metazoan Notch/CSL pathway also regulates cell-cycle progression and the expression of some periodic genes.Citation36-39 Furthermore, deregulation of the Notch pathway triggers catastrophic mitosis in mammalian cells, although any specific roles of CSL proteins in this process have not been investigated.Citation40,41 Detailed analysis of cut6 regulation by Cbf11 revealed that the responsiveness of the cut6 gene to Cbf11 is mediated by a promoter sequence motif (GTGGGAA) matching the canonical response element of metazoan CSL transcription factors (GTGRGAA).Citation42,43 Moreover, we found similar sequence motifs within the Cbf11-bound regions of the vht1 and bio2 promoters. These results document a striking degree of functional conservation among CSL family members across large evolutionary distances, and they may give valuable clues about cell cycle-related roles of CSL proteins in Metazoa.
The precise role of Cut6 in coordinating cytokinesis with mitosis is not known. Cut6 is an acetyl-CoA/biotin carboxylase, the rate-limiting enzyme of fatty acid biosynthesis, but a hypothetical regulatory role of Cut6 in cell-cycle progression has also been proposed.Citation6 During the closed mitosis of both S. pombe and the budding yeast, fatty acid synthesis is required for nuclear membrane expansion and, subsequently, spindle elongation and proper chromosome segregation.Citation44,45 Chemical inhibition of the fatty acid synthase Fas1 subunit results in the ‘cut’ phenotype in fission yeast.Citation45 The synthesis and/or addition of new phospholipids into the nuclear membrane is highly localized in the budding yeast.Citation46,47 Interestingly, Cut6 interacts with Sad1, a spindle pole body (SPB; yeast centrosome) component.Citation48 Thus, an SPB-localized Cut6 pool could hypothetically contribute to and/or regulate the production of new nuclear membrane, possibly in a Cbf11-dependent manner, to support proper mitotic progression. Recently, multiple Cbf11 target genes, including cut6, vht1 and bio2, were shown to be regulated by Mga2, a transcription factor (analogous to mammalian SREBP-1) controlling oxygen-responsive lipid homeostasis in S. pombe.Citation49 However, it is not yet clear whether or how Cbf11 and Mga2 cooperate to regulate lipid metabolism and/or cell-cycle progression. Intriguingly, it has recently been reported that mammalian cells remodel their membrane lipid composition during the cell cycle, and these periodic changes are important for proper cell-cycle progression.Citation50-52
Curiously, the mitotic defects of the Δcbf11 and cut6–621 mutants were alleviated when cells were grown in the minimal EMM medium ( and ref. Citation26). We have shown previously that Δcbf11 phenotypes can be suppressed by the deletion of pka1, which encodes the catalytic subunit of the PKA protein kinase, a positive regulator of cell proliferation in response to nutrients.Citation26 PKA counteracts the anaphase-promoting complex/cyclosome (APC/C), which is required for proper progression through mitosis. APC/C mutants also show the ‘cut’ phenotype. Notably, PKA inactivation or cultivation in EMM can also rescue APC/C ‘cut’ mutants.Citation5,53,54 It would be interesting to test directly whether Cbf11 and Cut6 are functionally linked to APC/C.
We show that Cbf11 regulates vht1 and bio2, the only known fission yeast biotin uptake and biosynthesis genes. Biotin (vitamin H) deficiency underlies several human diseases and is teratogenic in animal models.Citation35,55 Notably, Vht1 has recently been shown to play a role in the resistance to bortezomib, a proteasome inhibitor and an anti-cancer drug that causes aberrant mitosis and ‘cut’ phenotype in fission yeast, likely via inhibiting degradation of securin and cyclin B and thus blocking the metaphase–anaphase transition.Citation56,57 Our results show that complete removal of biotin from the medium leads to cell proliferation arrest without any noticeable mitotic defects (Fig. S4). However, this is a rather drastic disturbance, and it is still possible that less severe perturbations of biotin metabolism contribute to decreased mitotic fidelity seen in the Δcbf11 and Δvht1 Pcut6MUT strains ().
