ABSTRACT
Eukaryotic initiation factor 2A (eIF2A) is a 65-kDa protein that was first identified in the early 1970s as a factor capable of stimulating initiator methionyl-tRNAi (Met-tRNAMeti) binding to 40S ribosomal subunits in vitro. However, in contrast to the eIF2, which stimulates Met-tRNAMeti binding to 40S ribosomal subunits in a GTP-dependent manner, eIF2A didn't reveal any GTP-dependence, but instead was found to direct binding of the Met-tRNAMeti to 40S ribosomal subunits in a codon-dependent manner. eIF2A appears to be highly conserved across eukaryotic species, suggesting conservation of function in evolution. The yeast Saccharomyces cerevisae eIF2A null mutant revealed no apparent phenotype, however, it was found that in yeast eIF2A functions as a suppressor of internal ribosome entry site (IRES)-mediated translation. It was thus suggested that eIF2A my act by impinging on the expression of specific mRNAs. Subsequent studies in mammalian cell systems implicated eIF2A in non-canonical (non-AUG-dependent) translation initiation events involving near cognate UUG and CUG codons. Yet, the role of eIF2A in cellular functions remains largely enigmatic. As a first step toward characterization of the eIF2A function in mammalian systems in vivo, we have obtained homozygous eIF2A-total knockout (KO) mice, in which a gene trap cassette was inserted between eIF2A exons 1 and 2 disrupting expression of all exons downstream of the insertion. The KO mice strain is viable and to date displays no apparent phenotype. We believe that the eIF2A KO mice strain will serve as a valuable tool for researchers studying non-canonical initiation of translation in vivo.
Introduction
Initiation of protein synthesis in eukaryotes is a complex process requiring more than 12 different initiation factors, comprising over 30 polypeptide chains.Citation1,2 The function of many of these factors have been established in great detail;Citation1,2 however, the precise role of some of them and their mechanism of action are still not well understood.
eIF2A is a single chain 65 kDa protein that was initially believedCitation3,4 to serve as the functional homolog of prokaryotic IF2, since eIF2A and IF2 catalyze biochemically similar reactions, i.e. they stimulate initiator methionyl-tRNA (Met-tRNAMeti) binding to the small ribosomal subunit. However, subsequent identification of a heterotrimeric 126 kDa factor, eIF2(α,β,γ),Citation5-7 showed that this factor and not eIF2A is primarily responsible for the binding of Met-tRNAMeti to 40S ribosomal subunits in eukaryotes. In mammals, 4 stress-activated kinasesCitation8 reduce the level of active eIF2 by phosphorylating the eIF2α subunit and, consequently, reducing the global level of translation.Citation1-2 However, translation of many cellular and viral proteins appeared to be resistant to eIF2α phosphorylationCitation9 and Ref. therein, despite requiring Met-tRNAMeti. It was found that a subset of factors, i.e., Ligatin/eIF2D,Citation10,11 the oncogene MCT-1 and DENR (together)Citation10 as well as eIF5B (alone)Citation12-14 can promote efficient recruitment of Met-tRNAMeti to 40S/mRNA complexes under conditions of inhibition of eIF2 activity, or its absence.
eIF2A was originally suggested to function in the same pathway and promote eIF2-independent recruitment of Met-tRNAMeti to 40S ribosomal subunits in parallel with eIF2, or in the absence of eIF2.Citation3,4,15-18 The double deletion eIF2A/eIF5B and eIF2A/eIF4E-ts mutant Saccharomyces cerevisiae strains displayed a severe slow growth phenotype.Citation15,16 The phenotype of these mutants and the biochemical localization of the eIF2A on the 40S ribosomal subunits and 80S ribosomes further suggested that eIF2A participates in translation initiation.Citation15,16 Often confused with eIF2α, eIF2A was later implicated in non-AUG-dependent translation initiation events involving near cognate UUG and CUG codons.Citation19-21 eIF2A homologues were found in a wide range of eukaryotic species,Citation15 suggesting a conserved biological role. eIF2A was predicted to harbor a WD-repeat β-propeller domain,Citation15,16 the structure of which (for yeast Schizosaccharomyces pombe eIF2A protein) has been recently solved at 2.5 Å resolution.Citation22 The yeast S. cerevisiae eIF2A null strains however showed no apparent phenotype,Citation15,16 suggesting that the eIF2A does not function in major (key) steps in the initiation process but, might act at some minor/alternative initiation events such as reinitiation, internal initiation, or non-AUG initiation.Citation15,16 In yeast, eIF2A was found to function as a suppressor of internal ribosome entry site (IRES)-mediated translation.Citation16 Recently, eIF2A was reported to be involved in antigen presentation by major histocompatibility complex (MHC) class I molecules, the phosphatase and tensin homolog deleted on chromosome 10 (PTEN) protein expression and the integrated stress response.Citation19-21 All these events were affected by eIF2A silencing.Citation19-21 Yet, the precise role of eIF2A in higher eukaryotes and its precise mechanism of action remain largely enigmatic.
