ABSTRACT
We have carried out a haploinsufficiency (HI) screen in fission yeast using heterozygous deletion diploid mutants of a genome-wide set of cell cycle genes to identify genes encoding products whose level determines the rate of progression through the cell cycle. Cell size at division was used as a measure of advancement or delay of the G2-M transition of rod-shaped fission yeast cells. We found that 13 mutants were significantly longer or shorter (greater than 10%) than control cells at cell division. These included mutants of the cdc2, cdc25, wee1 and pom1 genes, which have previously been shown to play a role in the timing of entry into mitosis, and which validate this approach. Seven of these genes are involved in regulation of the G2-M transition, 5 for nuclear transport and one for nucleotide metabolism. In addition we identified 4 more genes that were 8–10% longer or shorter than the control that also had roles in regulation of the G2-M transition or in nuclear transport. The genes identified here are all conserved in human cells, suggesting that this dataset will be useful as a basis for further studies to identify rate-limiting steps for progression through the cell cycle in other eukaryotes.
Introduction
A more complete understanding of the eukaryotic cell cycle requires the global identification of gene functions necessary for cell cycle processes. To address this, near genome-wide gene deletion libraries constructed in yeastCitation1-4 and RNAi approaches in Metazoa,Citation5-12 have been used to identify genome wide sets of cell cycle genes. In fission yeast the rod-shaped cell and tip elongation growth pattern enables cell morphology to be used as an indicator of advancement or delay in progress through the cell cycle.Citation13-16 This has allowed a comprehensive genome-wide visual screen of 4844 haploid gene deletion mutants (Bioneer library) of both essential and non-essential genes for elongated and small cell phenotypes typical of cell cycle mutants in fission yeast Citation4 and has identified 538 genes as being required for cell cycle progression. Deletion mutants of 513 genes were either delayed or blocked in cell cycle progression and had an elongated cell phenotype while a further 25 deletion mutants advanced cells through the cell cycle and showed a small cell phenotype.Citation1,4
One of the key questions is which of these genes act as rate-limiting steps for cell cycle progression and so are involved in controlling this process. Genes giving a small cell phenotype when deleted are clearly candidates for contributing to control of the G2-M transition as absence of their gene function accelerates that transition.Citation17 Another class of genes that are candidates for being rate-limiting at the G2-M transition are those which delay or block entry into mitosis if the function is partly compromised.Citation17 In this study we use haploinsufficiency, exploiting the diploid heterozygous cell cycle gene deletion mutants to compromise gene expression, to identify regulatory genes that advance or delay cell cycle progression.
Screens for haploinsufficiency (HI), using mutants where one of two gene copies in a diploid strain has been deleted, provide a powerful way to identify gene products whose level is important for a biological process of interest.Citation1,17-20 Rate-limiting steps within a regulatory network can impose limits on biological processes such as cell cycle progression, and such steps are potentially susceptible to a reduction in gene copy number. In budding yeast, heterozygous gene deletion diploid mutants usually show gene expression equivalent to half the level of the homozygous wild-type diploid.Citation21,22 Therefore, screening heterozygous gene deletion diploid mutants for haploinsufficient genes is likely to identify rate-limiting steps for a process of interest. To identify genes that are HI for cell cycle progression we screened the heterozygous gene deletion diploid mutants of both essential and non-essential cell cycle genes for an increase or decrease in the cell size at septation, as a read-out of the timing at which cells enter mitosis.Citation14,17 Mutants showing a delay or advancement into mitosis have previously identified components of the CDK regulatory network acting at the G2-M transition.Citation16,23 CDK1-CyclinB activity (Cdc2-Cdc13 in fission yeast) is the key component of the mitotic control network and is regulated by Cdc2 tyrosine 15 phosphorylation by Wee1 kinase and dephosphorylation by Cdc25 phosphatase, respectively inhibiting or activating Cdc2 kinase activity.Citation24 Cells enter mitosis when they reach a certain cell size; changing the levels of Wee1 or Cdc25 increases or decreases this size threshold, demonstrating that the activity of Cdc2 is a major rate-limiting step for cell cycle progression.Citation25,26
Results and discussion
We have carried out a haploinsufficiency screen of the genome wide set of cell cycle genes Citation4 to identify genes that are rate-limiting for cell cycle progression. Based on our initial results a further 31 genes, which did not have a typical cell cycle deletion phenotype, were also screened (see Table S1A column D, Materials and Methods for details). We measured cell length at septation for each of the diploid heterozygous gene deletion mutants and found a total of 85 mutants that showed a statistically significant deviation in cell length at septation from the control using Analysis of Variance (ANOVA) and Tukey post hoc tests (see Materials and Methods, Table S1B). The majority of these strains showed only small differences compared to the control. To focus on genes exhibiting the most significant effects we set a cut off in cell length at division of 10% longer or shorter than the control both for the mean and median values and identified 13 such genes (10% gene set) (, , Table S1B). Using esyN (http://www.esyn.org) Citation27 and the high confidence physical interaction data set from Pombase (http://www.pombase.org) together with GO slim term annotations these 13 genes were categorised into the following functional groups: i) Regulation of the G2-M transition (7 genes), ii) Nuclear cytoplasmic transport (5 genes) and iii) Nucleotide metabolism (one gene). Given that the HI group of genes included 4 previously identified HI genes cdc2, cdc25, wee1 and pom1, all of which encode components of the rate-limiting CDK regulatory network,Citation17,28 we concluded that other genes in this set are also likely to be important for the timing of the G2-M transition.
Figure 1. Haploinsufficient genes. Cell length at septation of heterozygous gene deletion mutants plotted as mean cell length with SEM. The green line shows the mean cell length of the control. n=>289 cells in at least 3 biological repeats.
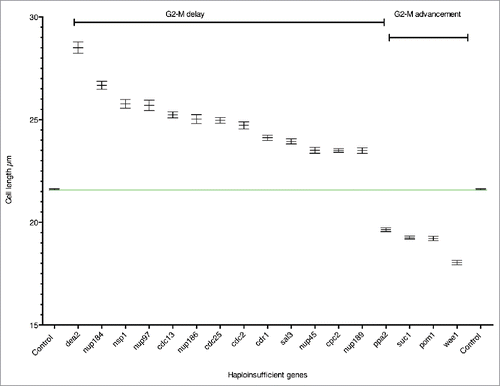
Table 1. Haploinsufficient gene set.