We also show that Cbf11 exerts its regulatory activity in a seemingly cell cycle-modulated manner, peaking at S phase/cytokinesis after cdc25–22 block-release (). However, when we followed Cbf11 target gene expression during 2 synchronized cell cycles, oscillations in mRNA levels were mostly present only in the first cycle. Previous transcriptomic studies identified cut6 and vht1 as cell-cycle periodic genes peaking at S and early G2 phase, respectively.Citation7-9,30 Nevertheless, the periodicity of cut6 expression was only claimed by one of these studies.Citation8 Our extensive testing of 3 other synchronization methods revealed that expression patterns of Cbf11 target genes were not consistent across different methods, with only the cdc10-M17 block-release results resembling the initial observations from cdc25–22 block-release (). As different synchronization methods can alter cell physiology and gene expression in different ways,Citation2,33 it is possible that any observed oscillations in mRNA levels of cut6, vht1 and bio2 are synchronization artifacts. Importantly, this regulation of gene activity is Cbf11-dependent (). A potential explanation is that Cbf11 targets might become transiently upregulated due to the heat shock employed in both cdc25–22 and cdc10-M17 block-release. Interestingly, cbf11 interacts genetically with sty1,Citation26 which encodes the main fission yeast stress-activated protein kinase (homolog of mammalian p38). Sty1 is essential for transcriptional response to heat shock,Citation58 and contributes to the regulation of cell-cycle progression.Citation59 The expression of cbf11 remains largely unchanged under various stresses, including heat shock.Citation58 It is therefore possible that any hypothetical stress-dependent control over Cbf11 activity would be achieved at the post-translational level. Indeed, 2 phosphorylation sites, S104 and S117, have been identified in Cbf11,Citation60-62 although it is not known whether their phosphorylation status changes after stress and which kinase(s) are involved. Another potential explanation for the oscillating expression of Cbf11 target genes relates to altered nuclear volume following some synchronization methods. The ratio between nuclear volume and the volume of chromosomes is important for mitotic fidelity,Citation44,45 and increased nuclear volume (with normal DNA content and compaction) found in the cdc25–22 mutant background can suppress chemically-induced catastrophic mitosis.Citation45 Thus, hypothetically, altered nuclear size during the first cell cycle following cdc25–22 (and possibly cdc10-M17) block-release synchronization might result in aberrant regulation of lipid biosynthesis genes. This misregulation would then be resolved in subsequent cell cycles, as cells attain normal proportions. In this scenario, Cbf11 would be needed for coupling the sensing of nuclear volume to the production of new membrane components.
In summary, we have shown that Cbf11 is an activator of several genes, including the Cut6 acetyl-CoA/biotin carboxylase, that are required for mitotic fidelity. Cbf11 regulates Cut6 via a canonical, metazoan-like CSL response element, suggesting functional conservation of the CSL protein family during evolution. Our results indicate potential mechanisms of how Cbf11 and its target genes act to prevent catastrophic mitosis, that will be addressed by future studies. Such research will shed further light into the intricacies of transcriptional control supporting cell-cycle progression.
Materials and methods
Yeast culture
Fission yeast cells were grown according to standard procedures at 30 or 32°C,Citation2,63 unless stated otherwise; temperature-sensitive strains were grown at 25°C. For cdc25–22 and cdc10-M17 block-release experiments, cell cultures grown in YES were synchronized at 36°C for 4 hours and released by rapid cooling to 25°C. For hydroxyurea block-release experiments, cell cultures grown in YES at 32°C were synchronized in the presence of 12 mM hydroxyurea for 4 hours and then released by washing and resuspending into fresh YES.Citation2 The protocol for cell synchronization by lactose gradient centrifugation was derived from ref. 2. Briefly, exponentially growing cells were centrifuged (200 × g, 3 min) through 2 consecutive 10–40% lactose gradients (volumes 45 and 10 ml, respectively; lactose was dissolved in YES). Approximately 109-1010 cells were layered on top of the first gradient. The collected fraction of small, early-G2 cells was washed and resuspended into fresh YES. Cell-cycle progression was monitored by flow cytometry and/or microscopy. Under standard cultivation conditions, the S phase coincides with the division septum formation in fission yeast.Citation2 Growth curves were measured in the VarioSkan Flash instrument (Thermo Scientific) using 12-well dishes and 1.4 ml culture volumes. For biotin depletion experiments, precultures were grown in YES and then cells were washed and resuspended into either standard EMM (1 mg/l biotin) or biotin-free EMM. The list of fission yeast strains used in this study is provided in Table S1.
Microscopy
Cells were fixed in 70% ethanol, rehydrated in water, stained with DAPI (1 μg/ml) and photographed using the Olympus CellR system. To quantify the occurrence of catastrophic mitosis, at least 200 cells (exponential growth phase) per sample were scored. Cell length measurements were performed in imageJCitation64 and at least 46 cells were scored per sample.