To continue the physical and functional characterization of a mammalian eIF2A we have obtained a homozygous eIF2A-total knockout (KO) mouse strain, in which a trap cassette was inserted between eIF2A exons 1 and 2 disrupting the expression of all exons downstream of the insertion. The KO mice strain is viable and displays no apparent phenotype. We believe that eIF2A KO mice strain will serve as a valuable tool for researchers studying non-canonical initiation of translation and the general function of eIF2A in vivo.
Results
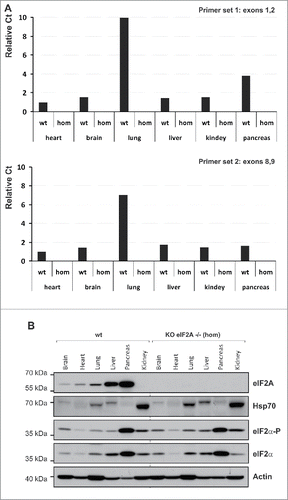
We report the construction and preliminary analysis of an eIF2A knockout mouse strain. We have used the OmniBankII gene trap library containing mutated embryonic stem (ES) cell clones, which employed gene trapping with retroviral vectors in mouse C57BL/6N ES cells to generate the library.Citation23 The ES cell clone IST13504C3 was identified from the library to contain a retroviral insertion (of a gene trap vector Omnibank Vector 74) in the eIF2A gene intron 1 (). Insertion of the retroviral vector into the eIF2A gene (a single-copy gene in the mouse genome) leads to the splicing of the endogenous upstream exons into this cassette to produce a fusion that terminates further transcription of the eIF2A endogenous exons downstream of the insertion.Citation23 Mice were generated from this cell line using standard procedures. A germline transmission event was scored when any chimera produced progeny with coat color. Chimeric males were bred to C57BL/6 females for germline transmission of the mutant eIF2A allele. The progeny were analyzed for transmission of the mutation. DNA was isolated from mouse tail and genotyped by PCR. Three genotypes wild-type +/+ (WT), heterozygous +/− (Het) and null (Null) −/− of eIF2A knockout (KO) mice were derived from heterozygous males and females (). The specific polymerase chain reaction (PCR) eIF2A-derived gene product of 357 bp in length was found only in wild-type and Het mice, while the Omnibank Gene Trap Vector 74 gene-derived product (216 bp) was found only in the Het and Null mice, thus confirming the absence of the functional eIF2A gene in the eIF2A knockout (KO) mice. The absence of eIF2A mRNA and protein expression was further confirmed by RT-PCR and Western blotting with anti-eIF2A specific antibodies (). We have analyzed 6 different mouse tissues (heart, brain, lung, liver, kidney and pancreas) for the presence of eIF2A mRNA and protein (). mRNA was detected by Real-Time (RT) PCR using 2 different sets of primers targeted to exons 1 and 2 and 8 and 9, respectively. In none of the cases we were able to detect eIF2A mRNA and eIF2A protein in the eIF2A knockout (KO) mice (). In wild-type mice, eIF2A mRNA was most abundant in lung tissues, however, the highest eIF2A protein abundance was observed in pancreas and liver. Interestingly, we could not detect any protein in kidneys (of the wild-type animal) even though a similar amount of eIF2A mRNA was present in this tissue in comparison with e.g. liver and pancreas. This suggests that eIF2A mRNA may be translationally silenced in kidneys. Abrogation of eIF2A expression didn't affect eIF2 expression or the level of eIF2 phosphorylation (). The eIF2A Null mice revealed no visible phenotype () being similar in size and morphology to wild-type mice. The eIF2A Null mice are also fertile, and exhibit no breeding abnormalities under standard growth conditions, when crossed to wild-type mice.
