ABSTRACT
Experimental evidence suggest that breast tumors originate from breast cancer stem cells (BCSCs), and that mitochondrial biogenesis is essential for the anchorage-independent clonal expansion and survival of CSCs, thus rendering mitochondria a significant target for novel treatment approaches. One of the recognized side effects of the FDA-approved drug, doxycycline is the inhibition of mitochondrial biogenesis. Here we investigate the mechanism by which doxycycline exerts its inhibitory effects on the properties of breast cancer cells and BCSCs, such as mammosphere forming efficiency, invasion, migration, apoptosis, the expression of stem cell markers and epithelial-to-mesenchymal transition (EMT) related markers of breast cancer cells. In addition, we explored whether autophagy plays a role in the inhibitory effect of doxycycline on breast cancer cells. We find that doxycyline can inhibit the viability and proliferation of breast cancer cells and BCSCs, decrease mammosphere forming efficiency, migration and invasion, and EMT of breast cancer cells. Expression of stem cell factors Oct4, Sox2, Nanog and CD44 were also significantly downregulated after doxycycline treatment. Moreover, doxycycline could down-regulate the expression of the autophagy marker LC-3BI and LC-3BII, suggesting that inhibiting autophagy may be responsible in part for the observed effects on proliferation, EMT and stem cell markers. The potent inhibition of EMT and cancer stem-like characteristics in breast cancer cells by doxycycline treatment suggests that this drug can be repurposed as an anti-cancer drug in the treatment of breast cancer patients in the clinic.
Introduction
Breast cancer (BC) is the leading site of new cancer cases and second in cancer deaths among women.Citation1-2 In 2016, it is estimated that among US women there will be 246,660 new cases of invasive breast cancer, which will result in 40,450 breast cancer deaths.Citation2 Despite effective early detection and anti-tumor therapies, approximately 10-15% of patients will experience tumor recurrence with distant metastatic disease,Citation3 the leading causes of death in breast cancer patients.Citation4
Cancer stem cells (CSCs) are a rare population of cells within a tumor that can give rise to tumors, and possess self-renewal ability and multi-lineage potency giving rise to the differentiated progeny that form the tumor bulk.Citation5 BCSCs are now thought to be key players in cancer initiation, growth, metastasis, and relapse.Citation6 In the past decades, a great deal of cancer research has focused on identifying CSCs in different types of cancers. In breast cancer, studies have identified the CD44+/CD24−/low/ESA+ subpopulation of breast cancer cells as being enriched for BCSCs.Citation6 Most importantly, BCSCs have been shown to have a quiescent phenotype and are resistant to current standard anti-cancer treatment modalities, such as chemotherapy and radiation therapy.Citation7-11 In addition, recent studies have found that 2 different and interchangeable populations of epithelial-like and mesenchymal-like CSCs exist in BCSCs.Citation12,13,14 Epithelial-like, BCSCs with high aldehyde dehydrogenase (ALDH) activity have high proliferative potential, while mesenchymal-like, CD44+CD24−/low BCSCs are quiescent and located at the tumor's invasive front.Citation14 Although great efforts have been put into identifying drugs that specifically target this important subpopulation of breast cancer cells, currently no treatments are approved in the clinic for their impact on the BCSC population. Therefore, new approaches that effectively eliminate this important subpopulation of breast cancer cells are needed.
Doxycycline is a tetracycline-derivative wide-spectrum antibiotic, which was first approved by FDA in the late 1960s.Citation15-17 Tetracyclines inhibit protein synthesis by interfering with the binding of activated aminoacyl-tRNAs on the A-site of the 30S subunit of bacterial ribosomes.Citation7 The 30S bacterial ribosome is homologous to the 28S mitochondrial ribosome in mammalian cells, leading to manageable side effects from tetracycline treatment, by inhibiting mitochondrial biogenesis in mammalian cells.Citation18,19 Interestingly, recent studies have demonstrated that BCSCs rely more on oxidative phosphorylation.Citation7,17 We found that BCSCs produce less lactate and have higher ATP content and mitochondrial labeling compared to their differentiated progeny.Citation20 Supporting a dependency of BCSCs on mitochondrial oxidative phosphorylation, more recently, Lamb et al. found that mammospheres (cell cultures enriched for BCSCs) highly upregulate mitochondrial proteins,Citation21 and that doxycycline can inhibit mitochondrial biogenesis and the “stem cell” phenotype of breast cancer cells.Citation21
However, although Lamb et al. demonstrated an effect of doxycycline on the CSC phenotype of a panel of different cancer cell lines (including 2 breast cancer lines), these observations were limited to only tumor-sphere forming assays. Therefore, the mechanism by which doxycycline exerts its inhibitory effects on the properties of breast cancer cells and BCSCs remain unclear. We build on the previous studies and in the present study investigated the inhibitory effect of doxycycline on the growth, migration, invasion, epithelial-to-mesenchymal transition (EMT), expression of stem-cell factors and autophagy in breast cancer cells in vitro.
Materials and methods
Cell culture
The human breast cancer cell lines MCF-7, and MDA-MB-468 were purchased from Chinese Academy of Sciences (Shanghai, People's Republic of China). The cells were grown in RPMI-1640 (Hyclone, Logan city, Utah, USA) containing 10% fetal bovine serum (FBS) (Hyclone), with penicillin (100 U/mL) and streptomycin (100 μg/mL). All cells were cultured in a 5% CO2 incubator at 37°C with 5% relative humidity.