Plasmids and strain construction
The lists of oligonucleotides and plasmids used in this study are provided in Tables S2 and S3, respectively. Transcription reporter plasmids were based on pREPORT-U (ref. 34) and had oligonucleotides (cut6r-fwd and cut6r-rev; cut6mutr-fwd and cut6mutr-rev) containing CSL response elements inserted upstream of the minimal promoter. The β-galactosidase assays were performed as described previously.Citation34
For vht1 overexpression, the vht1 ORF was amplified by PCR (primers mp165 and mp166), cloned into pGEM-T Easy (Promega), validated by sequencing, and subcloned into the BamHI/SalI sites of pDUAL-FFH51.Citation65 The resulting plasmid pMP118 was linearized with NotI and integrated into the leu1–32 locus. Vht1 functionality was tested by complementation of a Δvht1 strain.Citation28
For cut6 overexpression, the native cut6 promoter was replaced by the strong constitutive adh1 promoter by one-step integration of a PCR-amplified replacement cassette using the pFA6a-natMX6-PADH1–3HA vector template (primers mp222 and mp223).Citation66 Successful integration was verified by PCR (primers mp159 and mp160 and mp224) and cut6 overexpression was confirmed by RT-qPCR.
The procedure for the cut6 promoter mutagenesis was adapted from ref. 67. The ura4 gene (promoter, ORF, terminator) was amplified from wild-type (JB32) genomic DNA using primers mp161 and mp162 and inserted into the pCR-2.1 vector by TA cloning (Invitrogen). Subsequently, regions flanking the Cbf11 response element (located 28 bp upstream of the cut6 exon 1) were amplified using primers mp157 and mp158 and mp159 and mp160, respectively, and inserted in the pCR-2.1-ura4 plasmid in the HindIII/BamHI and NotI/XbaI sites, respectively, yielding plasmid pMP110. Primers mp158 and mp159 each contained an internal XhoI site, allowing ura4 gene excision from the pMP110 construct. Removing ura4 resulted in a DNA insert identical to the endogenous cut6 promoter region, except for the Cbf11 response element that was replaced with an XhoI site (pMP113). The HindIII/XbaI fragment of pMP110 was then used to knock-out the Cbf11 response element in the cut6 promoter in a ura- haploid background by homologous recombination, producing a ura+ strain (MP340). The integration was checked by PCR. Next, the HindIII/XbaI fragment of pMP113 was used to substitute the ura4 cassette in the genome for the mutated Cbf11 response element (i.e., XhoI site). Clones resistant to 5′-fluoroorotic acid (ura-) were further checked by PCR and sequencing (MP342).
Deletion of the bio2 gene was performed using the pCloneHyg1 system (primers mp198 and mp199; mp200 and mp201).Citation68 Deletion was verified by PCR (primers mp202 and mp144; mp33 and mp203).
All other strains used in this study were constructed by standard genetic crosses.
qPCR and RT-qPCR
Total RNA was extracted from cells using the MasterPure Yeast RNA Purification Kit (Epicentre) and converted to cDNA using random primers and the RevertAid™ Reverse Transcriptase kit (Fermentas). Quantitative PCR was performed using the MESA GREEN qPCR MasterMix Plus for SYBR (Eurogentec) and the LightCycler 480 II instrument (Roche). For RT-qPCR, act1 (actin) was used as a reference gene. The primers used are listed in Table S2.
Chromatin immunoprecipitation and deep sequencing (ChIP-seq)
Five hundred milliliter cultures of an untagged strain and strains expressing TAP-tagged Cbf11 were grown in YES, synchronized by cdc25–22 block-release and collected in S phase (coincides with peak of septation in both wild-type and Δcbf11 cells). Chromatin immunoprecipitation was performed as described previously;Citation26 2 loci not bound by Cbf11 were used as controls for ChIP-qPCR (see Table S2 for the respective chromosome coordinates). Sequencing libraries were prepared using the NEBNext ChIP-Seq Library Prep Master Mix Set for Illumina and NEBNext Multiplex Oligos for Illumina (New England Biolabs) according to manufacturer's instructions, and sequenced on an Illumina MiSeq instrument (12-fold multiplexing per run). Two independent biological repeats were performed. Sequencing reads were aligned to the reference fission yeast genome (release 12) using BWA 0.6.1,Citation69 processed with the samtools 0.1.18 package,Citation70 and inspected using IGV browser 2.0.23 and the igvtools package.Citation71 ChIP-seq data are available in the ArrayExpress database under accession number E-MTAB-2725.
