ABSTRACT
PPARs are a class of ligand-activated transcription factors belonging to the superfamily of receptors for steroid and thyroid hormones, retinoids and vitamin D that control the expression of a large number of genes involved in lipid and carbohydrate metabolism and in the regulation of cell proliferation, differentiation and death. The role of PPARs in the CNS has been primarily associated with lipid and glucose metabolism; however, these receptors are also implicated in neural cell differentiation and death, as well as neuronal maturation. Although it has been demonstrated that PPARs play important roles in determining NSCs fate, less is known about their function in regulating NSCs metabolism during differentiation. In order to identify the metabolic events, controlled by PPARs, occurring during neuronal precursor differentiation, the glucose and lipid metabolism was followed in a recognized model of neuronal differentiation in vitro, the SH-SY5Y neuroblastoma cell line. Moreover, PPARs distribution were also followed in situ in adult mouse brains. The concept of adult neurogenesis becomes relevant especially in view of those disorders in which a loss of neurons is described, such as Alzheimer disease, Parkinson disease, brain injuries and other neurological disorders. Elucidating the crucial steps in energetic metabolism and the involvement of PPARγ in NSC neuronal fate (lineage) may be useful for the future design of preventive and/or therapeutic interventions.
Introduction
Neurogenesis, the process by which new functional neurons are generated from precursors, was for a long time considered occur only during embryonic and perinatal stages in mammals.Citation1-2 Nowadays, we know that neurogenesis occurs in physiological conditions also in the adult mammalian brain in 2 main specific neurogenic niches, the subventricular zone (SVZ) of the lateral ventricles and the subgranular zone (SGZ) of the dentate gyrus (DG) in the hippocampal formation. Moreover, the production of new neurons in other adult brain areas, such as the subcallosal zone, and hippocampus,Citation3 neocortex, striatum, amygdala, and hypothalamus, has been reported, though with conflicting results.Citation4 The SGZ and the SVZ, are also present in the adult human brain. While the SVZ clearly has a prominent role during early childhood, its role in the adult remains elusive and available evidence indicates that this neurogenic niche may be largely quiescent in the adult human brain.Citation5 The Peroxisome Proliferator Activated receptors (PPARs) are a class of ligand-activated transcription factors belonging to the superfamily of receptors for steroid and thyroid hormones, retinoids and vitamin D. Several studies revealed that PPARs show distinct patterns of tissue distribution and control the expression of a large number of genes involved in lipid and carbohydrate metabolism and in the regulation of cell proliferation, differentiation and death. Given their crucial roles, PPARs have been widely studied in the brain, both in vivo and in vitro. PPAR isotypes (α, β/δ and γ) are all expressed in the central nervous system (CNS) of rodents both during embryonic development and in the adult. PPARβ/δ is widely distributed in the brain, PPARα and PPARγ are located in more restricted regions.Citation6-7 Recent findings have demonstrated that PPARs can directly regulate neural cell survival and differentiation.Citation8-16 The role of PPARs in the CNS has been primarily associated with lipid and glucose metabolism; however, these receptors, especially PPARγ, are also implicated in neural cell differentiation and death, as well as in inflammation and neurodegeneration.Citation9 It has also been speculated that the PPARα is involved in the metabolism of acetylcholine,Citation17 in excitatory amino acid neurotransmission and in the defense against oxidative stress.Citation6 PPARβ/δ expression and activation increase during neuronal maturation in vitroCitation8 and PPARβ/δ agonists trigger neuronal differentiation of SH-SY5Y, a human neuroblastoma cell line, and of primary rat neurons.Citation18-20,11
Recently PPARs have been also involved in the regulation of the proliferation, migration and differentiation of NSCs, through signaling pathways such as Wnt, STAT3 and NFkB.Citation21-23 Specifically, PPARγ seems to play important roles in controlling murine NSCs proliferation and survival21: when activated by low concentrations of specific agonists, it stimulates proliferation simultaneously inhibiting neuronal differentiation, while activation by high concentrations of agonists causes NSCs death. This dual role suggests that PPARγ regulates the expansion of NSC population in a concentration-dependent manner and indicates that precise concentrations of its agonists are crucial for the survival and proliferation of NSCs in vivo. It was recently shown that in neurospheres grown in vitro from adult mouse SVZ all 3 PPAR isotypes are expressed,Citation24-25 Although it has been demonstrated that PPARs play important roles in determining NSCs fate, less is known about their function in regulating NSCs metabolism during differentiation. In order to identify the metabolic events occurring during neuronal precursor differentiation, the glucose and lipid metabolism was followed in a recognized model of neuronal differentiation in vitro, the SH-SY5Y neuroblastoma cell line, which presents, according to literature, a subpopulation of stem-like cells expressing nestin and SOX2.Citation26 In the same model, considering its role in glucose and lipid metabolism and its involvement in NSCs maintenance, the PPARγ isotype was specifically investigated. Moreover, in order to gain more insight into PPARγ distribution also in situ, and considering the difficulties to obtain adult human normal brain specimens, the immunolocalization of PPARγ in adult mouse brain SVZ was also studied. The concept of adult neurogenesis becomes relevant especially in view of those disorders in which a loss of neurons is described, such as Alzheimer disease, Parkinson disease, brain injuries and other neurological disorders. Elucidating the crucial steps in energetic metabolism and the involvement of PPARγ in NSC neuronal fate (lineage) may be useful for the future design of preventive and/or therapeutic interventions.
Results
In vitro evaluation of PPARγ expression and energy metabolism during differentiation
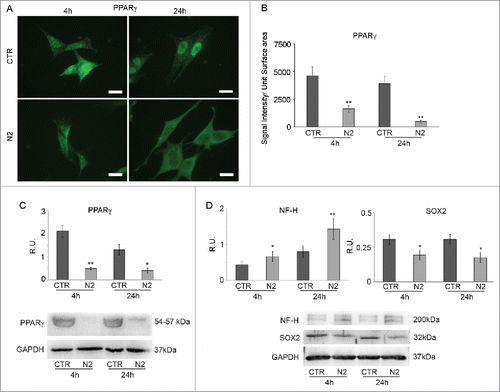
In order to get more insight into the cellular energetic metabolism during neuronal differentiation, undifferentiated (neuroblasts) and differentiated (neuronal-like) SH-SY5Y cells were used as a model. PPARγ localization, glycogen content and the presence of lipid droplets were studied together with the expression of key enzymes of glucose and lipid metabolism. Preliminary experiments were performed in order to set the specific time-points to better visualize the energetic metabolic shift (not shown). On this basis, 4 and 24 hours were chosen. shows PPARγ IF in untreated (CTR) and N2-treated (N2) cells at 4h and 24h. In undifferentiated cells, PPARγ has a strong nuclear localization both at 4h and 24h, while in differentiating ones PPARγ signal is less brilliant and the protein is mainly localized to the cytoplasm. This is more evident at 24h when PPARγ displays an exclusively cytoplasmic localization, thus suggesting loss of activity of the transcription factor. The results were further supported by the immunofluorescent signal quantification shown in B. The same figure confirms this behavior showing a strong decrease of PPARγ, analyzed by WB, in differentiating neurons both at 4h and 24h (C). In order to correlate PPARγ inactivation and decrease to neuronal differentiation, WB analysis for NF-H, a marker of neuronal terminal differentiation, and for SOX2, a marker of undifferentiated status, was performed. In D a significant increase of NF-H protein, paralleled by a significant decrease of SOX2 (D), both at 4h and 24h from N2 treatment is shown, suggesting that the down regulation of PPARγ is paralleled by the increase of the expression of the neuronal differentiation marker and a downregulation of the stemness marker.