Mammosphere cultures
Cells were plated in ultra-low-attachment 6-well plates (Corning, Acton, MA, USA) at a density of 1×104/mL, and cultured in serum-free DMEM/F12 media, supplemented with B27 (1:50; Invitrogen), 20 ng/mL recombinant EGF (Serotec, Raleigh, NC, USA), 20 ng/mL recombinant bFGF (PeproTech, Rocky Hill, NJ, USA), 0.4% bovine serum albumin (Sigma), and 4μg/mL insulin (Sigma). The medium containing the growth factors was replaced every 3 d. After the formation of the spheres (day 8), the cells were collected by gentle centrifugation for experiments. For the mammosphere forming efficiency experiments, 1000 cells were plated per well on 6-well plates and treated with doxycycline at the indicated concentrations or vehicle. 8-10 d later the number of mammospheres per well was counted and the percent of cells forming mammospheres was calculated.
Cell viability assay
Cells were seeded in 96-well plates at a density of 5000 cells/well. The cells were incubated with doxycycline at concentrations of 0, 1, 2, 10, 25, 50, 100 and 250 μM for 72 hours. After adding the solution of the Cell Counting Assay Kit-8 (Sigma-Aldrich) to the wells, the cells were incubated for another 2 hours. The absorbance was measured with a microplate reader at 450 nm. The amount of the formazan dye, generated by the activated dehydrogenases in the cells, was directly proportional to the number of living cells. Medium alone was used as the blank. The mammosphere proliferation assays were performed in a similar manner. % cell viability = Experimental group optical density (OD) value / Control group (OD) value ×100%. The drug concentration that inhibited 50% of the growth of control cells (IC50) was calculated by SPSS v19.0 software. All experiments were performed 3 times independently.
Flow cytometry analysis
Cells were treated with doxycycline for 72 hours in monolayer cultures. On day 4, the surviving cells were removed and stained for CD44 and CD24 expression on the cell surface, using FITC-conjugated anti-CD44 (BD PharMingen, CA, USA) and PE-conjugated anti-CD24 antibodies (BD PharMingen) according to the manufacturer's instructions. After incubation with antibodies for 30 min at 4°C, cells were analyzed by flow cytometry (BD Biosciences, Franklin Lakes, NJ, USA) and the CD44+CD24−/low BCSC population was estimated via flow cytometry.
Cell apoptosis analysis
Cells were seeded at 1×105 cells per well in 6-well plates and allowed to adhere overnight, then treated with different concentrations of doxycycline (0 µM, IC50 concentrations of doxycycline) for 72 hours. After washing with phosphate-buffered saline, the cells were resuspended in 500μL of binding buffer and incubated with annexin V–FITC/PI (BD Biosciences, San Jose, CA, USA) following the manufacturer's instructions. After incubation for 15 minutes at 4°C, the cells were analyzed using flow cytometry (BD Biosciences).
Colony formation assay
1,000 cells from MCF-7, and MDA-MB-486 cell lines were plated in 6-cm dishes and allowed to adhere overnight, then treated with doxycycline. 14 days later, the colonies were fixed with methanol and stained with 0.5% crystal violet. Colonies with over 50 cells were counted under an inverted microscope (Olympus, Japan). The surviving fraction was calculated according to the following formula: Surviving Fraction = number of colonies / number of seeded cells ×100%.
Three biologically independent experiments were performed.
Migration and invasion assays
MDA-MB-468 cells were pretreated for 72 hrs with doxycycline (IC50 concentration of 11.39uM) or vehicle. On day 4, cells were removed and counted with Trypan blue (Corning, USA). For the migration assay 30×104 viable cells were seeded on the upper layer of non-coated membrane trans-well inserts (pore size, 8.0um, Merck Millipore). Cells were allowed to adhere in starvation medium overnight and the next day the inserts were placed into the lower chambers filled with DMEM (10% FBS) incubated for 24h at 37°C at 5% CO2 atmosphere. 24 hours later cells on the top of the insert were carefully removed with a cotton swab. The invasion assay was performed as above, but Matrigel-coated membranes were used instead. Thereafter, the inserts were removed and the non-invading cancer cells remaining on the upper layer were scraped off. Cells that had migrated or invaded the matrigel and subsequently migrated to the bottom of the transwell were fixed with 3.7% paraformaldehyde for 30min, stained with 1% crystal violet in 2% ethanol for 30min, then viewed under a Nikon inverted light microscope and photographed. Three biological repeats were performed for each assay.
Quantitative real-time PCR assay
Total RNA was isolated using TRIzol reagent by the standard protocol (Ambion, Austin, Texas, USA), and the concentration was determined via 260/280 nm absorbance using a Nanodrop® ND-1000 spectrophotometer (Thermo Scientific). Reverse transcription reactions were performed using a Takara Kit (TaKaRa Bio.Inc.) following the manufacturer's instructions. The real-time PCR was performed on an ABI PRISM 7900HT Sequence Detection System. Amplification was performed in a total volume of 20 μL, which contained 10 μL of kit-supplied QuantiTect™ SYBR® Green RT-PCR Master mix (Applied-Biosystems), 0.4 μL of each primer (10 μM), 2μL of cDNA (50 ng RNA) and 7.2 μL ddH2O. The PCR primer sequences are shown in . The PCR cycling parameters were set as follows: 95°C for 5min followed by 40 cycles of PCR reacting at 95°C for 15s, 60°C for 60s, 72°C for 5min. All measurements were conducted in triplicate wells. GAPDH was used as an internal standard. The ΔΔCT values were calculated from differences between the targeted genes and GAPDH. Each experiment was repeated 3 times and averages were calculated. All PCR reactions were repeated 3 times independently. The relative quantification of the target mRNA was evaluated using 2−△△Ct calculations.
Table 1. Primer sequences.