Western blotting
Proteins were separated on a 7.5% Tris-glycine-SDS gel, transferred to a nitrocellulose membrane and probed with either a streptavidin-HRP polymer (S2438, Sigma; 1:1,000 dilution) or mouse monoclonal anti-PSTAIR antibody (anti-Cdc2 loading control; P7962, Sigma; 1:8,000 dilution), as appropriate. A goat-anti-mouse HRP-conjugated secondary antibody (sc-2031, Santa Cruz; 1:3,000) was used where required. Detection was performed using the ECL Prime Western Blotting Detection Reagent (Amersham) and the ImageQuant LAS 4000 instrument (GE Healthcare). ECL signals were quantified using ImageJ.Citation64 To determine the position of Cut6 on polyacrylamide gels, cells expressing tagged Cut6 from an integrated pDUAL-FFH1c plasmid were used.Citation72
Flow cytometry
For analysis of DNA content by flow cytometry, ethanol-fixed cells were treated with RNase and stained with 4 µg/ml propidium iodide as described previously,Citation26 and analyzed using the CytoFLEX instrument and CytExpert software version 1.2.11.0 (Beckman-Coulter). At least 1,000 singlet cells were measured per sample.
Data processing and statistics
The R software environment for statistical computing and graphics (https://www.r-project.org/) was used for data processing, statistical tests and preparation of graphs.
Abbreviations
| ChIP-seq | = | chromatin immunoprecipitation followed by deep sequencing |
| cut | = | cell untimely torn |
| EMM | = | Edinburgh minimal medium |
| RT-qPCR | = | reverse transcription followed by quantitative real-time PCR |
| YES | = | yeast extract with supplements |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplementary files
Download Zip (1.5 MB)Acknowledgments
The authors would like to thank Sandra Codlin for sequencing of ChIP-seq libraries, Káťa Strouhalová and Alice Škrlantová for help with ‘cut’ assays and strain constructions, Eva Krellerová for excellent technical assistance, Bähler and Folk lab members and Erik Boye and Beáta Grallert for help and discussions, and Eva Ničová for critical reading of this manuscript. The cut6–621 strain was provided by The Yeast Genetic Resource Center Japan. The 34/C12 plasmid clone containing the cut6 gene was provided by RIKEN BRC through the National Bio-Resource Project of the MEXT, Japan.
Funding
This work was funded by the Czech Science Foundation grant P305/12/P040, Charles University grants SVV 260083 and UNCE 204013, grant no. 640413 of the Charles University Grant Agency, and the Wellcome Trust Senior Investigator Award #095598/Z/11/Z.
References
- Gould KL. Protein kinases driving the cell cycle. In: Egel R, editor. The Molecular Biology of Schizosaccharomyces pombe. Berlin, Germany: Springer; 2004; p. 27-40.
- Sabatinos SA, Forsburg SL. Molecular genetics of Schizosaccharomyces pombe. Methods Enzymol 2010; 470:759-95; PMID:20946835; http://dx.doi.org/10.1016/S0076-6879(10)70032-X
- Carlson CR, Grallert B, Stokke T, Boye E. Regulation of the start of DNA replication in Schizosaccharomyces pombe. J Cell Sci 1999; 112:939-46; PMID:10036243
- Hirano T, Funahashi SI, Uemura T, Yanagida M. Isolation and characterization of Schizosaccharomyces pombe cut mutants that block nuclear division but not cytokinesis. EMBO J 1986; 5:2973-9; PMID:16453724
- Yanagida M. Fission yeast cut mutations revisited: control of anaphase. Trends Cell Biol 1998; 8:144-9; PMID:9695827; http://dx.doi.org/10.1016/S0962-8924(98)01236-7
- Saitoh S, Takahashi K, Nabeshima K, Yamashita Y, Nakaseko Y, Hirata A, Yanagida M. Aberrant mitosis in fission yeast mutants defective in fatty acid synthetase and acetyl CoA carboxylase. J Cell Biol 1996; 134:949-61; PMID:8769419; http://dx.doi.org/10.1083/jcb.134.4.949
- Rustici G, Mata J, Kivinen K, Lió P, Penkett CJ, Burns G, Hayles J, Brazma A, Nurse P, Bähler J. Periodic gene expression program of the fission yeast cell cycle. Nat Genet 2004; 36:809-17; PMID:15195092; http://dx.doi.org/10.1038/ng1377