Figure 1. PPARγ IF in SH-SY5Y during differentiation (A). Undifferentiated (CTR) and differentianted (N2) cells at 4h and 24h from N2 treatment Bar = 10 μm. B: PPARγ IF quantification expressed as Signal Intensity/Unit Surface Area. C: WB and relative densitometric analyses for PPARγ in undifferentiated (CTR) and differentiated (N2) cells at the indicated time-points. D: WB and relative densitometric analyses for NF-H and SOX2 in undifferentiated (CTR) and differentiating (N2) cells at the indicated time-points. The relative densities of the immunoreactive bands were determined and normalized with respect to GAPDH, using a semiquantitative densitometric analysis. Data are mean ± SE of 4 different experiments. *P ≤ 0.05 and **P ≤ 0.005.
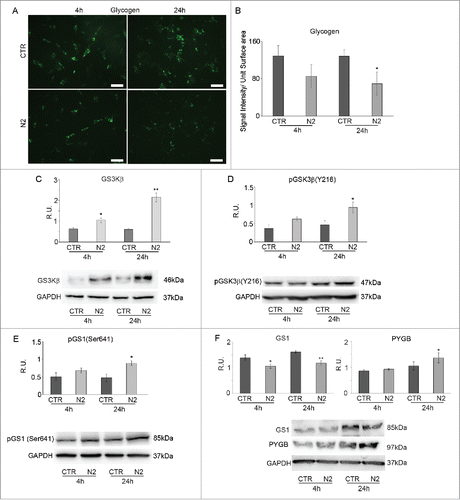
In , glycogen IF in undifferentiated (CTR) and differentiating (N2) cells at 4h and 24h is shown. Similarly to PPARγ, glycogen positive cells, abundant in undifferentiated cells decrease during differentiation. The quantification of the immunofluorescent signal further supports these findings (B).Since a decrease in glycogen storage may indicate a consumption of this energy store to supply glucose, protein levels for the enzymes regulating the formation and degradation of glycogen were analyzed. The same figure shows WB analysis for total GSK3β (C) and its active form pGSK3β (Y126) (D) and its substrate GS1in the inactive, phosphorylated, (E) and active form (F) together with PYGB (F) in undifferentiated and differentiating cells at the examined time points of differentiation. GSK3β is a serine-threonine kinase that, when active, inactivates GS1 through phosphorylation; inactive GS1 is unable to convert glucose residues into the polymeric chain of glycogen. Total GSK3β displays a trend to the increase in differentiating cells, which becomes significant at 24h. The western blotting analysis confirms that the increase regards the active form. Upregulation of this enzyme leads to inactivation of its substrate GS1, corresponds to an increase of the phosphorylated, inactive, form of the latter, paralleled by a decrease of the active form, thus suggesting decrease in glycogen synthesis once neuronal differentiation starts. At the same time PYGB levels were analyzed to assay glycogen degradation, since this enzyme breaks up α-1,4 glycosidic bonds of glycogen to release glucose monomers. The WB analysis for PYGB in undifferentiated and differentiated cells at the examined differentiation time-points shows a significant increase of PYGB protein levels at 24h in differentiating cells compared to control, suggesting high consumption of glycogen in agreement with the IF observations.
Figure 2. Glycogen immunolocalization in SH-SY5Y during differentiation (A). Undifferentiated (CTR) and differentianted (N2) cells at 4h and 24h from N2 treatment. Bar = 70 μm. B: Glycogen IF quantification expressed as Signal Intensity/Unit Surface Area (B). C: WB and relative densitometric analyses for GSK3β in undifferentiated (CTR) and differentiating (N2) cells at the indicated time-points. D: WB and relative densitometric analyses for pGSK3β(Y216) in undifferentiated (CTR) and differentiating (N2) cells at the indicated time-points. E: WB and relative densitometric analyses for pGS(Ser641) in undifferentiated (CTR) and differentiating (N2) cells at the indicated time-points. F: WB and relative densitometric analyses for GS1 and PYGB in undifferentiated (CTR) and differentiating (N2) cells at the indicated time-points. The relative densities of the immunoreactive bands were determined and normalized with respect to GAPDH, using a semiquantitative densitometric analysis. Data are mean ± SE of 4 different experiments. *P ≤ 0.05 and **P ≤ 0.005.
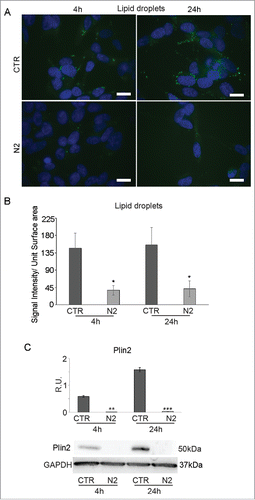
PPARγ is also involved in lipid storage as triglycerides and cholesteryl esters in lipid droplets (LDs). LDs in undifferentiated (CTR) and differentiating (N2) cells at 4h and 24h were detected by the BODIPY fluorescent dye (). Fluorescent images show marked depletion of LD in differentiating cells both at 4h and 24h compared to undifferentiated ones, also supported by the fluorescent signal quantification shown in B. This result is further supported by WB analysis for Plin2, a LD associated membrane protein, which appears significantly decreased in differentiating cells at 4h and 24h (). Thus, PPARγ inactivation and decrease seem to result also in the depletion of LDs during neuronal differentiation.
Figure 3. BODIPY staining in SH-SY5Y during differentiation (A). Undifferentiated (CTR) and differentianted (N2) cells at 4h and 24h from N2 treatment. Nuclei were counterstained with DAPI. Bar = 10 μm. B: BODIPY staining quantification expressed as Signal Intensity/Unit Surface Area. C: WB and relative densitometric analysis for Plin2 in undifferentiated (CTR) and differentiating (N2) cells at the indicated time-points. The relative densities of the immunoreactive bands were determined and normalized with respect to GAPDH, using a semiquantitative densitometric analysis. Data are mean ± SE of 4 different experiments. **P ≤ 0.005 and ***P ≤ 0.0005.