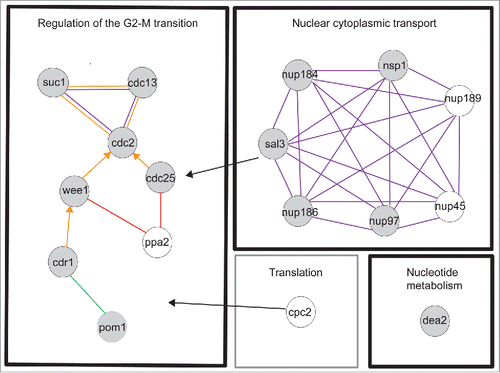
To ensure we did not miss any relevant genes just below the 10% cut-off, which were functionally related to the 10% gene set, we examined the biological function of genes 8–10% longer or shorter than the control at septation. Of the 7 genes in the 8–10% range (Table S1B) we identified 4 genes (8% gene set) that were greater than 8% longer or shorter than the control for both the mean and the median values and were directly related to Regulation of the G2-M Transition or Nuclear cytoplasmic transport (). We have considered both the 10% and 8% genes sets together as HI genes. The two categories Regulation of the G2-M transition and Nuclear cytoplasmic transport, included genes encoding products that physically interact in pathways or complexes (), further emphasizing the importance of these genes for cell cycle progression.
Figure 2. Functional groups and physical interactions for the HI genes. Evidence codes are orange edge = physical interaction, purple edge = within complex interaction, green edge = within pathway interaction, arrowhead = directed edge ie modification of a gene product by itself or another gene product, red edge = genetic interaction. Black arrows indicate genes involved in processes associated with more than one functional module. Gray filled circle = 10% gene set, unfilled circle = 8% gene set. The physical interactions were determined by esyN using Pombase high confidence interactions. The black boxes are the functional groups for the 10% gene set, the gray box is an additional functional group for the 8% gene set. This gene is also related to Regulation of the G2-M transition.
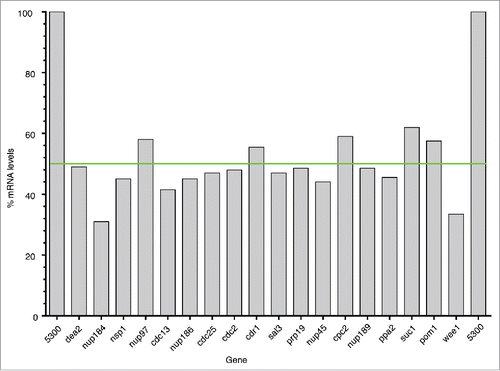
All 17 strains were checked by PCR to confirm they were deleted for the correct gene, and all non-essential gene deletion mutants were checked to confirm that they had not become homozygous at the deletion locus (Table S1C, D). We carried out qPCR to estimate the mRNA levels encoded by a single copy of each HI gene, and found that the transcript level in all cases was reduced by around 50% (Table S1E, ). This confirmed that for these genes there is no compensation by up-regulation of transcription from the remaining gene copy. For the 17 HI genes the mean and the median cell lengths were similar (Table S1B, compare columns H and K) and in the following discussion we have used the mean value when referring to cell length. The cell cycle gene set we used for the HI screen consisted of 368 (65.1%) essential genes and 197 (34.8%) non-essential genes (Table S1A). We found that the 17 HI genes had a similar distribution, with 10 (58.8%) essential genes and 7 (41.2%) non-essential genes (Fisher's Exact Test, Odds Ratio = 1.31, p > 0.05), suggesting that haploinsufficient cell cycle genes are no more likely to be essential or non-essential for cell cycle progression than the non-haploinsufficient genes.
Figure 3. mRNA expression levels for the HI gene set. Graph showing the mRNA expression level for each the HI gene in the heterozygous deletion diploid strain compared to the homozygous control strain (100%) normalized to act1 mRNA. The green line denotes 50% of the control mRNA level.
We analyzed all 565 mutants using flow cytometric analysis. All mutants showed a 4C DNA content profile similar to the control; there was no evidence for a 2C or 2–4C population that would indicate a delay in G1 or S phase (data not shown). The failure to find any genes affecting progression through G1 or S phase may mean that gene products related to these processes are in excess, or perhaps more likely, our analysis may not have been sufficiently sensitive to detect the small delays or advances in G1 or S phase to be expected in a haploinsufficiency screen.
The CDK1 mitotic control network
The timing of the G2-M transition is regulated by the conserved CDK1 mitotic control network Citation24 and we have identified 8 HI genes involved in this process (, ). The cell length at septation of the 8 heterozygous deletion diploid strains ranged between +11.6% and +16.8% (cdr1, cdc2, cdc25 and cdc13) and − 9.1% and −16.5% (ppa2, suc1, pom1 and wee1) longer (+) or shorter (−) than the control (, , Table S1B). The HI genes cdc2, cdc13, cdc25, and wee1 form the core of the mitotic control network.Citation24,29,30 The Suc1 protein forms a complex with Cdc2 in fission yeast,Citation31 and suc1 orthologues in budding yeast and frogs have been shown to affect the phosphorylation levels of a subset of CDK1 substrates.Citation30,32, 33 The fact that reduction of suc1 gene dosage in fission yeast advances cells into mitosis suggests that Suc1 normally delays mitotic entry. It has previously been shown that when the suc1 gene copy number in haploid cells is increased from one to two, cells are about 20% longer at cell division,Citation34 supporting the idea that the level of Suc1 acts as a rate-limiting inhibitor for mitotic entry. The two genes, pom1 and cdr1 Citation35-38 both act in the Pom1 cell geometry pathway that regulates CDK1 activity by inhibiting Wee1 activity, while Ppa2 directly influences the CDK1 regulatory network through both Wee1 and Cdc25 (, Table S1B). Ppa2 is a subunit of the protein phosphatase PP2A, and loss of its activity in fission yeast causes cells to enter mitosis at a small cell size. In other organisms PPA2 has been shown to be part of a CDK1 autoregulatory feedback loop required for entry into mitosisCitation39,40 and in fission yeast it is also involved in nutritional regulation of the G2-M size control.Citation41
In addition to the genes within the mitotic control network, we identified 2 genes, sal3 and cpc2 that affect localization and translation efficiency of Cdc25 and Wee1 respectively. The sal3 gene (+10.8%) is a β importin required for nuclear transport and plays a major role in Cdc25 nuclear localization, thus affecting the timing of the G2-M transition Citation42 (). The cpc2 gene (+ 8.8%) encodes the fission yeast ortholog of mammalian RACK1 (Receptor for activated C kinase 1), a conserved ribosome associated protein with a central role in signaling.Citation43 Cpc2 affects the efficient translation of a subset of proteins and may act as a scaffold for a number of signaling pathways in fission yeast.Citation44,45 In the absence of Cpc2 the level of Wee1 is increased, while the level of the Wee1 inhibitor Cdr2 is decreased, suggesting that the observed increased cell length at division of both the haploid gene deletion and diploid heterozygous gene deletion mutants could be due to a delay in activation of the Cdc2 kinase at the G2-M transition.Citation46 Cdr2 is a component of the Pom1 pathway and in our screen showed a statistically significant deviation in length at septation (+7.2%) to the control (Table S1B).