- Peng X, Karuturi RKM, Miller LD, Lin K, Jia Y, Kondu P, Wang L, Wong L-S, Liu ET, Balasubramanian MK, et al. Identification of cell cycle-regulated genes in fission yeast. Mol Biol Cell 2005; 16:1026-42; PMID:15616197; http://dx.doi.org/10.1091/mbc.E04-04-0299
- Oliva A, Rosebrock A, Ferrezuelo F, Pyne S, Chen H, Skiena S, Futcher B, Leatherwood J. The cell cycle-regulated genes of Schizosaccharomyces pombe. PLoS Biol 2005; 3:e225; PMID:15966770; http://dx.doi.org/10.1371/journal.pbio.0030225
- Garg A, Futcher B, Leatherwood J. A new transcription factor for mitosis: in Schizosaccharomyces pombe, the RFX transcription factor Sak1 works with forkhead factors to regulate mitotic expression. Nucleic Acids Res 2015; 43:6874-88; PMID:25908789; http://dx.doi.org/10.1093/nar/gkv274
- Zilahi E, Salimova E, Simanis V, Sipiczki M. The S. pombe sep1 gene encodes a nuclear protein that is required for periodic expression of the cdc15 gene. FEBS Lett 2000; 481:105-8; PMID:10996305; http://dx.doi.org/10.1016/S0014-5793(00)01990-6
- Bulmer R, Pic-Taylor A, Whitehall SK, Martin KA, Millar JBA, Quinn J, Morgan BA. The forkhead transcription factor Fkh2 regulates the cell division cycle of Schizosaccharomyces pombe. Eukaryot Cell 2004; 3:944-54; PMID:15302827; http://dx.doi.org/10.1128/EC.3.4.944-954.2004
- Buck V, Ng SS, Ruiz-Garcia AB, Papadopoulou K, Bhatti S, Samuel JM, Anderson M, Millar JBA, McInerny CJ. Fkh2p and Sep1p regulate mitotic gene transcription in fission yeast. J Cell Sci 2004; 117:5623-32; PMID:15509866; http://dx.doi.org/10.1242/jcs.01473
- Papadopoulou K, Chen J-S, Mead E, Feoktistova A, Petit C, Agarwal M, Jamal M, Malik A, Spanos A, Sedgwick SG, et al. Regulation of cell cycle-specific gene expression in fission yeast by the Cdc14p-like phosphatase Clp1p. J Cell Sci 2010; 123:4374-81; PMID:21098641; http://dx.doi.org/10.1242/jcs.073056
- Martín-Cuadrado AB, Dueñas E, Sipiczki M, Vázquez de Aldana CR, del Rey F. The endo-beta-1,3-glucanase eng1p is required for dissolution of the primary septum during cell separation in Schizosaccharomyces pombe. J Cell Sci 2003; 116:1689-98; http://dx.doi.org/10.1242/jcs.00377
- Lowndes NF, McInerny CJ, Johnson AL, Fantes PA, Johnston LH. Control of DNA synthesis genes in fission yeast by the cell-cycle gene cdc10+. Nature 1992; 355:449-53; PMID:1734281; http://dx.doi.org/10.1038/355449a0
- Aligianni S, Lackner DH, Klier S, Rustici G, Wilhelm BT, Marguerat S, Codlin S, Brazma A, de Bruin RAM, Bähler J. The fission yeast homeodomain protein Yox1p binds to MBF and confines MBF-dependent cell-cycle transcription to G1-S via negative feedback. PLoS Genet 2009; 5:e1000626; PMID:19714215; http://dx.doi.org/10.1371/journal.pgen.1000626
- de Bruin RAM, Kalashnikova TI, Aslanian A, Wohlschlegel J, Chahwan C, Yates JR, Russell P, Wittenberg C. DNA replication checkpoint promotes G1-S transcription by inactivating the MBF repressor Nrm1. Proc Natl Acad Sci U S A 2008; 105:11230-5; PMID:18682565; http://dx.doi.org/10.1073/pnas.0801106105
- Takayama Y, Takahashi K. Differential regulation of repeated histone genes during the fission yeast cell cycle. Nucleic Acids Res 2007; 35:3223-37; PMID:17452352; http://dx.doi.org/10.1093/nar/gkm213
- Vachon L, Wood J, Kwon E-JG, Laderoute A, Chatfield-Reed K, Karagiannis J, Chua G. Functional characterization of fission yeast transcription factors by overexpression analysis. Genetics 2013; 194:873-84; PMID:23695302; http://dx.doi.org/10.1534/genetics.113.150870
- Pursglove SE, Mackay JP. CSL: a notch above the rest. Int J Biochem Cell Biol 2005; 37:2472-7; PMID:16095948; http://dx.doi.org/10.1016/j.biocel.2005.06.013
- Převorovský M, Půta F, Folk P. Fungal CSL transcription factors. BMC Genomics 2007; 8:233; PMID:17629904; http://dx.doi.org/10.1186/1471-2164-8-233