PPARβ/δ and PPARα localization in vitro
Since it has been demonstrated that also PPARβ/δ and PPARα are involved in neuronal differentiation,Citation20-17 their localization was studied in undifferentiated and differentiating SH-SY5Y, at the same time-points. shows PPARβ/δ IF in undifferentiated (CTR) and differentiating (N2) cells at 4 h and 24 h from N2 treatment. In undifferentiated cells PPARβ/δ shows a diffuse localization in the cytoplasm and in the nucleus, while when differentiation starts PPARβ/δ moves to the nucleus, thus suggesting its transcriptional activation. This is further supported by the colocalization with DAPI nuclear staining. WB analysis for PPARβ/δ protein () shows that the transcription factor is upregulated in differentiating cells at 24h, indicating that during neuronal differentiation a significant increase of PPARβ/δ protein occurs together with its nuclear translocation. It is known that PPARβ/δ gene is regulated by β-cateninCitation27; WB () analysis reveals that β-catenin and PPARβ/δ proteins have a similar trend, thus suggesting that during neuronal differentiation PPARβ/δ increases and its nuclear translocation is induced by the transcriptional activity of β-catenin. To support these observations, IF of β-catenin in undifferentiated (CTR) and differentiating (N2) cells at 4h and 24h is presented in . In undifferentiated cells (CTR) β-catenin has a cytoplasmic localization. In differentiated cells (N2) at 4h β-catenin is still located in the cytoplasm but at 24h it is found in the nucleus, where it likely acts as transcription factor. Thus, at 24h of differentiation an increase of β-catenin protein levels together with its translocation to the nucleus is observed, strictly paralleling PPARβ/δ. Recently, it has been suggested that oxidative stress is transiently generated during adult neurogenesis,Citation28 leading to increase of ROS and derived products such as lipid peroxidation products as 4-hydroxynonenale (HNE), known to activate PPARβ/δ. In agreement within our experimental conditions, during neuronal differentiation, HNE levels increase, thus suggesting that the transcription factor is likely active ().
Figure 4. PPARβ/δ (A) and β-catenin (B) immunolocalization in SH-SY5Y during differentiation. Undifferentiated (CTR) and differentianted (N2) cells at 4h from N2 treatment. Bar = 10 μm. C: WB and relative densitometric analyses for PPARβ/δ and β-catenin in undifferentiated (CTR) and differentiating (N2) cells at the indicated time-points. D: WB and relative densitometric analyses for 4-HNE in undifferentiated (CTR) and differentiating (N2) cells at the indicated time-points. The relative densities of the immunoreactive bands were determined and normalized with respect to GAPDH using a semiquantitative densitometric analysis. Data are mean ± SE of 4 different experiments. *P ≤ 0.05; **P ≤ 0.005. E: PPARα IF in SH-SY5Y during differentiation. Undifferentiated (CTR) and differentianted (N2) cells at 4h and 24h from N2 treatment Bar = 10 μm. F: WB and relative densitometric analysis for PPARα in undifferentiated (CTR) and differentiating (N2) cells at the indicated time-points. The relative densities of the immunoreactive bands were determined and normalized with respect to GAPDH, using a semiquantitative densitometric analysis. Data are mean ± SE of 4 different experiments. *P ≤ 0.05.
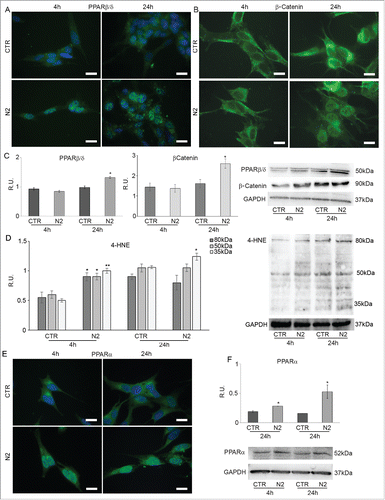
In the same PPARα IF in undifferentiated (CTR) and differentiated (N2) cells at 4h and 24h is shown. In undifferentiated cells (CTR) PPARα has cytoplasmic and perinuclear localization, while in differentiated cells (N2) it moves into the nucleus, as shown by the colocalization with DAPI, where it probably becomes an active transcription factor; this event is more apparent at 4h of differentiation. WB analysis of PPARα () reveals a significant change in protein levels both at 4h and at 24h, suggesting that during neuronal differentiation also the upregulation of this transcription factor is relevant.
PPARγ gene silencing (siRNA)
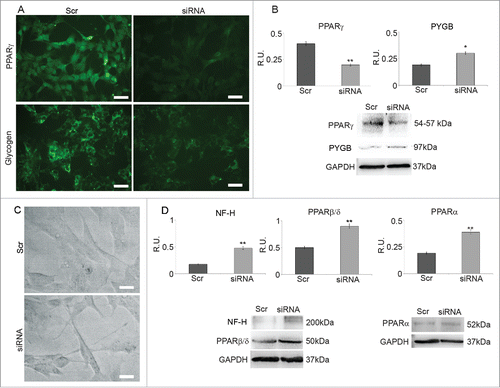
To validate the hypothezized correlation between PPARγ and the maintenance of undifferentiated status, PPARγ siRNA was performed and the silenced cells were analyzed by IF 48h. shows PPARγ IF in cells transfected with scrumbled sequence (Scr) and in silenced cells (siRNA) together with the relative WB analysis (). In IF images a strong decrease of PPARγ in silenced cells compared to Scr is observed, confirming that gene silencing has occurred. This result is further supported by WB analysis of PPARγ levels that are decreased to about 40% in silenced cells. shows also glycogen IF in Scr and in silenced cells (siRNA) together with WB analysis for PYGB (B). IF pictures indicate a strong decrease in glycogen content in silenced cells compared to Scr and, at the same time, an increase of PYGB levels are observed suggesting higher consumption of glycogen in silenced cells. These results encouraged to analyze neuronal differentiation in silenced cells. In agreement with the previous observations, phase contrast microscopy () shows that neurites in silenced cells are longer than in control ones, suggesting that PPARγ decrease is a condition for neuronal differentiation. WB analysis of NF-H protein levels () confirms this hypothesis since this neuronal marker is significantly increased in silenced cells. To further investigate the role of PPARγ in neuronal differentiation, PPARβ/δ and PPARα protein levels were also evaluated by WB in PPARγ silenced cells. shows that both transcription factors are significantly upregulated in silenced cells, to the same extent previously observed in N2 differentiated cells. These results further confirm previous observation on the involvement of PPARβ/δ in neuronal maturationCitation20 and agree with the suggested role for PPARα in the acquisition of the cholinergic phenotype.Citation17
Figure 5. A: PPARγ and glycogen IF in cells treated with scrambled sequence (Scr) and in PPARγ silenced cells (siRNA). Bar = 35 μm. B: WB and densitometric analysis for PPARγ and PYGP. The relative densities of the immunoreactive bands were determined and normalized with respect to GAPDH using a semiquantitative densitometric analysis. Data are mean ± SE of 4 different experiments. *P ≤ 0.05; **P ≤ 0.005. C: Phase contrast microscopy of cells treated with scrambled sequence (Src) and PPARγ silenced cells (siRNA); Bar = 20 μm. D: WB and densitometric analysis for NF-H, PPARβ/δ and PPARα. The relative densities of the immunoreactive bands were determined and normalized with respect to GAPDH using a semiquantitative densitometric analysis. Data are mean ± SE of 4 different experiments. **P ≤ 0.005.