Figure 1. The eIF2A gene. (A) Top: Mus musculus eukaryotic translation initiation factor 2A (eIF2A) (mouse Accession: NM_001005509) gene organization and the site of insertion of Omnibank Vector 74. Bottom: Mouse genomic sequence surrounding the gene trap insertion site identified in the C57BL/6 gene trap ES cell clone IST13504C3sA4 for eIF2A. The initiating ATG codon is in red and the coding part of exon 1 is in cyan. The insertion site is denoted with an asterisk *. (B) Top: Schematic of the insertion site and the genotyping strategy. Relative positions of the primers and the expected sizes of the PCR fragments are indicated. Bottom: Genotyping results (1.5% agarose gel).
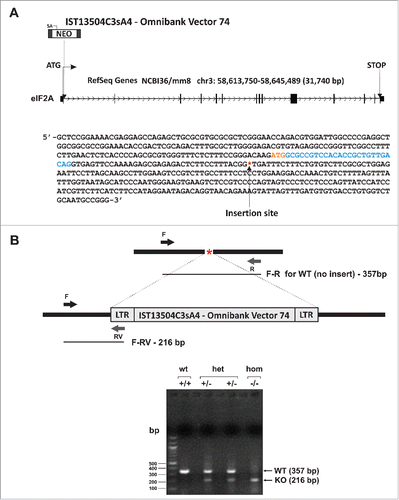
Figure 2. The eIF2A gene expression. (A) Real-time PCR. Relative Ct values are shown; Ct values were normalized to gapdh signal and are shown relative to wild-type brain sample. Two sets of primers were used: Primer set 1, recognizing sequences within exons 1 and 2 (top) and primer set 2 recognizing sequences within exons 8 and 9 (bottom). (B) Western blotting. mRNA and protein levels were analyzed in 6 different mouse tissues heart, brain, lung, liver, kidney and pancreas, respectively.
Discussion
eIF2A is often confused with the eIF2α subunit of the eukaryotic initiation factor 2 (eIF2). While eIF2 and eIF2A function similarly in model assays, the order of events is different between the 2: eIF2A binds Met-tRNAMeti to 40S ribosomal subunits in a codon-dependent manner, whereas eIF2 binds Met-tRNAMeti to 40S subunits in a GTP-dependent manner.Citation4 eIF2 is primarily responsible for the binding of Met-tRNAMeti to 40S ribosomal subunits in eukaryotes.Citation1,2,5-6 eIF2 is essential for translation initiation and defects in eIF2 are lethal. Phosphorylation of the eIF2α subunit attenuates protein synthesis in all eukaryotic cells and plays an important regulatory role.Citation1,2,8 Phosphorylation at eIF2α-Ser51 prevents the GDP/GTP exchange and thereby blocks further initiation.Citation1,2,8 Mice with a homozygous mutation at the eIF2α phosphorylation site (Ser51Ala) die within 18 h after birth.Citation24 Although the biochemical mechanism of translational inhibition mediated by eIF2α phosphorylation is well established, it appears that expression of some cellular and viral proteins appears to be resistant to eIF2α phosphorylation. It was found that in addition to eIF2A,Citation3,4,17-21 a subset of factors, i.e. Ligatin/eIF2D,Citation10,11 the oncogene MCT-1 and DENR (together)Citation10 as well as eIF5B (alone)Citation12-14 can promote efficient recruitment of Met-tRNAMeti to 40S/mRNA complexes under conditions of elevated eIF2α phosphorylation (in cellular systems), or its absence (in a reconstituted system in vitro). As of now, the role of eIF2A remains largely enigmatic, in spite of its high evolutionary conservation in eukaryotes. Data from yeast has shown that eIF2A is not essential for cell growth and viability and does not function in the major initiation pathway, but it was suggested that it may be implicated in minor events such internal initiation, or, potentially, non-AUG-dependent initiation.Citation15,16 It was later found using mammalian cell systems that eIF2A may help in the initiation of the alphaviruses Sindbis (SV) subgenomic 26S mRNA under conditions of eIF2α phosphorylation.Citation17 Similarly, eIF2A was found to facilitate translation of HCV mRNA under stress conditions.Citation18 Recently, eIF2A was shown to control tumor suppressor PTEN translation, which requires a CUG-centered palindromic motif for this process.Citation20 PTEN is involved in embryonic development, tissue homeostasis, metabolism, and tumor suppression.Citation25,26 PTENα is an N-terminally extended isoform of PTEN and is involved in electron transport through the induction of cytochrome c oxidase activity in mitochondria.Citation20 Disruption of PTENα impairs mitochondrial bioenergetics. Translation of PTENα is initiated from a CUG codon upstream of and in-frame with the canonical PTEN and was shown to require eIF2A for this process.Citation20 Similarly, translation of new polypeptides that supply antigenic precursors for loading the major histocompatability complex (MHC) class I molecules was found to be eIF2A-dependent.Citation19 Translation of antigenic precursors represents a distinct initiation pathway and is initiated from a CUG start codon.Citation19 Finally, eIF2A was found to function as a cis-acting regulatory element necessary for privileged BiP expression during stress.Citation21 The BiP 5′ UTR harbors several uORFs that are exclusively initiated by the non-AUG leucine codons UUG and CUG and regulate BiP expression under stress. Expression of the −190 UUG uORF from the BiP 5′ UTR was found to require eIF2A under these conditions.Citation21 Elevated levels of eIF2A were observed upon integrated stress response induction and thus eIF2A were suggested to protect BiP mRNA from translational shutdown under these conditions,Citation21 thus, implicating eIF2A in the control of cellular homeostasis and stress response.