- Převorovský M, Atkinson SR, Ptáčková M, McLean JR, Gould K, Folk P, Půta F, Bähler J. N-termini of fungal CSL transcription factors are disordered, enriched in regulatory motifs and inhibit DNA binding in fission yeast. PLoS One 2011; 6:e23650; PMID:21858190; http://dx.doi.org/10.1371/journal.pone.0023650
- Oravcová M, Teska M, Půta F, Folk P, Převorovský M. Fission Yeast CSL Proteins Function as Transcription Factors. PLoS One 2013; 8:e59435; http://dx.doi.org/10.1371/journal.pone.0059435
- Převorovský M, Groušl T, Staňurová J, Ryneš J, Nellen W, Půta F, Folk P. Cbf11 and Cbf12, the fission yeast CSL proteins, play opposing roles in cell adhesion and coordination of cell and nuclear division. Exp Cell Res 2009; 315:1533-47; PMID:19101542; http://dx.doi.org/10.1016/j.yexcr.2008.12.001
- Převorovský M, Oravcová M, Tvarůžková J, Zach R, Folk P, Půta F, Bähler J. Fission Yeast CSL Transcription Factors: Mapping Their Target Genes and Biological Roles. PLoS One 2015; 10:e0137820; PMID:26366556; http://dx.doi.org/10.1371/journal.pone.0137820
- Wood V, Harris MA, McDowall MD, Rutherford K, Vaughan BW, Staines DM, Aslett M, Lock A, Bähler J, Kersey PJ, et al. PomBase: a comprehensive online resource for fission yeast. Nucleic Acids Res 2012; 40:D695-9; PMID:22039153; http://dx.doi.org/10.1093/nar/gkr853
- Stolz J. Isolation and characterization of the plasma membrane biotin transporter from Schizosaccharomyces pombe. Yeast 2003; 20:221-31; PMID:12557275; http://dx.doi.org/10.1002/yea.959
- Phalip V, Lemoine Y, Jeltsch JM. Cloning of Schizosaccharomyces pombe bio2 by heterologous complementation of a Saccharomyces cerevisiae mutant. Curr Microbiol 1999; 39:348-0350; PMID:10525840; http://dx.doi.org/10.1007/s002849900470
- Marguerat S, Jensen TS, de Lichtenberg U, Wilhelm BT, Jensen LJ, Bähler J. The more the merrier: comparative analysis of microarray studies on cell cycle-regulated genes in fission yeast. Yeast 2006; 23:261-77; PMID:16544289; http://dx.doi.org/10.1002/yea.1351
- Santos A, Wernersson R, Jensen LJ. Cyclebase 3.0: a multi-organism database on cell-cycle regulation and phenotypes. Nucleic Acids Res 2015; 43:D1140-4; PMID:25378319; http://dx.doi.org/10.1093/nar/gku1092
- Fernandez Sarabia MJ, McInerny C, Harris P, Gordon C, Fantes P. The cell cycle genes cdc22+ and suc22+ of the fission yeast Schizosaccharomyces pombe encode the large and small subunits of ribonucleotide reductase. Mol Gen Genet 1993; 238:241-51; PMID:8479429
- Walker GM. Synchronization of yeast cell populations. Methods Cell Sci 1999; 21:87-93; PMID:10728641; http://dx.doi.org/10.1023/A:1009824520278
- Převorovský M. pREPORT: a multi-readout transcription reporter vector for fission yeast. Yeast 2015; 32:327-34; PMID:25395321; http://dx.doi.org/10.1002/yea.3055
- Pacheco-Alvarez D, Solórzano-Vargas RS, Del Río AL. Biotin in metabolism and its relationship to human disease. Arch Med Res 2002; 33:439-47; PMID:12459313; http://dx.doi.org/10.1016/S0188-4409(02)00399-5
- Joshi I, Minter LM, Telfer J, Demarest RM, Capobianco AJ, Aster JC, Sicinski P, Fauq A, Golde TE, Osborne BA. Notch signaling mediates G1/S cell-cycle progression in T cells via cyclin D3 and its dependent kinases. Blood 2009; 113:1689-98; PMID:19001083; http://dx.doi.org/10.1182/blood-2008-03-147967
- Sarmento LM, Huang H, Limon A, Gordon W, Fernandes J, Tavares MJ, Miele L, Cardoso AA, Classon M, Carlesso N. Notch1 modulates timing of G1-S progression by inducing SKP2 transcription and p27 Kip1 degradation. J Exp Med 2005; 202:157-68; PMID:15998794; http://dx.doi.org/10.1084/jem.20050559
- Ronchini C, Capobianco AJ. Induction of cyclin D1 transcription and CDK2 activity by Notch(ic): implication for cell cycle disruption in transformation by Notch(ic). Mol Cell Biol 2001; 21:5925-34; PMID:11486031; http://dx.doi.org/10.1128/MCB.21.17.5925-5934.2001