In situ analysis of PPARγ distribution in the SVZ
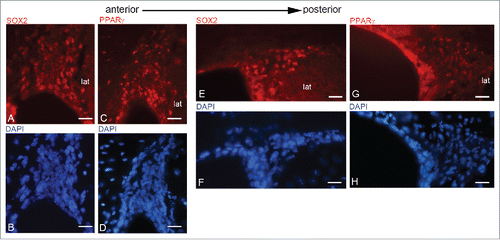
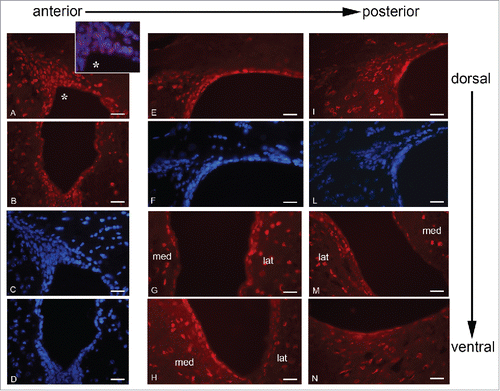
PPARγ expression was also investigated in adult mouse brain sections together with markers of different stages of neurogenesis.Citation29 This inquiry was conducted on 3 different levels of the SVZ taken along the antero-posterior axis (A-P) of the LV (), since it has been demonstrated that there is a rostro-caudal gradual loss of neurogenic potential within the periventricular germinal region.Citation30 The choice of the mouse as model to study the PPARγ and neurogenesis was due to the difficulties to obtain healthy human brain slices, particularly at subventricular level. shows PPARγ immunolocalization along the anterior-posterior and dorsal-ventral axes of LV. In A-D panels PPARγ localization in dorsal and ventral region of anterior LV is shown: PPARγ is expressed in almost all nuclei (inset A*) of ependyma, subependyma and migratory region, both in the dorsal and ventral portion of LV. In the dorso-lateral wall (E-F) PPARγ is present in ependymal, subependymal and migratory region, even if in the last PPARγ-positive nuclei are less numerous compared to the anterior LV; in the medial-lateral and ventral walls (G-H), PPARγ is observed in the nuclei of the ependyma and, limited to the lateral wall, of the subependyma. In caudal LV (I-N), PPARγ staining pattern is similar to that observed in the intermediate LV, but in the dorsal-lateral neurogenic region bright PPARγ IF is limited to some ependymal cells while the subependymal and migratory regions show only scattered positive nuclei. Therefore, it is apparent that PPARγ immunoreactivity decreases along the anterior-posterior axis together with the neurogenic potential.Citation30
Figure 6. PPARγ immunolocalization along the A-P axis of mouse brain LVs. A-D, dorsal (A, C) and ventral (B, D) neurogenic regions of rostral LV stained for PPARγ (A, B) and DAPI (C, D). E-H, intermediate region of LV, dorso-lateral neurogenic wall stained for PPARγ (E) and DAPI (F); medio-lateral and ventral walls stained for PPARγ (G-H). I-N, caudal LV; dorso-lateral neurogenic wall stained for PPARγ (I) and DAPI (L); medio-lateral and ventral walls stained for PPARγ (M-N). lat, lateral wall; med, medial wall. Bar = 40 μm.
In order to correlate the nuclear staining of PPARγ to the neurogenic niche, the co-expression of this transcription factor with markers of different stages of neurogenesis was studied ().
Figure 7. GFAP, Nestin and PPARγ in different rostro-caudal regions of the LVs. A-D, rostral LV. Dorsal neurogenic region immunostained for GFAP (A), PPARγ (B), merge (C), and DAPI (D). E-H, rostral LV. Dorsal neurogenic region immunostained for Nestin (E), PPARγ (F). Merge (G), and DAPI (H). I-N, caudal LV. Dorso-lateral neurogenic region immunostained for GFAP (I), PPARγ (L), Merge (M) and DAPI (N). O-R, caudal LV. Dorso-lateral neurogenic region immunostained for Nestin (O) and PPARγ (P). Merge (Q) and DAPI (R). lat, lateral wall; med, medial wall. Bar = 40 μm.
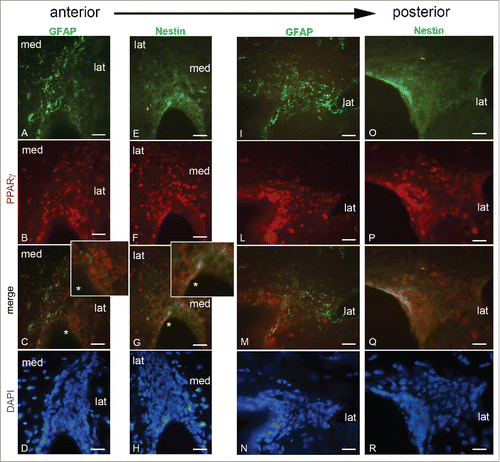
PPARγ/GFAP double immunostaining and DAPI staining (A-D) revealed that GFAP-positive cells are evident in the subependyma and migratory region; PPARγ-positive nuclei are mainly observed in the subependyma and migratory region, a colocalization with GFAP was not evident (C); however, in these regions clusters of PPARγ positive nuclei in faint GFAP-positive cells are present in the lateral wall (inset C*).
In the sections probed with PPARγ/Nestin/DAPI (E-H), Nestin is present in the cytoplasm of ependymal and subependymal cells while it is less expressed in the migratory region, where no NSCs are described. However in the ependymal-subependymal layer PPARγ colocalizes with Nestin in some cells highly nestin-positive (inset G*).
In the caudal region of LV GFAP-positivity (I) is higher in the subependymal and migratory region, where no apparent colocalization with PPARγ is observed (M). Nestin is particularly evident in the ependymal layer (O), where it colocalizes with PPARγ (Q), but it is also weakly expressed in the subependymal and migratory regions, even though a colocalization in the caudal LV is not evident.
In PPARγ and SOX2 immunofluorescence is presented; SOX2/DAPI (A-B) and PPARγ/DAPI (C-D) distribution are compared in anterior SVZ showing an evident overlap of these transcription factors in the whole neurogenic region. E-H show intermediate region of LV: here SOX2 (E) is present in the ependyma and subependyma, whose distribution also extends, in the migratory region with a nuclear localization. DAPI staining is shown in (F). PPARγ distribution in the LV intermediate region is shown in (G). AS in rostral LV, a pattern similar to SOX2 is evident.
In situ localization and possible role of Glycogen in the SVZ
Glycogen distribution in the neurogenic niche of LV was analyzed focusing on anterior SVZ since this is the region where neurogenesis is more active.Citation30 To evaluate which cellular types are able to store glycogen in the SVZ and the possible correlation between glucose metabolism and neurogenesis, glycogen was examined together with PPARγ, GFAP and βIII-tubulin, the latter 2 being markers of astrocyte and young neurons respectively.