To continue the functional characterization of a mammalian eIF2A we have obtained a homozygous eIF2A-total knockout (KO) mouse strain. eIF2A knockout mice are viable, further suggesting that eIF2A is not involved in key initiation pathway(s) vital for organism function. To our knowledge, the eIF2A knockout mice are the first mammalian model completely lacking a translation initiation factor. We believe that it will serve as a valuable tool for researchers studying non-canonical initiation of translation in vivo and will allow us to further establish, whether eIF2A is indispensable for the initiation events described above, and/or whether other proteins with similar functions (Ligatin/eIF2D, eIF5B, etc.) can act in its place in vivo under the above mentioned conditions.
Materials and methods
Generation of eIF2A null mice
Mutant eIF2A mice were generated using a gene-trapping technique.Citation23 Mice (strain C57BL/6) were cloned from an ES cell line (IST13504C3; Texas A&M Institute for Genomic Medicine, TIGM). The ES cell clone contained a retroviral insertion in the eIF2A gene (intron 1) identified from the TIGM gene trap database, and was microinjected into C57BL/6 host blastocysts to generate germline chimeras using standard procedures. The retroviral OmniBank Vector 74 contained a splice acceptor sequence () followed by a selectable neomycin resistance marker for identification of successful gene trap events further followed by a polyadenylation signal. Insertion of the retroviral vector into the eIF2A gene led to the splicing of the endogenous upstream exons into this cassette to produce a fusion that leads to termination of further transcription of the endogenous eIF2A exons downstream of the insertion. Chimeric males were bred to C57BL/6 females for germline transmission of the mutant eIF2A allele.
Genotyping
Mice were screened prior to weaning at the age of 12–17 d. For genotyping, tail snip DNA was extracted using tail lysis procedure with Proteinase K. The lysis (L) buffer contained 25 mM Tris-HCl pH 7.5, 100 mM NaCl, 50 mM EDTA, 1% Sarkosyl, 5 mM DTT, 0.05 mM Spermidine and 2 μl proteinase K (Sigma) per 100 μl of the buffer. Mice tails in buffer L + proteinase K (300–400 μl/tail) were incubated overnight at 65°C, diluted 1:40, heated at 95°C for 5 min and used for PCR reactions.
PCR was performed to determine the presence of the gene trap using LongAmp™ Taq 2X Master Mix (NEB, MA). The following pairs of primers were used to detect either the wild-type eIF2A gene Ffwd: 5′-GCCTTTCTTGAACTCTCACC-3′ and Rrev: 5′-GCAGACCACAGGTCACACAT-3′, giving raise to 357 bp product and/or its disrupted variant Ffwd: 5′-GCCTTTCTTGAACTCTCACC-3′ and RVrev: 5′- CCAATAAACCCTCTTGCAGTTGC −3′ (216 product).
RT-PCR and western blotting
RT-PCR and Western blotting were done following standard procedures. Two-step SYBR real-time RT-PCR was performed with 80 ng of RNA for both target gene and endogenous control using ABI StepOnePlus™ (Applied Biosystems, CA) instrument. ABI High-capacity cDNA RT kit and Power SYBR Green PCR Master Mix were used. CT (cycle threshold) values were analyzed using the comparative CT(ΔΔCT) method as described by the manufacturer (Applied Biosystems, CA). The amount of target(2−ΔΔCT) was obtained by normalization to an endogenous reference standard (GAPDH) and relative to a wild-type brain sample. The following primers were used:
GAPDH fwd: 5′-GGCATTGCTCTCAATGACAAC-3′, rev 5′-GCCATGTAGGCCATGAGGT-3′;
eIF2A primer set 1: fwd (exon 1) 5′-GCCTTTCTTGAACTCTCACC-3′, rev (exon 2) 5′-CCGTGCTTTCTGTGAAGTGT-3′
eIF2A primer set 2: fwd (exon 8) 5′-GGAGCCTCCTACTATGGAGA-3′, rev (exon 9) 5′-CCATAAACAGCGCAAAACTC-3′.