Western blots
The extracted proteins were separated using 10% SDS-PAGE gels. Blots were incubated at 4°C overnight with the primary antibodies against NANOG (#4903S, 1:1000, Cell Signaling Technology), OCT4 (#ab109183, 1:1,000, Abcam), SOX2 (#3579S, 1:1,000, Cell Signaling Technology), E-cadherin (#3195, 1:1000, Cell Signaling Technology), N-cadherin (#ab18203, 1:1000, Abcam), vimentin (#5741, 1:1000, Cell Signaling Technology), LC3B (#2775, 1:1000, Cell Signaling Technology), and GAPDH (#5714, 1:2000, Cell Signaling Technology), followed by incubation with secondary antibodies (Santa Cruz Biotechnology) for 1 h at room temperature, then detected using ECL Prime Western Blotting Detection Reagent (GE Healthcare). Images were obtained using Image Lab 4.1.
Results
Doxycycline inhibits cell viability of breast cancer cells and mammospheres
Cells from the luminal breast cancer line, MCF-7 and the triple-negative cell line, MDA-MB-468 were treated with increasing concentrations of doxycycline for 72 hours, and the effect of doxycycline on cell viability was measured. Doxycycline inhibited viability of breast cancer cells in a dose-dependent manner with IC50 values (concentration of drug that inhibits 50% of cell viability relative to untreated cells) for MCF-7 and MDA-MB-468 of 11.39 μM and 7.13 μM respectively (). In addition, treatment with doxycycline significantly inhibited proliferation of BCSC-enriched mammosphere cultures 72 hours after a single treatment (). However, in agreement with other studies demonstrating that BCSCs are in general more resistant to anticancer drugs,Citation22 the IC50 values for BCSC-enriched mammosphere cultures increased by more than 3-fold for both lines (37.5 μM and 29.1 μM for MCF7 and MDA-MB-468, respectively) ().
Figure 1. Doxycycline inhibits cell viability of breast cancer cells. (A) Breast cancer cells propagated as monolayers (differentiated cultures, solid line) or mammospheres (enriched in BCSCs, dotted line) were treated with the indicated concentrations of doxycycline for 72h. The % viability of cells at each doxycycline dose at the end of treatment was measured using a Cell Counting Assay Kit. The horizontal dotted line represents the value of IC50 (MCF7 and MDA-MB-468 was 11.39 and 7.13uM, respectively). (B) Doxycycline significantly reduced the number of colonies in both breast cancer cell lines, MCF7 and MDA-MB-468. (C-D) Doxycycline treatment induced both early (Annexin-Vpos/PIneg) and late (Annexin-Vpos/PIpos) apoptotic cell death in both breast cancer lines.
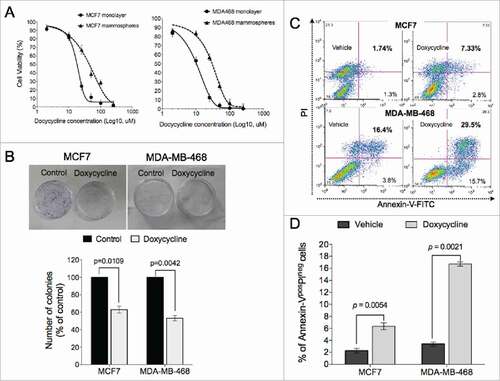
We further confirmed the toxic effect of doxycycline via clonogenic assays. The number of colonies formed in the presence of IC50 doses of doxycycline were significantly reduced compared to controls in MCF7 and MDA-MB-468 lines (p = 0.0109 and p = 0.0042, respectively, Students paired, 2-tailed t-test) (). The inhibition of proliferation of these cells by doxycycline treatment was in part due to the induction of apoptosis. Exposure to doxycycline for 72 hours resulted in accumulation of both early (Annexin-Vpos/PIneg) and late (Annexin-Vpos/PIpos) apoptotic cells in both cell lines tested (). Early apoptotic cells (Annexin-Vpos/PIneg) in the doxycycline treated group were significantly increased compared to vehicle treated groups in both cell lines (MCF7: p = 0.0054; MDA-MB-468: and p = 0.0021, Students paired, 2-tailed t-test) ().
Doxycycline inhibits stem cell marker expression and self-renewal in breast cancer stem cells
Next, we investigated the effect of doxycycline on the BCSC population using a surface marker combination for BCSCs. The CD44+CD24−/lowcell population has been shown to identify a subpopulation of cells in breast cancer enriched for BCSCs.Citation6 Treatment with a single IC50 dose of doxycycline for 72 hours significantly decreased the CD44+CD24−/lowcell population by 51% and 59%, in MCF7 and MDA-MB-468 respectively, compared to untreated cells (p < 0.05, Students paired, 2-tailed t-test) (). The effect of doxycycline on the putative CD44+CD24−/lowBCSC population was confirmed via functional mammosphere forming efficiency (MFE) assays, an assay that is widely accepted as a functional assay for measuring self-renewal capacity of stem cells.Citation23 We observed that in addition to inhibiting proliferation of breast cancer cells (), doxycycline treatment with IC50 doses also significantly inhibited the self-renewal capacity of BCSCs in both breast cancer lines as measured by the MFE assay (MFE for MCF7: vehicle, 2.96%, doxycycline, 1.28%, p = 0.0001; MFE for MDA-MB-468: vehicle, 4.14%, doxycycline, 1.41%, p = 0.0002, Students unpaired, 2-tailed t-test) ().