, Panel 1 shows glycogen distribution in the LV. Low magnification of the LV (A) illustrate that glycogen is mainly localized in the SVZ and is almost absent in the striatum where mature neurons reside; this is supported by higher magnifications of the different LV portions, showing that glycogen is localized in the subependymal layer of the dorsal (B) and migratory (C) region, in the lateral and medial walls (D) of LV. Merge with DAPI in the lateral wall (E) confirms the absence of glycogen from ependyma. In , Panel 2, glycogen double immunofluorescence with PPARγ, GFAP and βIII-tubulin in the LV is presented. Glycogen/PPARγ images evidentiate only in a small cluster of cells in the SVZ in ependymal-subependymal layer and in the migratory region (A) a colocalization of glycogen and PPARγ; moreover, they colocalize in many cells of the lateral wall of LV while glycogen is completely absent from the medial wall (B). Even if it is known that in the adult brain glycogen is stored mainly in astrocytes, these pictures indicate that glycogen and GFAP do not completely overlap, both in the dorso-lateral and migratory region (C), and in the lateral and medial walls of the LV (D). Glycogen is mainly localized in the dorsal and medial wall of the LV, and to a lesser extent in the migratory region and not all GFAP-positive cells are also glycogen-positive, especially in the lateral wall (D) where GFAP is poorly expressed.
Figure 9. Panel 1: Glycogen distribution in mouse LV. A: LV at low magnification immunostained for glycogen; B-D, dorsal wall (B), migratory region (C), lateral and medial wall (D) of LV at high magnification; E, lateral wall of LV counterstained with DAPI. cc, corpus callosum; lat, lateral wall; med, medial wall. Bar in (A) = 180 μm; Bar in B-E = 60 μm. Panel 2: Glycogen and PPARγ IF in the migratory region of LV (A) and in medial and lateral wall of LV (B); Glycogen and GFAP IF in migratory region of LV (C) and in medial and lateral wall of LV (D); E-F: glycogen and β-tubulin III IF in the migratory region of LV (E) and in medial and lateral wall of LV (F). Bar = 60 μm.
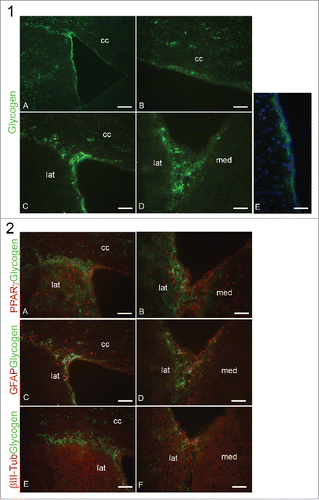
Finally in , glycogen and βIII-tubulin double immunolocalization in the LV is presented; in E, the migratory region of dorso-lateral SVZ is characterized by a weak βIII-tubulin positivity that overlaps with glycogen staining; βIII-tubulin is more evident in the lateral wall of LV (F), but colocalization with glycogen is observed only in few neurons.
Discussion
Neurogenesis occurs in 2 specialized niches in the adult brain, the SGZ of the dentate gyrus and the SVZ adjacent to the lateral ventricles. In both adult neurogenic niches, specific transcription factors have been shown to direct fate specification and lineage commitment.Citation31-32
Among the different factors involved in embryonic neurogenesis, great attention has been given to PPARs.Citation34
The metabolic requirements for neuronal progenitor maintenance in vitro and in vivo have been demonstrated by examining the metabolic adaptations that support progenitors and NSCs in their undifferentiated state.Citation3 These Authors demonstrated that glucose induces neuronal differentiation of neuronal progenitors. Many tissues are capable of metabolising glycogen including brain. In the brain, glycogen is found both in astrocytes and neurons and its degradation in the former is thought to spare blood glucose for neurons during high brain activation, whereas in the latter, it promotes neuronal tolerance to hypoxic stress.Citation35 The presence of glycogen in the brain implies that it has a definite role within the brain parenchyma. The cellular location of glycogen in brain is highly specialized, whereas glycogen is expressed in virtually all liver and skeletal muscle cells. While it is widely accepted that brain glycogen is almost exclusively expressed in astrocytes in the adult, in younger animals this is not the case and the expression of glycogen in specific populations of brain cells is a function of age. Brain glycogen is expressed in embryonic neuronal and glial cells but with maturity the degree of neuronal expression fades, and by adulthood only ependymal and choroid plexus cells, beyond astrocytes, express brain glycogen.Citation36 The function of brain glycogen in neural cells at birth is unknown, but this may imply that these cells have higher metabolic demand or a less secure glucose supply at that stage of development. This would correlate with the neonatal brain receiving about half of its energy requirements from non-glucose substrates, primarily in the form of lactate, which crosses the blood brain barrier via monocarboxylate transporters.Citation37
On the basis of these findings, and considering the importance of adult neurogenesis in brain plasticity and in neurodegenerative diseases, in this work we studied in vitro the energetic metabolism pathways controlled by PPARs and the in situ localization of PPARs together with early neuronal differentiation markers in adult mouse brain.
The in situ data demonstrate that PPARγ is localized at nuclear level in the rostral migratory stream (RMS), where it colocalizes with NP markers such SOX2 and nestin. In order to get more insight into the role/s of PPARs in adult neurogenesis, we used the human neuroblastoma cell lines SHSY5Y, as a model of neural precursor induced to differentiate to neuron. During the early phases of neuronal differentiation a significant downregulation of PPARγ is observed, paralleled by a change in its cellular localization, which becomes cytoplasmic, after the differentiation challenge. These data indicate that the active (nuclear) PPARγ is probably needed for the maintenance of NPs, as also previously described by us for neural stem cells.Citation25 The decrease of PPARγ is accompanied by a strong decrease of glycogen content in differentiating cells. This was also evident in the in situ model. In fact, the RMS, that was similarly characterized by the marked presence of glycogen and PPARγ in nestin-positive cells, shows that, in differentiated β-tubulin III-positive cells, glycogen content paralleled that of PPARγ, being significantly decreased in neurons. Since a decrease of glycogen indicates its degradation and utilization of glucose monomers via glycolysis, the enzymes involved in glycogenolysis were also assayed during differentiation in the in vitro model. In agreement with the observed decrease of glycogen, the enzyme negatively controlling glycogen synthase 1, GSK3β, appears increased and active, significantly at 24h, thus indicating inhibition of glycogen synthesis, as also indicated by the increase of the inactive form of GS1. This event was accompanied by a significant increase of PYGB strongly suggesting that glucose monomers are probably sent to the glycolytic pathway.
Since it is known that PPARγ is also involved in the control of lipid metabolism, mainly in lipid storage, the presence of lipid droplets (LD, containing triglyceride esters and cholesteryl esters) was also checked. In undifferentiated cells LDs are abundantly present and upon differentiation challenge, they appear strongly decreased. This is further supported by the decrease of the LD membrane protein Plin2 observed in this condition. Taken together, these results indicate that, when neuronal differentiation starts, energetic stores, namely glycogen and LDs, are consumed, thus releasing molecules necessary for the energy requirements of differentiating neurons. PPARγ downregulation seems crucial to this metabolic switch.
In order to unequivocally correlate PPARγ with the observed modulation of energetic metabolism, PPARγ gene silencing was performed and some parameters were re-assayed. Upon silencing, cells show a strong decrease of glycogen content, paralleled by a significant increase of PYGB.
Therefore it is possible to conclude that PPARγ is crucial for NPs maintenance and energetic storage. This is in agreement with the observation that NPs display a low energetic demand, while neurons are surely dependent on glucose metabolism for their activity. The presence of LDs, already described in neural stem cells,Citation24 and their disappearance when cells are induced to differentiate, agrees with this conclusion.