Previous studies, using reduction of function mutants of eIF4F subunits or the protein synthesis inhibitor cycloheximide, have also identified a link between translation efficiency and the translation of components of the CDK1 network; Cdc25, Wee1 and Cdc13.Citation47-51 To see if any of these genes were HI for cell cycle progression we measured cell size at septation of the heterozygous gene deletion diploid mutants of eIF4A (SPAC1006.07), eIF4E (tif45), eIF4G (tif471) and the RNA helicase sum3/ded1/moc2. None of the 4 mutants showed a statistically significant deviation in cell length at septation from the control. This suggests that a reduction of gene copy number did not reduce gene function sufficiently to affect the translation efficiency of cdc2, cdc13 or cdc25, and that these particular genes, although necessary for translation, are not HI for cell cycle progression. Regulating the translation efficiency of genes in the mitotic control network may work by directly regulating translation of genes regulating CDK activity or indirectly, for example through signaling pathways linking growth rate, monitored by translation efficiency, to the CDK network.
The 8 HI genes we have identified here all affect the timing of the G2-M transition and are rate-limiting for entry into mitosis. In addition Sal3 and Cpc2 are also potential candidates for linking other processes such as protein localization and translation efficiency to the regulation of the G2-M transition.
Nuclear transport
Six HI genes encoded nucleoporins, nup184 (+ 23.5%), nup97 (+18.9%), nsp1 (+19.3%) nup186 (+15.8%) nup45 (+ 8.8%) and nup189 (+ 8.7%) (, , , Table S1B). The nuclear pore complex (NPC) consists of around 30 subunits and studies have shown that its basic structure is very similar in different organisms including fission yeast. There are 3 major groups of nucleoporins; membrane nucleoporins which link the NPC to the inner and outer nuclear membranes, scaffold nucleoporins that form the structure of the pore and FG (phenylalanine glycine) nucleoporins, which are required for transport selectivity.Citation52-54 Five of the nucleoporins identified in this study, Nup186, Nup184, Nup97 (scaffold nucleoporins), Nsp1 and Nup45, (FG nucleoporins) are clustered together across the central core region of the nuclear pore.Citation53 Nsp1, Nup97 and Nup45 are subunits of the Nic96 sub-complex identified in humans and budding yeast.Citation55 This complex is required for nuclear pore assembly,Citation56 and haploid fission yeast mutants deleted for either nsp1 or nup97 cells arrest as ungerminated spores, probably because a number of different cellular processes dependent on nuclear cytoplasmic transport are affected. However, when the gene dosage of either of these genes is reduced in diploid cells, cells are viable but show a cell cycle delay. Nup45 is also a Nic96 subunit, but unlike Nsp1 and Nup97, the nup45 gene deletion mutant has a cell cycle phenotype in haploid cells as well as in the heterozygous gene deletion diploid mutant.Citation4,57 The remaining Nic96 complex subunit Nup44 also has a cell cycle deletion phenotype in haploid cells and in our study showed a statistically significant deviation in cell length at septation (+7.1%) compared to the control (Table S1B). These data suggest that when the Nic96 complex is compromised, as in diploid heterozygous gene deletion mutants of nsp1, nup97, nup45 or nup44, or in the haploid deletion mutants of nup45 and nup44, it is primarily the cell cycle function that is affected.
The only nucleoporin we identified that is not found in the NPC central core was Nup189,Citation58 which undergoes autocleavage to form 2 distinct nucleoporins (known as Nup189N or Nup98 and Nup189C or Nup96), although in fission yeast this cleavage is not essential for cell viability.Citation59 Nup189N/Nup98 is an FG nucleoporin located in the outer ring of the nuclear pore, while Nup189C/Nup96 is a scaffold nucleoporin.
In HeLa cells, nucleoporins have been implicated in the G2-M transition as a knockdown of Nsp1/Nup62 (Nic96 subunit) causes a G2-M arrest,Citation60 possibly as a result of altered localization of factors regulating the G2-M transition. It has also been shown that disassembly of the NPC at mitosis by CDK1 dependent phosphorylation of the GLFG repeats in Nup98 (Nup189N/Nup98 in S. pombe) is important for nuclear envelope breakdown and mitotic progression.Citation61 Nucleoporins may also have roles that are independent of their role in nuclear transport. The Nic96 complex, for example, also affects chromosome segregation and spindle orientationCitation56,60 and in budding yeast has a role ensuring equal segregation of nuclear pore complexes (NPCs) to daughter cells.Citation62,63 Other studies in human cells have shown that nucleoporins may function as transcription factors regulating genes with a cell cycle related function.Citation64 Nuclear transport also plays a central role during ribosome biogenesis,Citation65 so it is possible that these haploinsufficient nucleoporins mainly affect translation and have an indirect effect on cell cycle progression.
All the nuclear transport genes identified in this screen are conserved in other eukaryotes Citation66 (http://www.pombase.org/ ), and as mentioned earlier, Sal3 (+10.8%) (, , ) encodes a β importin regulating nuclear transport of Cdc25.Citation42 Based on these results for Sal3 we propose that the nucleopore proteins identified here affect nuclear import or export of components of the CDK regulatory network.