Figure 2. Doxycycline inhibits self-renewal of breast cancer cells. Doxycycline decreased the % of CD44+CD24-/low BCSC population in MCF7 and MDA-MB-468 cells (A), as well as mammosphere formation efficiency in both lines tested (B). (C) Relative mRNA expression levels of self-renewal related genes are significantly downregulated in doxycycline treated breast cancer cells. The dotted line represents mRNA expression of non-treated breast cancer cells. (D) Western blot analysis of the stem cell related genes. (Data are reported as means ± standard deviation, #p < 0.05, ##p < 0.01) MCF7 and MDA-MB-468 were treated with 11.39 μM and 7.13 μM doxycycline, respectively.
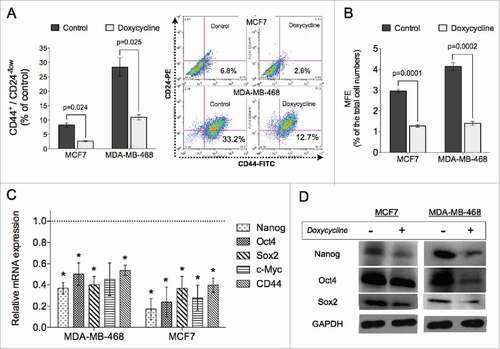
In order to further investigate the effect of doxycycline treatment on the BCSC population, we analyzed the gene and protein expression of stem cell-related factors. A single doxycycline treatment resulted in significant down-regulation of stem cell-related gene expression after 72 hours, such as nanog, oct4, sox2, and c-myc (). In addition, doxycycline treatment also inhibited the mRNA expression of cd44 (). The inhibition at the gene level of these stem cell factors was accompanied by lower protein levels after a single treatment with doxycycline compared to untreated controls ().
Doxycycline inhibits invasion, migration, and epithelial-to-mesenchymal transition of breast cancer cells
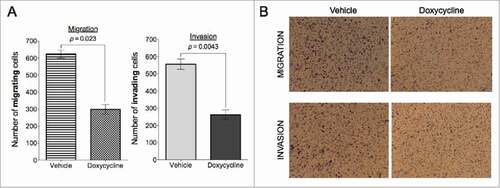
BCSCs have been shown to have an invading phenotypeCitation24 therefore, next we investigated whether the inhibition of viability by doxycycline treatment affected the invasion and migration capabilities of breast cancer cells. We performed transwell invasion and migration assays in the absence and presence of matrigel basement membrane.Citation25 MCF7 cells have relatively low migration and invasion abilitiesCitation26 therefore, we choose the MDA-MB-468 for these studies. Results showed that a 72-hour pre-treatment with doxycycline significantly inhibits their invading and migrating abilities (). Migration and invasion efficiencies were reduced by 52.08% (p = 0.023) and 52.88% (p = 0.0043, Students paired, 2-tailed t-test) respectively.
Figure 3. Doxycycline inhibits migration and invasion in MDA-MB-468 breast cancer cells. Doxycycline inhibits migration (A) and invasion (B) in MDA-MB-468 breast cancer cells in a transwell assay. MDA-MB-468 cells were treated with doxycycline for 72 h with a single dose of IC50.
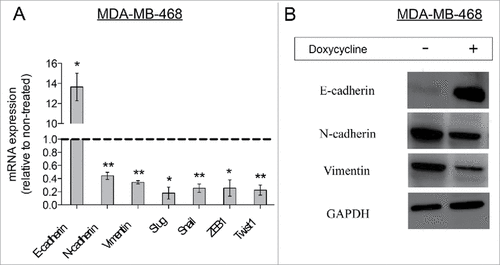
Invasion and subsequent migration of cancer cells is often preceded and mediated by an epithelial-mesenchymal transition.Citation27 Therefore, we analyzed mRNA expression levels of EMT-related genes in cells treated with doxycycline and their untreated controls. Doxycycline treatment resulted in a switch from expressing mesenchymal-related genes (N-cadherin, vimentin, snail, ZEB1, and Twist1) to expression of epithelial-related genes (such as E-cadherin) (), suggesting a doxycycline-induced conversion to a mesenchymal-to-epithelial phenotype, otherwise knows as MET.Citation28 The effect on the mRNA levels of vimentin, N-cadherin and E-cadherin were confirmed by analyzing the levels of their protein products. As expected, doxycycline treatment resulted in a suppression of mesenchymal-associated protein levels (), such as vimentin and N-cadherin, with a concomitant increase in the protein levels of E-cadherin, a protein associated with an epithelial phenotype.Citation29
Figure 4. Doxycycline decreased the EMT phenotype in MDA-MB-468 breast cancer cells. (A) Relative mRNA expression levels of EMT-related genes in doxycycline treated breast cancer cells. The dotted line represents mRNA expression of non-treated breast cancer cells. (Data are reported as means ± standard deviation, #p < 0.05, ##p < 0.01) (B) Western-blot analysis for EMT-related proteins. MDA-MB-468 cells were treated with doxycycline for 72 h with a single dose of IC50.
Doxycycline suppresses autophagy markers
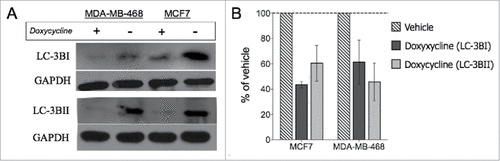
Autophagy has been shown to suppress tumor initiation at an early stage however, it can also help cancer cells survive under hypoxia, under-nutrition, antitumor therapies, and other stress conditionsCitation30 and is considered a general feature of solid tumors.Citation31,32 Earlier reports have also demonstrated an important role for autophagy in the maintenance of CSCs and metastasis.Citation32,33 Thus, we decided to analyze the effect of doxycycline on 2 autophagy-related proteins, LC-3BI and LC-3BII, as 2 of the most specific biomarkers of autophagy with broad tissue specificities and widely used in autophagy-related studies.Citation32,34 Treatment with a single dose of doxycycline resulted in suppression of protein levels of LC-3BI and LC-3BII in both cell lines tested (, Students unpaired, 2-tailed t-test), suggesting a potential mechanism by which doxycycline treatment mediates suppression of self-renewal in breast cancer stem cells.