- Rangarajan A, Talora C, Okuyama R, Nicolas M, Mammucari C, Oh H, Aster JC, Krishna S, Metzger D, Chambon P, et al. Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J 2001; 20:3427-36; PMID:11432830; http://dx.doi.org/10.1093/emboj/20.13.3427
- Curry CL, Reed LL, Broude E, Golde TE, Miele L, Foreman KE. Notch inhibition in Kaposi's sarcoma tumor cells leads to mitotic catastrophe through nuclear factor-kappaB signaling. Mol Cancer Ther 2007; 6:1983-92; PMID:17604336; http://dx.doi.org/10.1158/1535-7163.MCT-07-0093
- Lasagni L, Ballerini L, Angelotti ML, Parente E, Sagrinati C, Mazzinghi B, Peired A, Ronconi E, Becherucci F, Bani D, et al. Notch activation differentially regulates renal progenitors proliferation and differentiation toward the podocyte lineage in glomerular disorders. Stem Cells 2010; 28:1674-85; PMID:20680961; http://dx.doi.org/10.1002/stem.492
- Tun T, Hamaguchi Y, Matsunami N, Furukawa T, Honjo T, Kawaichi M. Recognition sequence of a highly conserved DNA binding protein RBP-J kappa. Nucleic Acids Res 1994; 22:965-71; PMID:8152928; http://dx.doi.org/10.1093/nar/22.6.965
- Del Bianco C, Vedenko A, Choi SH, Berger MF, Shokri L, Bulyk ML, Blacklow SC. Notch and MAML-1 complexation do not detectably alter the DNA binding specificity of the transcription factor CSL. PLoS One 2010; 5:e15034; PMID:21124806; http://dx.doi.org/10.1371/journal.pone.0015034
- Yam C, He Y, Zhang D, Chiam K-H, Oliferenko S. Divergent strategies for controlling the nuclear membrane satisfy geometric constraints during nuclear division. Curr Biol 2011; 21:1314-9; PMID:21802294; http://dx.doi.org/10.1016/j.cub.2011.06.052
- Takemoto A, Kawashima SA, Li J-J, Jeffery L, Yamatsugu K, Elemento O, Nurse P. Nuclear envelope expansion is crucial for proper chromosomal segregation during a closed mitosis. J Cell Sci 2016; 129:1250-9; PMID:26869222; http://dx.doi.org/10.1242/jcs.181560
- Arnone JT, Walters AD, Cohen-Fix O. The dynamic nature of the nuclear envelope: lessons from closed mitosis. Nucleus 2013; 4:261-6; PMID:23873576; http://dx.doi.org/10.4161/nucl.25341
- Witkin KL, Chong Y, Shao S, Webster MT, Lahiri S, Walters AD, Lee B, Koh JLY, Prinz WA, Andrews BJ, et al. The budding yeast nuclear envelope adjacent to the nucleolus serves as a membrane sink during mitotic delay. Curr Biol 2012; 22:1128-33; PMID:22658600; http://dx.doi.org/10.1016/j.cub.2012.04.022
- Miki F, Kurabayashi A, Tange Y, Okazaki K, Shimanuki M, Niwa O. Two-hybrid search for proteins that interact with Sad1 and Kms1, two membrane-bound components of the spindle pole body in fission yeast. Mol Genet Genomics 2004; 270:449-61; PMID:14655046; http://dx.doi.org/10.1007/s00438-003-0938-8
- Burr R, Stewart EV, Shao W, Zhao S, Hannibal-Bach HK, Ejsing CS, Espenshade PJ. Mga2 Transcription Factor Regulates an Oxygen-Responsive Lipid Homeostasis Pathway in Fission Yeast. J Biol Chem 2016; 291:12171-83; PMID:27053105; http://dx.doi.org/10.1074/jbc.M116.723650
- Atilla-Gokcumen GE, Muro E, Relat-Goberna J, Sasse S, Bedigian A, Coughlin ML, Garcia-Manyes S, Eggert US. Dividing cells regulate their lipid composition and localization. Cell 2014; 156:428-39; PMID:24462247; http://dx.doi.org/10.1016/j.cell.2013.12.015
- Scaglia N, Tyekucheva S, Zadra G, Photopoulos C, Loda M. De novo fatty acid synthesis at the mitotic exit is required to complete cellular division. Cell Cycle 2014; 13:859-68; PMID:24418822; http://dx.doi.org/10.4161/cc.27767
- Sanchez-Alvarez M, Zhang Q, Finger F, Wakelam MJO, Bakal C. Cell cycle progression is an essential regulatory component of phospholipid metabolism and membrane homeostasis. Open Biol 2015; 5:150093; PMID:26333836; http://dx.doi.org/10.1098/rsob.150093
- Yamada H, Kumada K, Yanagida M. Distinct subunit functions and cell cycle regulated phosphorylation of 20S APC/cyclosome required for anaphase in fission yeast. J Cell Sci 1997; 110:1793-804; PMID:9264466