Other genes affecting the timing of entry into mitosis
The HI gene dea2, encodes an adenine deaminase (+31.9%)Citation67 (, , Table S1B). Fission yeast adenine deaminases are related to the eukaryotic adenosine deaminases, which play a key role in the adenine salvage pathway (http://www.ebi.ac.uk/interpro/entry/IPR006330), and are involved in adenine catabolism.Citation67 Because of its role in adenine catabolism in fission yeast a reduction in the level of Dea2 may lead to an imbalance between de novo synthesis, salvage and degradation of purines, which could affect both DNA replication and repair directly or indirectly via nucleotide/deoxynucleotide levels. A number of other genes involved in nucleotide metabolism (adk1, hpt1, dut1, dcd1, tmp1, and dfr1), although not haploinsufficient, have been identified as cell cycle genes,Citation4 supporting the idea that levels of nucleotide intermediates are important for progress through the cell cycle. In HeLa cells the impairment of adenosine deaminase leads to high levels of dATP, which is an inhibitor of ribonucleotide reductase (RNR).Citation68,69 In fission yeast the large subunit of RNR, encoded by cdc22 and the small subunit encoded by suc22, are both essential for DNA replication and cell viability.Citation70 However neither gene was identified in this study as HI either for the G2-M transition or progression through G1 or S phase (data not shown) nor did either gene show a statistically significant deviation in cell length at septation from the control. Flow cytometry showed that cells with reduced dea2 gene dosage did not cause an obvious delay in G1 or S phase progression (Fig. S1), suggesting that the dea2 heterozygous gene deletion strain is delayed in the G2 phase of the cell cycle. Perhaps when expression of dea2 is only partially compromised (), RNR has sufficient activity to allow a doubling of DNA during S phase but that error prone DNA synthesis occurs due to increased levels of dATP, leading to activation of the DNA damage checkpoint and a delay of entry into mitosis. Alternatively S phase maybe extended with DNA replication continuing at a low level below our level of detection, due to reduced activity of RNR and activation of the DNA replication checkpoint to delay onset of mitosis.
Rate-limiting steps
The idea of major rate-limiting steps controlling the rate of progress through a pathway was first postulated over 100 years ago.Citation71 Since then work on metabolic control has led to the idea that many enzymes within a metabolic pathway contribute to flux through the pathway rather than there being a major step.Citation72 For the rate of progression through the cell cycle, 2 major rate-limiting steps have been identified, one at the G1-S and one at the G2-M transition.Citation73-76 For both of these transitions the key step is activation of CDK1 activity.Citation74,75,77 In this study we have identified a number of genes that regulate Cdc2 activity, most of which act within the Cdc2 mitotic control network (). These results suggest that regulation of Cdc2 kinase activity by the mitotic control network occurs at a number of different steps each contributing to the overall timing of entry into mitosis. Other HI genes, for example cpc2, dea2 and sal3 affect other processes as well as cell cycle progression and these genes could be acting as major rate-limiting steps linking processes such as translation efficiency, nucleotide levels and localization of cell cycle regulators with cell cycle progression. Our results suggest that regulation of cell cycle progression depends not only on the major rate-limiting steps but is distributed further to a number of different steps, integrating multiple input signals feeding into the major cell cycle control genes.
Our haploinsufficiency screening approach has identified the canonical rate-limiting gene functions regulating the CDK1 mitotic control network involved in the G2 to mitosis transition, cdc2, cdc13, wee1 and cdc25. In addition we have identified a further 4 genes, suc1, cdr1, ppa2 and pom1, all previously shown to interact with components of the mitotic control network. Of these 4 genes, suc1, cdr1 and ppa2 have not previously been shown to be rate-limiting when their expression is reduced in a limited way. Translation efficiency has previously been implicated in cell cycle progression, and we have identified cpc2 that influences translation efficiency and may be rate-limiting for levels of the CDK1 mitotic control network components. In addition we have identified 6 genes encoding nuclear pore complex subunits, Nup184, Nup186, Nup189N/Nup98, Nup189C/Nup96, Nsp1, Nup97, Nup45 and Sal3 a β importin which imports Cdc25 into the nucleus and regulates its nuclear localization.Citation42 We propose that nuclear transport plays a key rate-limiting role in the regulation of the CDK1 mitotic control network, which has not been fully recognized. Finally we have identified Dea2 that, unexpectedly, implicates nucleotide metabolism in regulation of the G2-M transition, perhaps via a defect in DNA replication and activation of the DNA damage/replication checkpoint controls.
As the number of identified open reading frames increases in S. pombe (currently 5118 protein coding genes, http://www.pombase.org/status/statistics ), more genes important for cell cycle regulation can be expected to be identified. For example new open reading frames of less than 100 amino acids identified in fission yeast and deletion of genes not included in the Bioneer library have revealed a further 2 cell cycle genes.Citation78,79 Further analysis of the Bioneer non-essential gene collection of ∼3000 haploid deletion mutants has also identified additional cell cycle genes. A screen for genes generating a small (wee) cell deletion phenotype identified 18 genes (an additional 8 genes).Citation80 A multiprocess screen uncovered 25 genes in addition to the 538 genes previously identified, which affected the duration of the cell cycle phases but which did not have a long cell deletion phenotype, and a further 9 genes that when deleted result in an increased cell volume at cell division.Citation81
As genes required for the cell cycle and its regulation are highly conserved in eukaryotes Citation24 it is likely that the haploinsufficient genes identified here as new rate-limiting components of the CDK1 mitotic network and nuclear transport, are also important for the timing of progression through the cell cycle in other eukaryotes. Further investigation of the role of these genes and the identification of new cell cycle genes in both fission yeast and other eukaryotes is likely to reveal new levels of cell cycle control and illuminate how this is linked to other cellular processes such as translation and cellular localization.
Materials and methods
Media and cell growth conditions
These were as described by Moreno, Klar and NurseCitation82 unless otherwise stated.
Gene set and cell length measurements
All strains were grown in YES liquid media at 32°C to mid exponential growth (∼2 × 106 − 8 × 106 cells /ml). Live cells were examined using a Zeiss Axioskop 40 with a X63 Plan APOCHROMAT 1.4 oil immersion objective and photographed using a Zeiss Axiocam. Cell length for ∼30 septated cells for the initial analysis and at least 100 septated cells for each biological repeat was measured using pointpicker in Image J, (Table S1B). The subset of heterozygous diploid mutants used for this study was from the genome-wide set constructed by The South Korean consortium including Bioneer Citation1 and used by Hayles et al.Citation4 The majority of strains in all the versions of the Bioneer haploid libraries are derived from this diploid heterozygous gene deletion library. The 17 HI strains identified in this study were the originally constructed diploids, except for 3 strains heterozygous for deletions of cdc25, wee1 or sal3, which were host exchanged diploids to remove a background ‘miss’ mutation.Citation1 The working code suffix −0 or −1 (Table S1A column C) denotes host exchange or reconstructed deletion respectively.