Finally, since it has been previously reported that also the other PPAR isotypes, PPARβ/δ and PPARα, are crucial for neuronal differentiation,Citation8,17 these transcription factors were also investigated. In agreement with our previous observation, during differentiation PPARβ/δ moves from the cytoplasm to the nucleus, where it probably becomes active. As it is known that PPARβ/δ transcription is dependent on β-catenin transcriptional activity, also this protein was evaluated. At 24h from differentiation challenge β-catenin increases, and moves to the nucleus, thus suggesting its activation as transcription factor. Finally, it has been hypothesized that the energetic demands of highly proliferative progenitors generates localized oxidative stress. In an in vitro cell culture model of primary hippocampal NSCs, the induction of differentiation in primary NSCs resulted in an immediate increase in total mitochondria number and overall ROS production, suggesting that oxidative stress is generated during a transient period of elevated neurogenesis accompanying normal neurogenesis.Citation28 For this reason, and considering that one of the major product of oxidative stress 4-HNE is a PPARβ/δ ligand, the 4-HNE protein adducts were also assayed in our cultures. The observed increase of 4-HNE during differentiation, strongly points and confirms the neuronal differentiation activity of PPARβ/δ, as previously indicated by us.Citation11
It is worth-mentioning that unliganded PPARβ/δ may modulate the transcriptional activity of the other PPARs also by modulating their localization and transcriptional activity,Citation38 therefore it may be conceivable that during neuronal differentiation, PPARβ/δ increases and modulates the transcriptional activity and localization of the other PPARs. In fact, also PPARα changes its subcellular localization during differentiation, from cytoplasmic to nuclear, suggesting its activation and its possible role in the lipid utilization observed in this condition.
In conclusion, the data obtained in this work point again toward specific role/s for PPARs in the control of the different phases of adult neurogenesis and demonstrate for the first time that the energetic metabolic shift occurring during NPs differentiation is regulated by PPARs.
Getting more insights into the molecular regulation of adult neurogenesis appears of particular interest in view of the prevention and/or treatment of neurodegenerative diseases, suggesting a potential use of PPARs agonist/antagonist for inducing NPs proliferation and/or differentiation.
Materials and methods
Cell line and reagents
Neuroblastoma cell line SH-SY5Y purchased from ATCC (Manassas, VA, USA). Adult male C57BL/6 mice purchased from Taconic Farms, Inc. (Germantown, NY, USA). RPMI-1640, Fetal Bovine Serum (FBS), penicillin/streptomycin, glutamine, formaldehyde, paraformaldehyde, triton X-100, Phosphate Buffered Saline (PBS), Bovine Serum Albumin (BSA), poly-L-lysine, dapi, trypan blue, ethanol, propidium iodide, Nonidet-P40, sodium deoxycholate, Sodium Dodecyl Sulfate (SDS), Tween 20, Igepal CA 630, Ethylenediamine tetraacetic acid (EDTA), phosphatase inhibitor cocktail 2, protease inhibitor cocktail, ethylenediamine tetraacetic acid (EDTA), acrylamide/bis-acrylamide, Tris(hydroxymethyl)aminomethane (Tris), hydrogen chloride (HCl), sodium chloride (NaCl), xylazine hydrochloride, were all purchased from Sigma Chemical CO (St. Louis, Mo, USA). N2 supplement were from Gibco Life Technologies, 4,4-difluoro-1,3,5,7,8-pentamethyl 4-bora-3a,4a-diaza-s-indacene (BODIPY 493/503) Molecular Probes, Life Technologies, target-specific siRNA for PPARγ and scrambled sequence Ambion, Life Technologies, Lipofectamine, Invitrogen Life Technologies (Lofer, Salzburg, Austria Life Technologies). Primary antibodies: PPARα (rabbit), PPARγ (rabbit), PPARβ/δ (rabbit) were from Thermo Scientific (Rockford, IL, USA). GFAP (mouse) was from Immunological Sciences (Rome, Italy). Nestin (chicken), SOX2 (rabbit), GSK3β (rabbit), GS1 (rabbit), PYGB (mouse) pGSK3β(Y216) (rabbit) were from Abcam (Cambridge, UK). βIII-tubulin (mouse) was from Promega (Madison, WI, USA). β-catenin (mouse) was from Zymed (Carlsbad, CA, USA. Now sold as Life Technologies). Glycogen (mouse) Generous gift of Prof. Otto Baba, Ohu University, Japan. NF-H (mouse) and Plin2 (rabbit) were purchased from Sigma Chemical CO (St. Louis, Mo, USA). pGS(Ser641) (rabbit) was purchased from Cell Signaling technology (Danvers, MA, USA). GAPDH was from Santa Cruz (Dallas, Texas, USA). PPARγ (mouse) was from Chemicon (Temecula, CA, USA). PPARα (mouse) was from Novus Biologicals (Littleton, CO, USA). anti-mouse IgG Alexa Fluor 488/546 conjugated or anti-chicken IgG Alexa Fluor 488 secondary antibodies were purchased from Molecular Probes (Life Technology, Carlsbad, CA, USA) and peroxidase conjugated anti-mouse or anti-rabbit IgG secondary antibodies were from KPL (Gaithersburg, USA). Bicinchoninic acid (BCA) protein assay kit from Pierce (Rockford, IL, USA). Vectashield mounting medium was required to Vector Laboratories (Burlingame, CA, USA), non-fat dry milk was from Bio-Rad Laboratories (Hercules, CA, USA), SuperSignal West Pico Chemiluminescent Substrate was purchased from Thermo Scientific (Rockford, IL, USA). Immobilon-P Transfer Membrane (PVDF) was required to Millipore Corporation (Billerica, MA, USA), O.C.T. was from Sakura (St. Torrance, CA, USA). All chemicals were of the highest analytical grade.
Cell culture and differentiation
Neuroblastoma cell line SH-SY5Y was used as an in vitro model of neuronal differentiation. Cells were seeded at 1×104 cells/cm2 and maintained in a monolayer culture in RPMI-1640 medium supplemented with 10% FBS, 1% penicillin/streptomycin and 1% glutamine. To induce neuronal differentiation cell culture medium was replaced with RPMI-1640 FBS-free containing N2 Supplement and cell samples for immunofluorescence and western blotting analysis were analyzed at 4h and 24h of differentiation. N2 supplement is a serum-free, chemically defined, concentrated media supplement formulated to provide optimal growth conditions for neuronal cells N2 is composed of Bovine Insulin, Human Transferrin, Putrescine, Selenite, and Progesterone.
Animals
Adult male C57BL/6 mice were used to perform morphological analysis. The experiments were performed in accordance with the European Communities Council Directive of 24 November 1986 (86/609/EEC). Formal approval to conduct the experiments described was obtained from the Italian Ministry of Health. All efforts were made to minimize the number of animals used and their suffering.