Discussion
An increasing body of evidence demonstrates that breast cancer cell populations enriched for cells that express “stem cell” markers have significantly higher tumor-forming capacity,Citation6,35,36 and we have recently shown that this subpopulation of breast cancer cells is important not only for tumor initiation, but also propagation.Citation37 It is now believed that elimination of BCSCs is necessary to achieve long-term tumor control.
These findings have launched an effort for identifying the Achilles heel of CSCs with the goal of developing anti-cancer drugs that not only eliminate the more differentiated cells within tumors, but also effective against the CSC population. Recently, Lamb et al. used an unbiased quantitative proteomic profiling to identify the global phenotypic properties of cancer stem cells (CSCs) that could be targeted across multiple tumor types. They found that mitochondrial biogenesis was essential for the anchorage-independent clonal expansion and survival of CSCs, so this common feature could be utilized to target CSCs and treat cancer effectively as a single disease of “stemness”.Citation21 Although contradicting evidence exists in the literatures,Citation38,39 in agreement with the above studies, CSC have been shown to depend more on mitochondrial oxidative metabolism compared to their differentiated progeny in breast cancer and glioblastoma multiforme.Citation20,40 Interestingly, doxycycline, a member of the tetracycline family of broad-spectrum antibiotics, is known to interfere with mitochondrial metabolism by interfering with mitochondrial biogenesis,Citation7 thus leading to manageable side-effects.Citation7 Therefore, doxycycline has been considered as a candidate for targeting the populations of cells, which rely on mitochondrial metabolism, such as BCSCs.Citation7,20,21 To this effect, Lamb et al. demonstrated an effect of doxycycline on the ability of breast cancer cells to form mammospheres,Citation7 but these studies did not investigate additional effects of doxycycline on BCSCs and its mechanism of action. Here, we have extended these findings and demonstrate that doxycycline efficiently inhibits cell proliferation, clonogenicity, invasion, migration and the expression of stem cell markers as well as epithelial-to-mesenchymal (EMT) transition markers.
In particular, doxycycline significantly inhibits proliferation and BCSC self-renewal ability in MCF7 and MDA-MB-468 cell lines, with IC50 values similar to those reported by Lamb et al.Citation7 Of note, and in agreement with previous findingsCitation22 serum-free mammosphere cultures enriched in BCSCs were more resistant to doxycycline compared to the more differentiated monolayer cultures propagated in serum-supplemented media (). Furthermore, we demonstrated that this old drug was very efficient at eliminating the CD44+CD24−/lowBCSC population as shown in . The observation that doxycycline can eliminate the BCSC population identified by the surface marker profile (CD44+CD24−/low) was confirmed by functional mammophere formation assays (). Finally, the inhibition of self-renewal of BCSCs by doxycycline was accompanied by a significant downregulation of stem cell-associated factors, such as Oct4, Sox2, Nanog, c-myc and CD44 at the gene transcriptional level () as well as protein level (). It should be emphasized, that since CD44+CD24−/lowBCSCs display a mesenchymal state with high invasive capacity,Citation14 our results on the effect of doxycycline on BCSCs mainly apply to the mesenchymal-like BCSCs, and that these conclusions do not necessarily apply to epithelial-like, ALDH-pos BCSCs.Citation12,13,14
BCSC and EMT contributes to metastasis in breast cancer.Citation27 EMT refers to the initial stage of invasion and metastasis, during the process of metastasis, during which the polarized epithelial cells convert into motile mesenchymal cells.Citation27 The decrease expression of E-cadherin, which often occurs concurrently with increased expression of N-cadherin, is a critical step in the EMT process. Expression of vimentin represents the completely dedifferentiated state of tumor cells.Citation29 Recently, Qin Y et al. reported that doxycycline could reverse EMT and suppress the proliferation and metastasis of lung cancer cells.Citation41 Wan L et al. reported that combination of aspirin, lysine, mifepristone and doxycycline have an inhibitory effect on the expression of cell adhesion molecules and cancer metastasis.Citation42 In the present study, we found that doxycycline treated breast cancer cells have higher expression levels of E-cadherin, accompanied with lower expression levels of N-cadherin and vimentin compared to the nontreated cells, indicating a reversal of EMT (). These molecular changes were also associated by changes in migration and invasion ability, which were significantly inhibited by a single treatment with doxycycline ().
Evidence supports that autophagy plays double roles in cancer development, it acts not only as a kind of cell death form, but also as a protective mode for the survival of cancer cells, and can even enhance resistance to antitumor treatment.Citation43,44 In mammals, the expression of membrane-bound microtubule–associated protein chain 3 (LC3) has 3 isoforms, A, B, and C. The B isoform, LC3B, which has 2 subtypes, LC3B-I and LC3B-II, is one of the most specific biomarkers of autophagy.Citation32,34 LC3B expression as a common feature of solid tumors is associated with proliferation, metastasis, and poor outcome.Citation32 Recently, Chen et al. found that LC3B acts as a potential prognostic marker in local advanced breast cancer patients after neoadjuvant chemotherapy.Citation31 In addition, a growing number of studies suggest a link between autophagy and BCSCs.Citation43,46,47 Guan et al. has reported that BCSCs have a higher autophagic flux than non-CSC cellsCitation48,49 and Cufí et al. has demonstrated that autophagy positively regulates the CD44+CD24− breast cancer stem-like phenotype,Citation43 and Maycotte et al. reported that autophagy supports BCSC maintenance by modulating IL6 secretion, and that inhibition of autophagy decreases cell survival, as well as mammosphere forming efficiency.Citation46 Here, we report that doxycycline down-regulates the autophagy-related protein levels of LC-3BI and LC-3BII, suggesting a role for autophagy in the doxycycline-induced suppression of proliferation, invasion, and self-renewal of breast cancer cells.