Four genes from the genome wide, 538 gene set were omitted from the study for technical reasons e.g. strains were a mixed culture or haploidised at high frequency and we included 31 genes that when deleted do not have the classical cell cycle phenotype of long or small cells. These genes were included as they were known to be required for the cell cycle (4 genes), were related to genes we identified as HI genes (23 nuclear transport related genes) or were required for the initiation of translation (4 genes) (Table S1A column D).
Flow cytometry analysis
Flow cytometry analysis was carried out as described in,Citation83 using a Becton Dickinson FACScan.
PCR and primer sequences
Total RNA was isolated from S. pombe cells using the hot phenol RNA extraction methodCitation84 and purified using RNeasy columns (Qiagen). Contaminating genomic DNA was removed by DNase 1 treatment (Ambion rDNase1). cDNA was synthesized using a Superscript III Invitrogen kit (Catalog no. 18080–051) with Oligo(dT)20 primers. qPCR reactions were carried out using EXPRESS SYBR® GreenER™ (Invitrogen) according to manufacturer's recommendations in an ABI 7500 Fast qPCR machine (annealing temp of 56°C). Levels of transcript were quantified using a standard curve method. Two biological repeats were carried out for each strain.
Primers used to confirm that the strain was deleted for the correct gene are shown in Table S1C and to confirm the strain was heterozygous for non-essential gene deletions are shown in Table S1D. Primers used for qPCR to measure the transcript level are shown in Table S1E. All primers were selected using ApE-A plasmid editor v1.17.
Statistical analyses
Cell lengths from each mutant strain and wildtype measurements were analyzed using R software with the core statistics package (see Table S1B for summaries). The dataset was fitted with an Analysis of Variance (ANOVA) model, which identified significant differences between the strains (F = 29.54, df = 565, p <2e-16). To identify which strains contributed to this difference, a post-hoc Tukey test was applied in order to identify strains with significantly different cell length measurements from the control strain. To determine which genes could confidently be designated as HI we used 2 criteria: a) The distribution of cell length measurements showed a statistically significant deviation from the control cell length measurements and, b) The average cell length was ≥10% longer or ≤10% shorter than that of the wildtype control. The implementation of this second criterion was in order to minimise type I errors (false-positives) and ensure that all genes designated as HI were also likely to be biologically significant. While the 10% cut-off was somewhat arbitrary, the range is similar to the batch-variability of the wildtype strain (141 batches, median = 21.416 µm, lowest batch median = 19.11 µm (10.7% shorter), highest batch median = 23.53 µm (9.8% longer)). Additional genes functionally related to the HI genes are referred to within the text when they show average lengths ≥8% significantly longer or ≤8% significantly shorter than the control, in order to highlight similar behavior within classes of genes.
Batch effects between different experiments of the same strain were also tested by ANOVA to validate the reproducibility of data sets and are shown in Table S1, column W.
Abbreviations
| HI | = | haploinsufficiency or haploinsufficient |
| CDK1 | = | cyclin dependent kinase 1 |
| RACK | = | Receptor for activated C kinase |
| nup | = | nucleoporin |
| NPC | = | nuclear pore complex |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
1242535_Supplemental_Material.zip
Download Zip (378.8 KB)Acknowledgments
We thank everyone in the Cell Cycle laboratory, particularly Fran Navarro, Helena Cantwell, Scott Curran and Jessica Greenwood for reading the manuscript and for helpful and insightful comments. We would also like to thank Val Wood at Pombase for her invaluable help and for reading the manuscript and Giorgio Favrin for his help with esyN.
Funding
This work was supported by The Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001121), The Wellcome Trust Grant 093917 and The Breast Cancer Research Foundation.
References
- Kim DU, Hayles J, Kim D, Wood V, Park HO, Won M, Yoo HS, Duhig T, Nam M, Palmer G, et al. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol 2010; 28:617-23; PMID:20473289; http://dx.doi.org/10.1038/nbt.1628
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature 2002; 418:387-91; PMID:12140549; http://dx.doi.org/10.1038/nature00935
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 1999; 285:901-6; PMID:10436161; http://dx.doi.org/10.1126/science.285.5429.901
- Hayles J, Wood V, Jeffery L, Hoe KL, Kim DU, Park HO, Salas-Pino S, Heichinger C, Nurse P. A genome-wide resource of cell cycle and cell shape genes of fission yeast. Open biology 2013; 3:130053; PMID:23697806; http://dx.doi.org/10.1098/rsob.130053
- Pau G, Walter T, Neumann B, Heriche JK, Ellenberg J, Huber W. Dynamical modelling of phenotypes in a genome-wide RNAi live-cell imaging assay. BMC Bioinformatics 2013; 14:308; PMID:24131777; http://dx.doi.org/10.1186/1471-2105-14-308
- Lee J, Nam S, Hwang SB, Hong M, Kwon JY, Joeng KS, Im SH, Shim J, Park MC. Functional genomic approaches using the nematode Caenorhabditis elegans as a model system. J Biochem Mol Biol 2004; 37:107-13; PMID:14761308; http://dx.doi.org/10.5483/BMBRep.2004.37.1.107