Immunofluorescence (IF)
For morphological studies in vivo, C57BL/6 mice were deeply anesthetized with urethane (1 g/ kg body weight, injected i.p.), before rapid killing by transcardial perfusion. Mice were perfusion-fixed with 4% paraformaldehyde solution. Brains were removed, postfixed in the same solution at 4°C overnight and then cryoprotected in PBS solution containing 30% sucrose. Samples were embedded in OCT freezing medium and frozen at −20°C; subsequently 12μm coronal sections were obtained in a cryostat and collected on Superfrost Plus slides. Frozen sections were then processed for IF: slides were thawed at room temperature and then permeabilized and blocked in PBS containing 4% BSA and 0,05% Tween-20 for 2 h at RT; sections were then incubated at 4°C overnight with primary antibodies PPARγ (rabbit) 1:300, GFAP (mouse) 1:500, Nestin (chicken) 1:200, SOX2 (rabbit) 1:200, βIII-tubulin (mouse) 1:500, Glycogen (mouse) 1:200 diluted in PBS containing 4% BSA for single and double staining. After 24h, slides were rinsed in PBS and then incubated with goat anti-rabbit IgG Alexa Fluor 488/546 conjugated, anti-mouse IgG Alexa Fluor 488/546 conjugated or anti-chicken IgG Alexa Fluor 488 secondary antibodies diluted 1:2000 in PBS containing 4% BSA for 2 h at RT; for glycogen immunolocalization, anti-mouse IgM FITC conjugated secondary antibody (1:200) was used (Sigma-Aldrich). Nuclei were counterstained with DAPI 5 min at RT. After several washes in PBS slides were mounted with Vectashield mounting medium and then observed at Zeiss Axioplan 2 fluorescence microscope (Zeiss, Oberkochen, Germany). Images were acquired with Leica IM 500 software. Controls were performed in parallel by omitting the primary antibody.
For morphological studies in vitro, cells grown on coverslips were fixed in 4% paraformaldehyde in PBS for 10 min at RT and permeabilized in PBS containing 0.1% Triton X-100 for 5–10 min at RT. Cells were then blocked with PBS containing 4% BSA for 20 min and incubated with primary antibodies PPARγ (rabbit), 1:300, Glycogen (mouse) 1:200, PPARα (rabbit) 1:100, PPARβ/δ (rabbit) 1:100, β-catenin (mouse) 1:100 diluted in the blocking solution overnight at 4°C. The following day cells were rinsed in PBS several times before incubation with the same secondary antibodies as above described for the in situ experiments. Also glycogen was immunolocalized as above. After extensive washing coverslips were mounted with Vectashield mounting medium with DAPI and then observed at Zeiss Axioplan 2 fluorescence microscope. Images were acquired with Leica IM 500 software. Controls were performed in parallel by omitting the primary antibody.
For quantitative evaluation of immunofluorescent signals 4 photomicrographs for each condition were analyzed by the ImageJ software (National Institutes of Health, Bethesda, MD) according to.Citation39 Briefly, the pictures were converted in gray levels, and arbitrary units were assigned; 0 = black (i.e. absence of signal) and 255 = white. The signal was first measured in an area devoid of signal at visual inspection and assumed as background; the threshold was then set at 1.5 times the background and the surface area and mean gray intensity were measured for all areas above the threshold. To get the signal intensity (in arbitrary units), the background was subtracted from the mean gray intensity and the result was multiplied for the surface area above threshold. This value was divided by the surface area to calculate the signal intensity per unit surface area.
BODIPY staining
Cells grown on coverslips were fixed for 10 min at RT in 4% paraformaldehyde in PBS and permeabilized in PBS containing 0.1% Triton X-100 for 10 min at RT. Cells were incubated with 4,4-difluoro-1,3,5,7,8-pentamethyl 4-bora-3a,4a-diaza-s-indacene for 10 min at RT. A stock solution of BODIPY 493/503 1 mg/ml in ethanol was prepared and then stored at −20 in the dark until required. Before use, BODIPY stock solution was diluted 1:1000 in PBS and used for incubating coverslips in the dark for 10 min. After incubation the cells were washed with PBS, mounted with Vectashield mounting medium with DAPI and then observed at Zeiss Axioplan 2 fluorescence microscope. Images were acquired with Leica IM 500 software.
Protein assay
Control and treated cells were washed in ice-cold PBS and harvested in ice-cold RIPA buffer (phosphate buffer saline pH 7.4 containing 0.5% sodium deoxycolate, 1% Igepal, 0.1% SDS, 5mM EDTA, 1% phosphatase inhibitor cocktail, 1% protease inhibitor cocktail. After 1h incubation at 4°C, cell lysates were centrifuged at 14000 rpm for 30 min and the supernatants were assayed for protein content with BCA kit. Briefly, this assay is a detergent-compatible formulation based on BCA for the colorimetric detection and quantification of total protein. The method combines the reduction of Cu+2 to Cu+1 by protein in alkaline medium (the biuret reaction) with the highly sensitive and selective colorimetric detection of the cuprous cation using a BCA-containing reagent. The purple-stained reaction product, formed by the chelation of 2 molecules of BCA with one cuprous ion, exhibits a strong absorbance at 562 nm. Samples were then diluted 3:4 in 200 mM Tris–HCl, pH 6.8, containing 40% glycerol, 20% β-mercaptoethanol, 4% sodium dodecyl phosphate (SDS), bromophenol blue.
Western blotting (WB)
SDS-PAGE was performed running samples (30 μg protein) on 8–15% polyacrylamide denaturing gels. Protein bands were transferred onto polyvinylidene fluoride (PVDF) sheets by wet electrophoretic transfer. Non-specific binding sites were blocked for 1 h at room temperature (RT) with 5% non-fat dry milk in Tris-buffered saline containing 0.1% Tween 20 (TBS-T). Membranes were incubated overnight at 4˚C with primary antibodies: PPARα (mouse) 1:500, PPARγ (mouse) 1:200, PPARβ/δ (rabbit) 1:1000, GSK3β (rabbit) 1:7000, GS1 (rabbit) 1:3000, PYGB (mouse) 1:4000, Plin2 (rabbit) 1:2000, β-catenin (mouse) 1:500, NF-H (mouse) 1:1000, SOX2 (rabbit) 1:2000, pGSK3β (Y216) (rabbit) 1:1000, pGS(Ser641) (rabbit) 1:1000 appropriately diluted in blocking solution (for pGS(Ser641) antibody the blocking solution used was 5%BSA). After extensive washings in TBS-T membranes were incubated with HRP-conjugated anti-rabbit or anti-mouse IgG secondary antibody (KPL, Gaithersburg, MD, USA) diluted 1:10000 in blocking solution, for 1 h at 4˚C. After rinsing, immunoreactive bands were visualized by enhanced chemiluminescence (ECL) according to the manufacturer's instructions and acquired by Alliance 7 imaging instrument (UVItec Limited, Cambridge, UK). The relative densities of the immunoreactive bands were determined and normalized with respect to GAPDH using a semi quantitative densitometric analysis.