Repositioning of old drugs to new indications is an attractive approach as it has the potential to save considerable effort and time involved in the drug development process. Doxycycline is an FDA-approved antibiotic which has been used to treat a wide variety of bacterial and parasitic infections for nearly 50 years, and importantly without significant side effects.Citation7,15,16 It has been found that doxycycline can down-regulate DNA-PK and radiosensitize tumor initiating cellsCitation50 and Pulvino et al. have reported that doxycycline could inhibit tumor growth of diffuse large B-cell lymphoma.Citation51 In combination with the study presented here, these studies make doxycycline an attractive candidate for repurposing for cancer treatment. In the clinic, doxycycline has favorable pharmacokinetics with a long serum half-life (18-22h), when a standard dose of 200mg per day is administered.Citation50
In summary, our findings support that doxycycline, a FDA approved drug, can inhibit proliferation of breast cancer cells, as well as self-renewal of BCSCs. Part of the mechanism of inhibition of self-renewal is due to the suppression of key developmental stem cell factors, such as Oct4, Sox2, Nanog and c-myc. Importantly, treatment with doxycycline results in inhibition of EMT, invasion and migration of breast cancer cells, as EMT-related markers are significantly down-regulated. Finally, doxycycline treatment suppresses autophagy-related proteins, suggesting a role autophagy in the doxycycline-induced effects. In light of this study it is plausible to envisage clinical treatment strategies for breast cancer patients that include this old drug to prevent relapse and metastasis, and improve prognosis of breast cancer patients.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by China council scholarship.
Reference
- Wang D, Lu P, Zhang H, Luo M, Zhang X, Wei X, Gao J, Zhao Z, Liu C. Oct-4 and Nanog promote the epithelial-mesenchymal transition of breast cancer stem cells and are associated with poor prognosis in breast cancer patients. Oncotarget 2014; 5(21):10803; PMID:25301732; http://dx.doi.org/10.18632/oncotarget.2506
- Siegel RL MK, Jemal A. Cancer statistics. CA Cancer J Clin 2016; 66(1):7–30; PMID:26742998; http://dx.doi.org/10.3322/caac.21332
- Weigelt B, Peterse JL, Van't Veer LJ. Breast cancer metastasis: markers and models. Nat Rev Cancer 2005; 5(8):591–602; PMID:16056258; http://dx.doi.org/10.1038/nrc1670
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA: Cancer J Clin 2011; 61(2):69–90; PMID:21296855
- Jordan CT, Guzman ML, Noble M. Cancer stem cells. New Engl J Med 2006; 355(12):1253–61; PMID:16990388; http://dx.doi.org/10.1056/NEJMra061808
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci 2003; 100(7):3983–8; http://dx.doi.org/10.1073/pnas.0530291100
- Lamb R, Ozsvari B, Lisanti CL, Tanowitz HB, Howell A, Martinez-Outschoorn UE, Sotgia F, Lisanti MP. Antibiotics that target mitochondria effectively eradicate cancer stem cells, across multiple tumor types: Treating cancer like an infectious disease. Oncotarget 2015; 6(7): 4569–4584; PMID:25625193; http://dx.doi.org/10.18632/oncotarget.3174
- Vlashi E, McBride WH, Pajonk F. Radiation responses of cancer stem cells. J Cell Biochem 2009; 108(2):339–342; PMID:19623582; http://dx.doi.org/10.1002/jcb.22275
- Phillips TM, McBride WH, Pajonk F. The response of CD24−/low/CD44+ breast cancer–initiating cells to radiation. J Natl Cancer Inst 2006; 98(24):1777–1785; PMID:17179479; http://dx.doi.org/10.1093/jnci/djj495
- Lagadec C, Vlashi E, Della Donna L, Meng Y, Dekmezian C, Kim K, Pajonk F. Survival and self-renewing capacity of breast cancer initiating cells during fractionated radiation treatment. Breast Cancer Res 2010; 12(1):1; http://dx.doi.org/10.1186/bcr2479
- Lagadec C, Vlashi E, Della Donna L, Dekmezian C, Pajonk F. Radiation-induced reprogramming of breast cancer cells. Stem Cells 2012; 30(5):833–44; PMID:22489015; http://dx.doi.org/10.1002/stem.1058
- Martin-Castillo B, Lopez-Bonet E, Cuyàs E, Viñas G, Pernas S, Dorca J, Menendez JA. Cancer stem cell-driven efficacy of trastuzumab (Herceptin): towards a reclassification of clinically HER2-positive breast carcinomas. Oncotarget 2015; 6(32):32317
- Sarrio D, Franklin CK, Mackay A, Reis-Filho JS, Isacke CM. Epithelial and mesenchymal subpopulations within normal basal breast cell lines exhibit distinct stem cell/progenitor properties. Stem Cells 2012; 30(2):292–303; PMID:22102611; http://dx.doi.org/10.1002/stem.791