Western blotting was performed using the following antibodies: anti-eIF2A (Abcam, MA) #ab169528 (1:2500), anti-Hsp70 (Enzo Life Sciences, NY) #ADI-SPA-810 (1:1000); anti- eIF2α-P (Abcam, MA) #ab32157 (1:3000); anti-eIF2α (Santa Cruz, CA) #sc-133227 (1:3000); anti-Actin (Abcam, MA) #ab3280 (1:2000).
Animals and diets
Animals were maintained following standard procedures using the guidelines approved by Public Health Service and the Institutional Animal Care and Use Committee at Texas A&M University and Cleveland State University.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Amy Gonzales, John Adams Jr., Johnathan Ballard, and Huiping Guo for the excellent technical assistance.
Funding
This work was supported by CSU Faculty Research Development grant award (KOMARFRD) to A.A.K., CSU's Office of Sponsored Program and Research Bridge Funding award (BRIMAZU) to B.M., funds from the Center for Gene Regulation in Health and Disease (GRHD) at CSU and NIH grants DK53307 and DK60596 to M.H.
References
- Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 2010; 11:113-27; PMID:20094052; http://dx.doi.org/10.1038/nrm2838
- Hinnebusch AG. The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem 2014; 83:779-812; PMID:24499181; http://dx.doi.org/10.1146/annurev-biochem-060713-035802
- Merrick WC, Anderson WF. Purification and characterization of homogeneous protein synthesis initiation factor M1 from rabbit reticulocytes. J Biol Chem 1975; 250:1197-206; PMID:1112800
- Adams SL, Safer B, Anderson WF, Merrick WC. Eukaryotic initiation complex formation. Evidence for two distinct pathways. J Biol Chem 1975; 250:9083-89; PMID:1194278
- Benne R, Brown-Luedi ML, Hershey JW. Protein synthesis initiation factors from rabbit reticulocytes: purification, characterization, and radiochemical labeling. Methods Enzymol 1979; 60:15-35; PMID:459896; http://dx.doi.org/10.1016/S0076-6879(79)60005-8
- Majumdar A, Dasgupta A, Chatterjee B, Das HK, Gupta NK. Purification and properties of rabbit reticulocyte protein synthesis initiation factors EIF-1, EIF-2, and EIF-3. Methods Enzymol 1979; 60:35-52; PMID:459906; http://dx.doi.org/10.1016/S0076-6879(79)60006-X
- Staehelin T, Erni B, Schreier MH. Purification and characterization of seven initiation factors for mammalian protein synthesis. Methods Enzymol 1979; 60:136-65; PMID:459895; http://dx.doi.org/10.1016/S0076-6879(79)60013-7
- Donnelly N, Gorman AM, Gupta S, Samali A. The eIF2α kinases: their structures and functions. Cell Mol Life Sci 2013; 70:3493-511; PMID:23354059; http://dx.doi.org/10.1007/s00018-012-1252-6
- Andreev DE, O'Connor PB, Fahey C, Kenny EM, Terenin IM, Dmitriev SE, Cormican P, Morris DW, Shatsky IN, Baranov PV. Translation of 5′ leaders is pervasive in genes resistant to eIF2 repression. Elife 2015; 4:e03971; PMID:25621764
- Skabkin MA, Skabkina OV, Dhote V, Komar AA, Hellen CU, Pestova TV. Activities of Ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Dev 2010; 24(16):1787-801; PMID:20713520; http://dx.doi.org/10.1101/gad.1957510
- Dmitriev SE, Terenin IM, Andreev DE, Ivanov PA, Dunaevsky JE, Merrick WC, Shatsky IN. GTP-independent tRNA delivery to the ribosomal P-site by a novel eukaryotic translation factor. J Biol Chem 2010; 285:26779-87; http://dx.doi.org/10.1074/jbc.M110.119693
- Choi SK, Lee JH, Zoll WL, Merrick WC, Dever TE. Promotion of met-tRNAiMet binding to ribosomes by yIF2, a bacterial IF2 homolog in yeast. Science 1998; 280:1757-60; PMID:9624054; http://dx.doi.org/10.1126/science.280.5370.1757