- Bjorklund M, Taipale M, Varjosalo M, Saharinen J, Lahdenpera J, Taipale J. Identification of pathways regulating cell size and cell-cycle progression by RNAi. Nature 2006; 439:1009-13; PMID:16496002; http://dx.doi.org/10.1038/nature04469
- Neumann B, Walter T, Heriche JK, Bulkescher J, Erfle H, Conrad C, Rogers P, Poser I, Held M, Liebel U, et al. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature 2010; 464:721-7; PMID:20360735; http://dx.doi.org/10.1038/nature08869
- Gonczy P, Echeverri C, Oegema K, Coulson A, Jones SJ, Copley RR, Duperon J, Oegema J, Brehm M, Cassin E, et al. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature 2000; 408:331-6; PMID:11099034; http://dx.doi.org/10.1038/35042526
- Billmann M, Horn T, Fischer B, Sandmann T, Huber W, Boutros M. A genetic interaction map of cell cycle regulators. Mol Biol Cell 2016; 27:1397-407; PMID:26912791; http://dx.doi.org/10.1091/mbc.E15-07-0467
- Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 2000; 408:325-30; PMID:11099033; http://dx.doi.org/10.1038/35042517
- Somma MP, Ceprani F, Bucciarelli E, Naim V, De Arcangelis V, Piergentili R, Palena A, Ciapponi L, Giansanti MG, Pellacani C, et al. Identification of Drosophila mitotic genes by combining co-expression analysis and RNA interference. PLoS Genet 2008; 4:e1000126; PMID:18797514; http://dx.doi.org/10.1371/journal.pgen.1000126
- Bonatti S, Simili M, Abbondandola A. Isolation of temperature sensitive mutants of Schizosaccharomyces pombe. J Bact 1972; 109:484-91; PMID:4110142
- Mitchison JM. The growth of single cells. I. Schizosaccharomyces pombe. Exp Cell Res 1957; 13:244-62; PMID:13480293; http://dx.doi.org/10.1016/0014-4827(57)90005-8
- Nurse P. Genetic control of cell size at cell division in yeast. Nature 1975; 256:547-51; PMID:1165770; http://dx.doi.org/10.1038/256547a0
- Nurse P, Thuriaux P, Nasmyth K. Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet 1976; 146:167-78; PMID:958201; http://dx.doi.org/10.1007/BF00268085
- Nurse P, Thuriaux P. Regulatory genes controlling mitosis in the fission yeast Schizosaccharomyces pombe. Genetics 1980; 96:627-37; PMID:7262540
- Deutschbauer AM, Jaramillo DF, Proctor M, Kumm J, Hillenmeyer ME, Davis RW, Nislow C, Giaever G. Mechanisms of haploinsufficiency revealed by genome-wide profiling in yeast. Genetics 2005; 169:1915-25; PMID:15716499; http://dx.doi.org/10.1534/genetics.104.036871
- Chuang CH, Wallace MD, Abratte C, Southard T, Schimenti JC. Incremental genetic perturbations to MCM2-7 expression and subcellular distribution reveal exquisite sensitivity of mice to DNA replication stress. PLoS Genet 2010; 6:e1001110; PMID:20838603; http://dx.doi.org/10.1371/journal.pgen.1001110
- Hafen E, Dickson B, Brunner T, Raabe T. Genetic dissection of signal transduction mediated by the sevenless receptor tyrosine kinase in Drosophila. Philos Trans R Soc Lond B Biol Sci 1993; 340:273-8; PMID:8103929; http://dx.doi.org/10.1098/rstb.1993.0068
- Springer M, Weissman JS, Kirschner MW. A general lack of compensation for gene dosage in yeast. Mol Syst Biol 2010; 6:368; PMID:20461075; http://dx.doi.org/10.1038/msb.2010.19
- Torres EM, Springer M, Amon A. No current evidence for widespread dosage compensation in S. cerevisiae. elife 2016; 5.e10996.
- Thuriaux P, Nurse P, Carter B. Mutants altered in the control co-ordinating cell division with cell growth in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet 1978; 161:215-20; PMID:672898
- Nurse P. Universal control mechanism regulating onset of M-phase. Nature 1990; 344:503-8; PMID:2138713; http://dx.doi.org/10.1038/344503a0
- Russell P, Nurse P. Negative regulation of mitosis by wee1+, a gene encoding a protein kinase homolog. Cell 1987; 49:559-67; PMID:3032459; http://dx.doi.org/10.1016/0092-8674(87)90458-2
- Russell P, Nurse P. cdc25+ functions as an inducer in the mitotic control of fission yeast. Cell 1986; 45:145-53; PMID:3955656; http://dx.doi.org/10.1016/0092-8674(86)90546-5
- Bean DM, Heimbach J, Ficorella L, Micklem G, Oliver SG, Favrin G. esyN: network building, sharing and publishing. PLoS One 2014; 9:e106035; PMID:25181461; http://dx.doi.org/10.1371/journal.pone.0106035
- Bhatia P, Hachet O, Hersch M, Rincon SA, Berthelot-Grosjean M, Dalessi S, Basterra L, Bergmann S, Paoletti A, Martin SG. Distinct levels in Pom1 gradients limit Cdr2 activity and localization to time and position division. Cell Cycle 2014; 13:538-52; PMID:24316795; http://dx.doi.org/10.4161/cc.27411
- Morgan D. Cell Cycle: Principles of Control. London: New Science Press, 2006.
- McGrath DA, Balog ER, Koivomagi M, Lucena R, Mai MV, Hirschi A, Kellogg DR, Loog M, Rubin SM. Cks confers specificity to phosphorylation-dependent CDK signaling pathways. Nat Struct Mol Biol 2013; 20:1407-14; PMID:24186063; http://dx.doi.org/10.1038/nsmb.2707
- Brambilla P, Ducommun B, Draetta G. Cdc2 protein kinase: interactions with cyclins and Suc1. Cold Spring Harbor symposia on quantitative biology 1991; 56:515-21; PMID:1840262; http://dx.doi.org/10.1101/SQB.1991.056.01.058
- Koivomagi M, Ord M, Iofik A, Valk E, Venta R, Faustova I, Kivi R, Balog ER, Rubin SM, Loog M. Multisite phosphorylation networks as signal processors for Cdk1. Nat Struct Mol Biol 2013; 20:1415-24; PMID:24186061; http://dx.doi.org/10.1038/nsmb.2706
- Ha SH, Kim SY, Ferrell JE, Jr. The Prozone Effect Accounts for the Paradoxical Function of the Cdk-Binding Protein Suc1/Cks. Cell Rep 2016; 14:1408-21; PMID:26854218; http://dx.doi.org/10.1016/j.celrep.2016.01.033