- Liu S, Cong Y, Wang D, Sun Y, Deng L, Liu Y, Martin-Trevino R, Shang L, McDermott SP, Landis MD, et al. Breast cancer stem cells transition between epithelial and mesenchymal states reflective of their normal counterparts. Stem Cell Rep 2014; 2(1):78–91; PMID:24511467; http://dx.doi.org/10.1016/j.stemcr.2013.11.009
- Federici TJ. The non-antibiotic properties of tetracyclines: clinical potential in ophthalmic disease. Pharmacol Res 2011; 64(6): 614–623; PMID:21843641; http://dx.doi.org/10.1016/j.phrs.2011.06.013
- Griffin MO, Ceballos G, Villarreal FJ. Tetracycline compounds with non-antimicrobial organ protective properties: possible mechanisms of action. Pharmacol Res 2011; 63(2): 102–107; PMID:20951211; http://dx.doi.org/10.1016/j.phrs.2010.10.004
- Garrido-Mesa N, Zarzuelo A, Galvez J. ( 2013) Minocycline: far beyond an antibiotic. Br J Pharmacol 2013; 169(2): 337–352; PMID:23441623; http://dx.doi.org/10.1111/bph.12139
- Saikali Z, Singh G. Doxycycline and other tetracyclines in the treatment of bone metastasis. Anti-Cancer Drugs 2003; 14(10):773–8; PMID:14597870
- Newby AC. Matrix metalloproteinase inhibition therapy for vascular diseases. Vasc Pharmacol 2012; 56(5): 232–244; PMID:22326338
- Vlashi E, Lagadec C, Vergnes L, Reue K, Frohnen P, Chan M, Alhiyari Y, Dratver MB, Pajonk F. Metabolic differences in breast cancer stem cells and differentiated progeny. Breast Cancer Res Treatment 2014; 146(3):525–534; PMID:25007966; http://dx.doi.org/10.1007/s10549-014-3051-2
- Lamb R, Harrison H, Hulit J, Smith DL, Lisanti MP, Sotgia F. Mitochondria as new therapeutic targets for eradicating cancer stem cells: Quantitative proteomics and functional validation via MCT1/2 inhibition. Oncotarget 2014; 5(22): 11029–11037; PMID:25415228; http://dx.doi.org/10.18632/oncotarget.2789
- Vinogradov S, Wei X. Cancer stem cells and drug resistance: the potential of nanomedicine. Nanomedicine 2012; 7(4):597–615; http://dx.doi.org/10.2217/nnm.12.22
- Ponti D, Costa A, Zaffaroni N, Pratesi G, Petrangolini G, Coradini D, Pilotti S, Pierotti MA, Daidone MG. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res 2005; 65(13):5506–5511; PMID:15994920; http://dx.doi.org/10.1158/0008-5472.CAN-05-0626
- Bozorgi A, Khazaei M, Khazaei MR. New findings on breast cancer stem cells: a review. J Breast Cancer 2015; 18(4):303–312; PMID:26770236; http://dx.doi.org/10.4048/jbc.2015.18.4.303
- Shaw LM. Tumor cell invasion assays. Cell Migration: Dev Methods Protocols 2005:97–105.
- Kim MJ, Kim HS, Lee SH, Yang Y, Lee MS, Lim JS. NDRG2 controls COX-2/PGE2-mediated breast cancer cell migration and invasion. Mol Cells 2014; 37(10): 759–765; PMID:25256221; http://dx.doi.org/10.14348/molcells.2014.0232
- Hayashida T, Jinno H, Kitagawa Y, Kitajima M. Cooperation of cancer stem cell properties and epithelial-mesenchymal transition in the establishment of breast cancer metastasis. J Oncol 2011; 2011:591427; http://dx.doi.org/10.1155/2011/591427
- Meng J, Sun B, Zhao X, Zhang D, Zhao X, Gu Q, Dong X, Zhao N, Liu P, Liu Y. Doxycycline as an Inhibitor of the Epithelial-to-Mesenchymal Transition and Vasculogenic Mimicry in Hepatocellular Carcinoma. Mol Cancer Ther 2014; 3(12): 3107–3122; http://dx.doi.org/10.1158/1535-7163.MCT-13-1060
- Ding Y, Yu AQ, Li CL, Fang J, Zeng Y, Li DS. TALEN-mediated Nanog disruption results in less invasiveness, more chemosensitivity and reversal of EMT in Hela cells. Oncotarget 2014; 5(18): 8393–8401; PMID:25245189; http://dx.doi.org/10.18632/oncotarget.2298
- Ogier-Denis E, Codogno P. Autophagy: a barrier or an adaptive response to cancer. Biochimica et Biophysica Acta (BBA)-Reviews on Cancer 2003; 1603(2):113–28; http://dx.doi.org/10.1016/S0304-419X(03)00004-0
- Chen S, Jiang YZ, Huang L, Zhou RJ, Yu KD, Liu Y, Shao ZM. The residual tumor autophagy marker LC3B serves as a prognostic marker in local advanced breast cancer after neoadjuvant chemotherapy. Clin Cancer Res 2013; 19(24): 6853–6862; http://dx.doi.org/10.1158/1078-0432.CCR-13-1617
- Lazova R, Camp RL, Klump V, Siddiqui SF, Amaravadi RK, Pawelek JM. Punctate LC3B expression is a common feature of solid tumors and associated with proliferation, metastasis, and poor outcome. Clin Cancer Res 2012; 18(2): 370–379; PMID:22080440; http://dx.doi.org/10.1158/1078-0432.CCR-11-1282
- Yang MC, Wang HC, Hou YC, Tung HL, Chiu TJ, Shan YS. Blockade of autophagy reduces pancreatic cancer stem cell activity and potentiates the tumoricidal effect of gemcitabine. Mol Cancer 2015; 14(1):1; PMID:25560632; http://dx.doi.org/10.1186/1476-4598-14-1