- Pestova TV, de Breyne S, Pisarev AV, Abaeva IS, Hellen CU. eIF2-dependent and eIF2-independent modes of initiation on the CSFV IRES: a common role of domain II. EMBO J 2008; 27:1060-72; PMID:18337746; http://dx.doi.org/10.1038/emboj.2008.49
- Terenin IM, Dmitriev SE, Andreev DE, Shatsky IN. Eukaryotic translation initiation machinery can operate in a bacterial-like mode without eIF2. Nat Struct Mol Biol 2008; 15:836-41; http://dx.doi.org/10.1038/nsmb.1445
- Zoll WL, Horton LE, Komar AA, Hensold JO, Merrick WC. Characterization of mammalian eIF2A and identification of the yeast homolog. J Biol Chem. 2002; 277:37079-87; PMID:12133843; http://dx.doi.org/10.1074/jbc.M207109200
- Komar AA, Gross SR, Barth-Baus D, Strachan R, Hensold JO, Goss Kinzy T, Merrick WC. Novel characteristics of the biological properties of the yeast Saccharomyces cerevisiae eukaryotic initiation factor 2A. J Biol Chem 2005; 280:15601-11; PMID:15718232; http://dx.doi.org/10.1074/jbc.M413728200
- Ventoso I, Sanz MA, Molina S, Berlanga JJ, Carrasco L, Esteban M. Translational resistance of late alphavirus mRNA to eIF2alpha phosphorylation: a strategy to overcome the antiviral effect of protein kinase PKR. Genes Dev 2006; 20:87-100; PMID:16391235; http://dx.doi.org/10.1101/gad.357006
- Kim JH, Park SM, Park JH, Keum SJ, Jang SK. eIF2A mediates translation of hepatitis C viral mRNA under stress conditions. EMBO J 2011; 30:2454-64; PMID:21556050; http://dx.doi.org/10.1038/emboj.2011.146
- Starck SR, Jiang V, Pavon-Eternod M, Prasad S, McCarthy B, Pan T, Shastri N. Leucine-tRNA initiates at CUG start codons for protein synthesis and presentation by MHC class I. Science 2012; 336:1719-23; PMID:22745432; http://dx.doi.org/10.1126/science.1220270
- Liang H, He S, Yang J, Jia X, Wang P, Chen X, Zhang Z, Zou X, McNutt MA, Shen WH, et al. PTENα, a PTEN isoform translated through alternative initiation, regulates mitochondrial function and energy metabolism. Cell Metab 2014; 19:836-48; PMID:24768297; http://dx.doi.org/10.1016/j.cmet.2014.03.023
- Starck SR, Tsai JC, Chen K, Shodiya M, Wang L, Yahiro K, Martins-Green M, Shastri N, Walter P. Translation from the 5′ untranslated region shapes the integrated stress response. Science 2016; 351:aad3867; PMID:26823435; http://dx.doi.org/10.1126/science.aad3867
- Kashiwagi K, Ito T, Yokoyama S. Crystal structure of the eukaryotic translation initiation factor 2A from Schizosaccharomyces pombe. J Struct Funct Genomics 2014; 15:125-30; PMID:24569939; http://dx.doi.org/10.1007/s10969-014-9177-y
- Hansen GM, Markesich DC, Burnett MB, Zhu Q, Dionne KM, Richter LJ, Finnell RH, Sands AT, Zambrowicz BP, Abuin A. Large-scale gene trapping in C57BL/6N mouse embryonic stem cells. Genome Res 2008; 18:1670-9; PMID:18799693; http://dx.doi.org/10.1101/gr.078352.108
- Scheuner D, Song B, McEwen E, Liu C, Laybutt R, Gillespie P, Saunders T, Bonner-Weir S, Kaufman RJ. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol Cell 2001; 7:1165-76; PMID:11430820; http://dx.doi.org/10.1016/S1097-2765(01)00265-9
- Bermúdez Brito M, Goulielmaki E, Papakonstanti EA. Focus on PTEN Regulation. Front Oncol 2015; 5:166; PMID:26284192
- Leslie NR, Kriplani N, Hermida MA, Alvarez-Garcia V, Wise HM. The PTEN protein: cellular localization and post-translational regulation. Biochem Soc Trans 2016; 44:273-8; PMID:26862215; http://dx.doi.org/10.1042/BST20150224