- Yamashita YM, Nakaseko Y, Samejima I, Kumada K, Yamada H, Michaelson D, Yanagida M. 20S cyclosome complex formation and proteolytic activity inhibited by the cAMP/PKA pathway. Nature 1996; 384:276-9; PMID:8918880; http://dx.doi.org/10.1038/384276a0
- Watanabe T. Teratogenic effects of biotin deficiency in mice. J Nutr 1983; 113:574-81; PMID:6827377
- Takeda K, Mori A, Yanagida M. Identification of genes affecting the toxicity of anti-cancer drug bortezomib by genome-wide screening in S. pombe. PLoS One 2011; 6:e22021; PMID:21760946; http://dx.doi.org/10.1371/journal.pone.0022021
- Kawashima SA, Takemoto A, Nurse P, Kapoor TM. Analyzing fission yeast multidrug resistance mechanisms to develop a genetically tractable model system for chemical biology. Chem Biol 2012; 19:893-901; PMID:22840777; http://dx.doi.org/10.1016/j.chembiol.2012.06.008
- Chen D, Toone WM, Mata J, Lyne R, Burns G, Kivinen K, Brazma A, Jones N, Bähler J. Global transcriptional responses of fission yeast to environmental stress. Mol Biol Cell 2003; 14:214-29; PMID:12529438; http://dx.doi.org/10.1091/mbc.E02-08-0499
- Shiozaki K, Russell P. Cell-cycle control linked to extracellular environment by MAP kinase pathway in fission yeast. Nature 1995; 378:739-43; PMID:7501024; http://dx.doi.org/10.1038/378739a0
- Koch A, Krug K, Pengelley S, Macek B, Hauf S. Mitotic substrates of the kinase aurora with roles in chromatin regulation identified through quantitative phosphoproteomics of fission yeast. Sci Signal 2011; 4:rs6; PMID:21712547; http://dx.doi.org/10.1126/scisignal.2001588
- Carpy A, Krug K, Graf S, Koch A, Popic S, Hauf S, Macek B. Absolute proteome and phosphoproteome dynamics during the cell cycle of fission yeast. Mol Cell Proteomics 2014; 13:1925-36; PMID:24763107; http://dx.doi.org/10.1074/mcp.M113.035824
- Kettenbach AN, Deng L, Wu Y, Baldissard S, Adamo ME, Gerber SA, Moseley JB. Quantitative phosphoproteomics reveals pathways for coordination of cell growth and division by the fission yeast DYRK kinase Pom1. Mol Cell Proteomics 2015; 14:1275-87; PMID:25720772; http://dx.doi.org/10.1074/mcp.M114.045245
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol 1991; 194:795-823; PMID:2005825; http://dx.doi.org/10.1016/0076-6879(91)94059-L
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 2012; 9:671-5; PMID:22930834; http://dx.doi.org/10.1038/nmeth.2089
- Matsuyama A, Shirai A, Yoshida M. A series of promoters for constitutive expression of heterologous genes in fission yeast. Yeast 2008; 25:371-6; PMID:18437702; http://dx.doi.org/10.1002/yea.1593
- Van Driessche B, Tafforeau L, Hentges P, Carr AM, Vandenhaute J. Additional vectors for PCR-based gene tagging in Saccharomyces cerevisiae and Schizosaccharomyces pombe using nourseothricin resistance. Yeast 2005; 22:1061-8; PMID:16200506; http://dx.doi.org/10.1002/yea.1293
- Rodríguez-Sánchez L, Rodríguez-López M, García Z, Tenorio-Gómez M, Schvartzman JB, Krimer DB, Hernández P. The fission yeast rDNA-binding protein Reb1 regulates G1 phase under nutritional stress. J Cell Sci 2011; 124:25-34; http://dx.doi.org/10.1242/jcs.070987
- Gregan J, Rabitsch PK, Rumpf C, Novatchkova M, Schleiffer A, Nasmyth K. High-throughput knockout screen in fission yeast. Nat Protoc 2006; 1:2457-64; PMID:17406492; http://dx.doi.org/10.1038/nprot.2006.385
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009; 25:1754-60; PMID:19451168; http://dx.doi.org/10.1093/bioinformatics/btp324
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009; 25:2078-9; PMID:19505943; http://dx.doi.org/10.1093/bioinformatics/btp352
- Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief Bioinform 2013; 14:178-92; http://dx.doi.org/10.1093/bib/bbs017
- Matsuyama A, Arai R, Yashiroda Y, Shirai A, Kamata A, Sekido S, Kobayashi Y, Hashimoto A, Hamamoto M, Hiraoka Y, et al. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol 2006; 24:841-7; PMID:16823372; http://dx.doi.org/10.1038/nbt1222