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature 2008; 451(7182):1069–75; PMID:18305538; http://dx.doi.org/10.1038/nature06639
- Patrawala L, Calhoun T, Schneider-Broussard R, Zhou J, Claypool K, Tang DG. Side population is enriched in tumorigenic, stem-like cancer cells, whereas ABCG2+ and ABCG2− cancer cells are similarly tumorigenic. Cancer Res 2005; 65(14):6207–6219; PMID:16024622; http://dx.doi.org/10.1158/0008-5472.CAN-05-0592
- Ponti D, Costa A, Zaffaroni N, Pratesi G, Petrangolini G, Coradini D, Pilotti S, Pierotti MA, Daidone MG. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res 2005; 65(13):5506–5511; PMID:15994920; http://dx.doi.org/10.1158/0008-5472.CAN-05-0626
- Vlashi E, Lagadec C, Chan M, Frohnen P, McDonald AJ, Pajonk F. Targeted elimination of breast cancer cells with low proteasome activity is sufficient for tumor regression. Breast Cancer Res Treatment 2013; 141(2): 197–203; PMID:24013708; http://dx.doi.org/10.1007/s10549-013-2688-6
- Ciavardelli D, Rossi C, Barcaroli D, Volpe S, Consalvo A, Zucchelli M, De Cola A, Scavo E, Carollo R, D'Agostino D, et al. Breast cancer stem cells rely on fermentative glycolysis and are sensitive to 2-deoxyglucose treatment. Cell Death Dis 2014; 5(7): e1336
- Feng W, Gentles A, Nair RV, Huang M, Lin Y, Lee CY, Cai S, Scheeren FA, Kuo AH, Diehn M. Targeting unique metabolic properties of breast tumor initiating cells. Stem Cells 2014; 32(7): 1734–1745; PMID:24497069; http://dx.doi.org/10.1002/stem.1662
- Vlashi E, Lagadec C, Vergnes L, Matsutani T, Masui K, Poulou M, Popescu R, Della Donna L, Evers P, Dekmezian C, et al. Metabolic state of glioma stem cells and nontumorigenic cells. Proc Nat Acad Sci 2011; 108(38): 16062–16067; http://dx.doi.org/10.1073/pnas.1106704108
- Qin Y, Zhang Q, Lee S, Zhong WL, Liu YR, Liu HJ, Zhao D, Chen S, Xiao T, Meng J, et al. Doxycycline reverses epithelial-to-mesenchymal transition and suppresses the proliferation and metastasis of lung cancer cells. Oncotarget 2015; 6(38):40667; PMID:26512779
- Wan L, Dong H, Huo Xu JM, Zhu Y, Lu Y, Wang J, Zhang T, Li T, Xie J, Xu B, et al. Aspirin, lysine, mifepristone and doxycycline combined can effectively and safely prevent and treat cancer metastasis: prevent seeds from gemmating on soil. Oncotarget 2015; 6(34):35157; PMID:26459390
- Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gélinas C, Fan Y, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 2006; 10(1): 51–64; PMID:16843265; http://dx.doi.org/10.1016/j.ccr.2006.06.001
- Lum JJ, Bauer DE, Kong M, Harris MH, Li C, Lindsten T, Thompson CB. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell 2005; 120(2): 237–248; PMID:15680329; http://dx.doi.org/10.1016/j.cell.2004.11.046
- Cufí S, Vazquez-Martin A, Oliveras-Ferraros C, Martin-Castillo B, Vellon L, Menendez JA. Autophagy positively regulates the CD44+ CD24-/low breast cancer stem-like phenotype. Cell Cycle 2011; 10(22): 3871–3885; http://dx.doi.org/10.4161/cc.10.22.17976
- Maycotte P, Jones KL, Goodall ML, Thorburn J, Thorburn A. Autophagy Supports Breast Cancer Stem Cell Maintenance by Regulating IL6 Secretion. Mol Cancer Res 2015; 13(4):651–658; http://dx.doi.org/10.1158/1541-7786.MCR-14-0487
- Gong C, Bauvy C, Tonelli G, Yue W, Delomenie C, Nicolas V, Zhu Y, Domergue V, Marin-Esteban V, Tharinger H, et al. Beclin 1 and autophagy are required for the tumorigenicity of breast cancer stem-like/progenitor cells. Oncogene 2013; 32(18): 2261–2272; PMID:22733132; http://dx.doi.org/10.1038/onc.2012.252
- Guan JL, Simon AK, Prescott M, Menendez JA, Liu F, Wang F, Wang C, Wolvetang E, Vazquez-Martin A, Zhang J. Autophagy in stem cells. Autophagy 2013; 9(6):830–49; PMID:23486312; http://dx.doi.org/10.4161/auto.24132
- Gong C, Song E, Codogno P, Mehrpour M. The roles of BECN1 and autophagy in cancer are context dependent. Autophagy 2012; 8:1853–5; PMID:22960473; http://dx.doi.org/10.4161/auto.21996
- Lamb R, Fiorillo M, Chadwick A, Ozsvari B, Reeves KJ, Smith DL, Clarke RB, Howell SJ, Cappello AR, Martinez-Outschoorn UE, et al. Doxycycline down-regulates DNA-PK and radiosensitizes tumor initiating cells: Implications for more effective radiation therapy. Oncotarget 2015; 6(16): 14005; PMID:26087309; http://dx.doi.org/10.18632/oncotarget.4159
- Pulvino M, Chen L, Oleksyn D, Li J, Compitello G, Rossi R, Spence S, Balakrishnan V, Jordan C, Poligone B, et al. Inhibition of COP9-signalosome (CSN) deneddylating activity and tumor growth of diffuse large B-cell lymphomas by doxycycline. Oncotarget 2015; 6(17):14796; PMID:26142707; http://dx.doi.org/10.18632/oncotarget.4193