- Hayles J, Aves S, Nurse P. suc1 is an essential gene involved in both the cell cycle and growth in fission yeast. Embo J 1986; 5:3373-9; PMID:16453733
- Martin SG, Berthelot-Grosjean M. Polar gradients of the DYRK-family kinase Pom1 couple cell length with the cell cycle. Nature 2009; 459:852-6; PMID:19474792; http://dx.doi.org/10.1038/nature08054
- Moseley JB, Mayeux A, Paoletti A, Nurse P. A spatial gradient coordinates cell size and mitotic entry in fission yeast. Nature 2009; 459:857-60; PMID:19474789; http://dx.doi.org/10.1038/nature08074
- Russell P, Nurse P. The mitotic inducer Nim1+ functions in a regulatory network of protein kinase homologs controlling the initiation of mitosis. Cell 1987; 49:569-76; PMID:3453113; http://dx.doi.org/10.1016/0092-8674(87)90459-4
- Young PG, Fantes PA. Schizosaccharomyces pombe mutants affected in their division response to starvation. J Cell Sci 1987; 88 (Pt 3):295-304; PMID:3448096
- Yu J, Zhao Y, Li Z, Galas S, Goldberg ML. Greatwall kinase participates in the Cdc2 autoregulatory loop in Xenopus egg extracts. Molecular cell 2006; 22:83-91; PMID:16600872; http://dx.doi.org/10.1016/j.molcel.2006.02.022
- Mochida S, Maslen SL, Skehel M, Hunt T. Greatwall phosphorylates an inhibitor of protein phosphatase 2A that is essential for mitosis. Science 2010; 330:1670-3; PMID:21164013; http://dx.doi.org/10.1126/science.1195689
- Chica N, Rozalen AE, Perez-Hidalgo L, Rubio A, Novak B, Moreno S. Nutritional Control of Cell Size by the Greatwall-Endosulfine-PP2A.B55 Pathway. Curr Biol 2016; 26:319-30; PMID:26776736; http://dx.doi.org/10.1016/j.cub.2015.12.035
- Chua G, Lingner C, Frazer C, Young PG. The sal3(+) gene encodes an importin-beta implicated in the nuclear import of Cdc25 in Schizosaccharomyces pombe. Genetics 2002; 162:689-703; PMID:12399381
- Nilsson J, Sengupta J, Frank J, Nissen P. Regulation of eukaryotic translation by the RACK1 protein: a platform for signalling molecules on the ribosome. EMBO reports 2004; 5:1137-41; PMID:15577927; http://dx.doi.org/10.1038/sj.embor.7400291
- Shor B, Calaycay J, Rushbrook J, McLeod M. Cpc2/RACK1 is a ribosome-associated protein that promotes efficient translation in Schizosaccharomyces pombe. J Biol Chem 2003; 278:49119-28; PMID:12972434; http://dx.doi.org/10.1074/jbc.M303968200
- McLeod M, Shor B, Caporaso A, Wang W, Chen H, Hu L. Cpc2, a fission yeast homologue of mammalian RACK1 protein, interacts with Ran1 (Pat1) kinase To regulate cell cycle progression and meiotic development. Mol Cell Biol 2000; 20:4016-27; PMID:10805744; http://dx.doi.org/10.1128/MCB.20.11.4016-4027.2000
- Nunez A, Franco A, Soto T, Vicente J, Gacto M, Cansado J. Fission yeast receptor of activated C kinase (RACK1) ortholog Cpc2 regulates mitotic commitment through Wee1 kinase. J Biol Chem 2010; 285:41366-73; PMID:20974849; http://dx.doi.org/10.1074/jbc.M110.173815
- Daga RR, Jimenez J. Translational control of the Cdc25 cell cycle phosphatase: a molecular mechanism coupling mitosis to cell growth. J Cell Sci 1999; 112 (Pt 18):3137-46; PMID:10462529
- Suda M, Yamada S, Toda T, Miyakawa T, Hirata D. Regulation of Wee1 kinase in response to protein synthesis inhibition. FEBS letters 2000; 486:305-9; PMID:11119724; http://dx.doi.org/10.1016/S0014-5793(00)02299-7
- Grallert B, Kearsey SE, Lenhard M, Carlson CR, Nurse P, Boye E, Labib K. A fission yeast general translation factor reveals links between protein synthesis and cell cycle controls. J Cell Sci 2000; 113 (Pt 8):1447-58; PMID:10725227
- Forbes KC, Humphrey T, Enoch T. Suppressors of cdc25p overexpression identify two pathways that influence the G2/M checkpoint in fission yeast. Genetics 1998; 150:1361-75; PMID:9832516
- Liu HY, Nefsky BS, Walworth NC. The Ded1 DEAD box helicase interacts with Chk1 and Cdc2. J Biol Chem 2002; 277:2637-43; PMID:11711540; http://dx.doi.org/10.1074/jbc.M109016200
- Bilokapic S, Schwartz TU. 3D ultrastructure of the nuclear pore complex. Current opinion in cell biology 2012; 24:86-91; PMID:22244612; http://dx.doi.org/10.1016/j.ceb.2011.12.011
- Asakawa H, Yang HJ, Yamamoto TG, Ohtsuki C, Chikashige Y, Sakata-Sogawa K, Tokunaga M, Iwamoto M, Hiraoka Y, Haraguchi T. Characterization of nuclear pore complex components in fission yeast Schizosaccharomyces pombe. Nucleus 2014; 5:149-62; PMID:24637836; http://dx.doi.org/10.4161/nucl.28487
- Ibarra A, Hetzer MW. Nuclear pore proteins and the control of genome functions. Genes Dev 2015; 29:337-49; PMID:25691464; http://dx.doi.org/10.1101/gad.256495.114
- Schrader N, Stelter P, Flemming D, Kunze R, Hurt E, Vetter IR. Structural basis of the nic96 subcomplex organization in the nuclear pore channel. Mol Cell 2008; 29:46-55; PMID:18206968; http://dx.doi.org/10.1016/j.molcel.2007.10.022
- Vollmer B, Antonin W. The diverse roles of the Nup93/Nic96 complex proteins – structural scaffolds of the nuclear pore complex with additional cellular functions. Biol Chem 2014; 395:515-28; PMID:24572986; http://dx.doi.org/10.1515/hsz-2013-0285
- Chen XQ, Du X, Liu J, Balasubramanian MK, Balasundaram D. Identification of genes encoding putative nucleoporins and transport factors in the fission yeast Schizosaccharomyces pombe: a deletion analysis. Yeast 2004; 21:495-509; PMID:15116432; http://dx.doi.org/10.1002/yea.1115
- Tange Y, Hirata A, Niwa O. An evolutionarily conserved fission yeast protein, Ned1, implicated in normal nuclear morphology and chromosome stability, interacts with Dis3, Pim1/RCC1 and an essential nucleoporin. J Cell Sci 2002; 115:4375-85; PMID:12376568; http://dx.doi.org/10.1242/jcs.00135
