Shapes of the cell have fascinated our curiosity since the discovery of the cell by Robert Hooke in 1665Citation1 and by Anton van Leeuwenhoek slightly laterCitation2 using the state-of-the-art microscopes of the time. In medicine, cell shapes have long been exploited to diagnose disease types and conditions, such as cancer types and malignant vs. benign tumors, respectively. Cell division orientation is also influenced by cell shape.Citation3 A number of other cellular functions, ranging from growth to intracellular signals, are also controlled by cell shape.Citation4 While these previous studies clearly indicated the importance of cell shapes in controlling cell functionality, it remained unclear if and how the shape of a parent cell can influence the fate of its daughter cells, considering that mitotic cells round up and thus loose their shape during cell division.
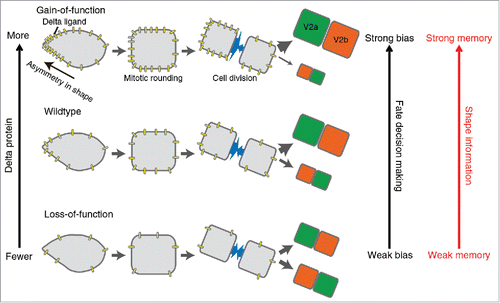
We have recently addressed this question in a study of cell-fate decision-making during the asymmetric division of the V2 neural progenitor cell in the developing zebrafish.Citation5 Each V2 cell divides into two daughter cells - one acquiring the V2a fate (Vsx1+, Chx10+) and the other the V2b fate (Gata2+, Scl+). V2 cells exhibit a variety of shapes – some relatively round and the others with a “spiky” end causing asymmetry in shape. After observing the behavior of V2 cells using confocal live-imaging microscopy, we noticed that the shapes immediately preceding mitotic rounding correlated with the fates of the daughter cells. To further analyze this initial observation, we devised a mathematical method to quantify the shapes prior to cell division, and used these to study how cell shape prior to division correlated with the cell fates immediately after division. This analysis suggested that the daughter cell derived from the spiky-end of the V2 cell preferentially acquires the V2a fate. This observation suggests that V2 cell shape information prior to mitotic rounding is stored as a “memory” to influence the daughter cell fate, and this mechanism is referred to as the “shape-memory” system in our studyCitation5 (). The causal shape-fate relation for the “shape-memory” system was demonstrated by a shape-manipulation experiment in which we show the location of the new spike, but not the old spike, generated by femtosecond laser irradiation, becomes the best predictor for the fate of the daughter cells.
Figure 1. The “shape-memory” system mediated by the biased DeltaC protein localization. The cellular asymmetry causes polarized localization of DeltaC protein and biases the daughter cell fate despite mitotic rounding and division. Our integrative approach combining both experimental and computational methods demonstrates that this “shape-memory” system is quantitatively influenced by the amount of DeltaC protein and the degree of cellular asymmetry.
The critical role of Delta-Notch signaling pathway in the binary asymmetric fate-decision-making of V2 cells was previously reported.Citation6 Hence, we performed a computational study to investigate whether Delta-Notch signals could quantitatively transduce the shape information to the shape-dependent biased fate-decision-making and to generate an experimentally testable model. Our in silico model assumed that the Delta molecules localize preferentially toward the spiky-end and induce the V2a fate, while preventing the adjacent cell from entering the same fate as well. Simulations of this model suggested that this could suffice for pushing the daughter cell on the formed spiking cell into the V2a fate, even despite the observation that Delta molecules diffuse along the cell membrane during mitotic rounding, thus smoothening out the initial shape information. Computational parameter studies of the model suggested that the degree of the spikiness, the abundance of Delta molecules in the cell, the diffusion rate of Delta molecules relative to the duration of mitotic rounding quantitatively influences the fate bias.
The critical importance of these parameters was experimentally validated by combining live-imaging microscopy, genetic and laser manipulations of V2 cells in developing zebrafish (). Among the four Delta molecular species, we found that DeltaC is the player. The femtosecond laser-mediated alteration of the spikiness of the V2 cells showed that the degree of the spikiness quantitatively influences the polarized localization of Delta molecules which causally biases the asymmetric fate-decision-making. The abundance of Delta molecules in individual V2 cells was altered using morpholino-mediated knockdown of the Delta molecule level and also by the re-expression of Delta molecules in the Delta-deficient V2 cells. The results from these experiments showed that the abundance of Delta molecules quantitatively influences the biased asymmetric fate-decision-making of V2 cell as predicted by the computational simulations ().
Our study effectively combined live-imaging of cellular behavior, model generation and quantitative parameter testing by computational simulation, followed by experimental validation of the model predictions. This integrative approach allowed us to show that the past cell shape information remains influential on the future fate despite the loss of cellular morphology during mitotic rounding (). Our approach also unveiled the quantitative and mechanistic underpinnings of this “shape-memory” system ().
While the current study suggests a role of Delta-Notch signals in the “shape-memory” system, other signals are possibly exploited by other cell types and/or in other situations. Discoveries of such other molecular pathways and their detailed mechanisms, together with our current finding, contribute to gaining further quantitative insights into the molecular mechanisms of cell lineage, morphogenesis, disease development and progression, and other diverse ranges of biological and disease processes in the dynamically changing environments. Furthermore, the methodology developed in this study could be exploited to develop a system to diagnose the future disease types and/or to assess the current and future states of cells based on their current shapes.Citation7
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- Hooke R. Micrographia: or, Some physiological descriptions of minute bodies aade by magnifying glasses. (J. Martyn and J. Allenstry, 1665).
- Leeuwenhoek AV. De natis e semine genitali animalculis. Philosophical Transactions of the Royal Society of London 1678; 12:1040-3; http://dx.doi.org/10.1098/rstl.1677.0068
- Hertwig O. Das Problem der Fefrunchtung une der isotropie des Eies, eine Theory der Vererbung. Jenaische Zeitschrift für Naturwissenschaft 1884; 18:276-318.
- Schmick M, Bastiaens PI. The interdependence of membrane shape and cellular signal processing. Cell 2014; 156:1132-8; PMID:24630717; http://dx.doi.org/10.1016/j.cell.2014.02.007
- Akanuma T, Chen C, Sato T, Merks RM, Sato TN. Memory of cell shape biases stochastic fate decision-making despite mitotic rounding. Nat Commun 2016; 7:11963; PMID:27349214; http://dx.doi.org/10.1038/ncomms11963
- Kimura Y, Satou C, Higashijima S. V2a and V2b neurons are generated by the final divisions of pair-producing progenitors in the zebrafish spinal cord. Development 2008; 135:3001-5; PMID:18684740; http://dx.doi.org/10.1242/dev.024802
- Kozawa S. et al. Real-time prediction of cell division timing in developing zebrafish embryo. Scientific Reports 2016; 6:32962; PMID:27597656; http://dx.doi.org/10.1038/srep32962