- Asakawa H, Mori C, Ohtsuki C, Iwamoto M, Hiraoka Y, Haraguchi T. Uncleavable Nup98-Nup96 is functional in the fission yeast Schizosaccharomyces pombe. FEBS open bio 2015; 5:508-14; PMID:26137436; http://dx.doi.org/10.1016/j.fob.2015.06.004
- Hashizume C, Moyori A, Kobayashi A, Yamakoshi N, Endo A, Wong RW. Nucleoporin Nup62 maintains centrosome homeostasis. Cell Cycle 2013; 12:3804-16; PMID:24107630; http://dx.doi.org/10.4161/cc.26671
- Laurell E, Beck K, Krupina K, Theerthagiri G, Bodenmiller B, Horvath P, Aebersold R, Antonin W, Kutay U. Phosphorylation of Nup98 by multiple kinases is crucial for NPC disassembly during mitotic entry. Cell 2011; 144:539-50; PMID:21335236; http://dx.doi.org/10.1016/j.cell.2011.01.012
- Colombi P, Webster BM, Frohlich F, Lusk CP. The transmission of nuclear pore complexes to daughter cells requires a cytoplasmic pool of Nsp1. J Cell Biol 2013; 203:215-32; PMID:24165936; http://dx.doi.org/10.1083/jcb.201305115
- Makio T, Lapetina DL, Wozniak RW. Inheritance of yeast nuclear pore complexes requires the Nsp1p subcomplex. J Cell Biol 2013; 203:187-96; PMID:24165935; http://dx.doi.org/10.1083/jcb.201304047
- Kalverda B, Pickersgill H, Shloma VV, Fornerod M. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell 2010; 140:360-71; PMID:20144760; http://dx.doi.org/10.1016/j.cell.2010.01.011
- Nazar RN. Ribosomal RNA processing and ribosome biogenesis in eukaryotes. IUBMB life 2004; 56:457-65; PMID:15545225; http://dx.doi.org/10.1080/15216540400010867
- Terry LJ, Wente SR. Flexible gates: dynamic topologies and functions for FG nucleoporins in nucleocytoplasmic transport. Eukaryot Cell 2009; 8:1814-27; PMID:19801417; http://dx.doi.org/10.1128/EC.00225-09
- Ribard C, Rochet M, Labedan B, Daignan-Fornier B, Alzari P, Scazzocchio C, Oestreicher N. Sub-families of alpha/beta barrel enzymes: a new adenine deaminase family. J Mol Biol 2003; 334:1117-31; PMID:14643670; http://dx.doi.org/10.1016/j.jmb.2003.10.005
- Lin AL, Elford HL. Adenosine deaminase impairment and ribonucleotide reductase activity and levels in HeLa cells. J Biol Chem 1980; 255:8523-8; PMID:6997299
- Jonna VR, Crona M, Rofougaran R, Lundin D, Johansson S, Brannstrom K, Sjoberg BM, Hofer A. Diversity in Overall Activity Regulation of Ribonucleotide Reductase. J Biol Chem 2015; 290:17339-48; PMID:25971975; http://dx.doi.org/10.1074/jbc.M115.649624
- Fernandez Sarabia MJ, McInerny C, Harris P, Gordon C, Fantes P. The cell cycle genes cdc22+ and suc22+ of the fission yeast Schizosaccharomyces pombe encode the large and small subunits of ribonucleotide reductase. Mol Gen Genet 1993; 238:241-51; PMID:8479429
- Blackman FF. Optima and limiting factors. Ann Bot 1905; 19:281-98
- Kacser H, Burns JA. The control of flux. Biochemical Society transactions 1995; 23:341-66.
- Pardee AB. G1 events and regulation of cell proliferation. Science 1989; 246:603-8; PMID:2683075; http://dx.doi.org/10.1126/science.2683075
- Nurse P, Bissett Y. Gene required in G1 for commitment to cell cycle and in G2 for control of mitosis in fission yeast. Nature 1981; 292:558-60; PMID:7254352; http://dx.doi.org/10.1038/292558a0
- Johnston GC, Pringle JR, Hartwell LH. Coordination of growth with cell division in the yeast Saccharomyces cerevisiae. Exp Cell Res 1977; 105:79-98; PMID:320023; http://dx.doi.org/10.1016/0014-4827(77)90154-9
- Evans T, Rosenthal ET, Youngblom J, Distel D, Hunt T. Cyclin: a protein specified by maternal mRNA in sea urchin eggs that is destroyed at each cleavage division. Cell 1983; 33:389-96; PMID:6134587; http://dx.doi.org/10.1016/0092-8674(83)90420-8
- Reed SI, Dulic V, Lew DJ, Richardson HE, Wittenberg C. G1 control in yeast and animal cells. Ciba Found Symp 1992; 170:7-15; discussion −9; PMID:1483351
- Bitton DA, Wood V, Scutt PJ, Grallert A, Yates T, Smith DL, Hagan IM, Miller CJ. Augmented annotation of the Schizosaccharomyces pombe genome reveals additional genes required for growth and viability. Genetics 2011; 187:1207-17; PMID:21270388; http://dx.doi.org/10.1534/genetics.110.123497
- Spirek M, Benko Z, Carnecka M, Rumpf C, Cipak L, Batova M, Marova I, Nam M, Kim DU, Park HO, et al. S. pombe genome deletion project: an update. Cell Cycle 2010; 9:2399-402; PMID:20519959; http://dx.doi.org/10.4161/cc.9.12.11914
- Navarro FJ, Nurse P. A systematic screen reveals new elements acting at the G2/M cell cycle control. Gen Biol 2012; 13:R36; PMID:22624651; http://dx.doi.org/10.1186/gb-2012-13-5-r36
- Graml V, Studera X, Lawson JL, Chessel A, Geymonat M, Bortfeld-Miller M, Walter T, Wagstaff L, Piddini E, Carazo-Salas RE. A genomic Multiprocess survey of machineries that control and link cell shape, microtubule organization, and cell-cycle progression. Dev Cell 2014; 31:227-39; PMID:25373780; http://dx.doi.org/10.1016/j.devcel.2014.09.005
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol 1991; 194:795-823; PMID:2005825; http://dx.doi.org/10.1016/0076-6879(91)94059-L
- Sazer S, Sherwood SW. Mitochondrial growth and DNA synthesis occur in the absence of nuclear DNA replication in fission yeast. J Cell Sci 1990; 97 (Pt 3):509-16; PMID:2074269
- Lyne R, Burns G, Mata J, Penkett CJ, Rustici G, Chen D, Langford C, Vetrie D, Bahler J. Whole-genome microarrays of fission yeast: characteristics, accuracy, reproducibility, and processing of array data. BMC genomics 2003; 4:27; PMID:12854975; http://dx.doi.org/10.1186/1471-2164-4-27