Small interference RNA (siRNA)
Gene silencing by siRNA was performed by transfecting cells with target-specific siRNA or scrambled sequence for PPARγ, at the final concentration of 50 nM, according to the manufacturer's instructions. Briefly, SH-SY5Y cells were seeded one day before transfection at 1×105 cells/ml in RPMI medium with 10% FBS, 1% glutamine and without antibiotics. The transfection complex was made as following: target-specific siRNA for PPARγ (Ambion, Austin, TX, USA) or scrambled sequence were diluted into RPMI medium and combined with the transfection agent (Lipofectamine) previously prepared according to manufacturer's instructions. The mixture so obtained was incubated for 15 min at RT to allow the formation of transfection complex. This complex was then added to the well together with the cell suspension prepared in the previous step and incubated for 24 h at 37°C in a humidified 95% air-5% CO2 atmosphere. Subsequently, cells were washed with PBS and cultured for additional 24h in the 10% FBS medium. Controls were performed using siRNA containing a scrambled sequence that will not allow the specific degradation of PPARγ mRNA.
Statistical analysis
Statistical analysis was performed by t test using the SPSS software SPSS software (Statistical Package for Social Sciences, v. 11.0, Tokyo, Japan). For all statistical analyses, P < 0.05 was considered as statistically significant. Data were expressed as Mean ± SE of 4 separate experiments.
Abbreviations
| 4-HNE | = | 4-Hydroxynonenal |
| CNS | = | Central Nervous System |
| DAPI | = | 4′,6-diamidino-2-phenylindole |
| DG | = | Dentate gyrus |
| GFAP | = | Glial fibrillary acidic protein |
| GSK3β | = | Glycogen synthase kinase-3 beta |
| IF | = | Immuno Fluorescence |
| LDs | = | Lipid Droplets |
| mESCs | = | mouse Embryonic Stem Cells |
| NF-H | = | Neurofilament heavy polypeptide |
| NFkB | = | Nuclear factor of kappa light polypeptide gene enhancer in B-cells |
| NPs | = | Neural Precursors |
| NSCs | = | Neural Stem Cells |
| Plin2 | = | Perilipin-2 |
| PPARs | = | Peroxisome Proliferator Activated receptors |
| PYGB | = | Phosphorylase glycogen brain |
| RXR | = | 9cis-Retinoic acid receptor |
| SGZ | = | Subgranular zone |
| SOX2 | = | Sex determining region Y-box 2 |
| STAT3 | = | Signal transducer and activator of transcription 3 |
| SVZ | = | Subventricular zone |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the RIA funds (Proff Cimini, Benedetti, Ippoliti). The Authors thank the Human Health Foundation for the support. Many of the experiments have been performed in the Research Center for Molecular Diagnostics and Advanced Therapies granted by the Abruzzo Earthquacke Relief Fund (AERF).
References
- Ming GL, Song H. Adult neurogenesis in the mammalian central nervous system. Annu.Rev.Neurosci 2005; 28:223-50; PMID:16022595; http://dx.doi.org/10.1146/annurev.neuro.28.051804.101459
- Ming G, Song H. Adult Neurogenesis in the Mammalian Brain: significant answers and significant questions. Neuron 2011; 70:687-702; PMID:21609825; http://dx.doi.org/10.1016/j.neuron.2011.05.001
- Seri B, Herrera DG, Gritti A, Ferron S, Collado L, Vescovi A, Garcia-Verdugo JM, Alvarez-Buylla A. Composition and organization of the SCZ: a large germinal layer containing neural stem cells in the adult mammalian brain. Cerebral Cortex 2006; 16:i103-11; PMID:16766696; http://dx.doi.org/10.1093/cercor/bhk027
- Balu DT, Lucki I. Adult hippocampal neurogenesis: regulation, functional implications, and contribution to disease pathology. Neurosci Biobehav Rev 2009; 33:232-52; PMID:18786562; http://dx.doi.org/10.1016/j.neubiorev.2008.08.007
- Butti E, Cusimano M, Bacigaluppi M, Martino G. Neurogenic and non-neurogenic functions of endogenous neural stem cells. Front Neurosci 2014; 8:92
- Moreno S, Farioli-vecchioli S, Cerù MP. Immunolocalization of peroxisome proliferator-activated receptors and retinoid X receptors in the adult rat CNS. Neuroscience 2004; 123:131-45; PMID:14667448; http://dx.doi.org/10.1016/j.neuroscience.2003.08.064
- Woods JW, Tanen M, Figueroa DJ, Biswas C, Zycband E, Moller DE, Austin CP, Berger JP. Localization of PPARδ; in murine central nervous system: expression in oligodendrocytes and neurons. Brain Res 2003; 975:10-21; PMID:12763589; http://dx.doi.org/10.1016/S0006-8993(03)02515-0
- Cimini A, Benedetti E, Cristiano L, Sebastiani P, D'Amico MA, D'Angelo B, Di Loreto S. Expression of peroxisome proliferator-activated receptors (PPARs) and retinoic acid receptors (RXRs) in rat cortical neurons. Neuroscience 2005; 130:325-37; PMID:15664689; http://dx.doi.org/10.1016/j.neuroscience.2004.09.043
- Heneka MT, Klockgether T, Feinstein DL. Peroxisome proliferator-activated receptor-γ ligands reduce neuronal inducible nitric oxide synthase expression and cell death in vivo. J Neurosci 2000; 20:6862-7; PMID:10995830
- Smith SA, Monteith GR, Robinson JA, Venkata NG, May FJ, Roberts-Thomson SJ. Effect of the peroxisome proliferator-activated receptor β activator GW0742 in rat cultured cerebellar granule neurons. J Neurosci Res 2004; 77:240-9; PMID:15211590; http://dx.doi.org/10.1002/jnr.20153
- Benedetti E, Di Loreto S, D'Angelo B, Cristiano L, d'Angelo M, Antonosante A, Fidoamore A, Raffaella G, Cinque B, Cifone MG, et al. The PPARβ/δ Agonist GW0742 Induces Early Neuronal Maturation of Cortical Post-Mitotic Neurons: Role of PPARβ/δ in Neuronal Maturation. J Cell Physiol 2015; 23:597-606
- Saluja I, Granneman JG, Skoff RP. PPAR delta agonists stimulate oligodendrocyte differentiation in tissue culture. Glia 2001; 33:191-204; PMID:11241737; http://dx.doi.org/10.1002/1098-1136(200103)33:3%3c191::AID-GLIA1018%3e3.0.CO;2-M
- Cimini A, Bernardo A, Cifone G, Di Muzio L, Di Loreto S. TNFα downregulates PPARδ expression in oligodendrocyte progenitor cells: implications for demyelinating diseases. Glia 2003; 41:3-14; PMID:12465041; http://dx.doi.org/10.1002/glia.10143
- Bernardo A, Minghetti L. PPAR-gamma agonists as regulators of microglial activation and brain inflammation. Cur Pharm Des 2006; 12:93-109; http://dx.doi.org/10.2174/138161206780574579
- Cristiano L, Cimini A, Moreno S, Ragnelli A.M, Paola Cerù M. Cerù Peroxisome proliferator-activated receptors (PPARs) and related transcription factors in differentiating astrocyte cultures. Neuroscience 2005; 131:577-87; PMID:15730864; http://dx.doi.org/10.1016/j.neuroscience.2004.11.008
- Cullingford TE, Bhakoo K, Peuchen S, Dolphin CT, Patel R, Clark JB. Clark Distribution of mRNAs encoding the peroxisome proliferator-activated receptor α, β, and γ and the retinoid X receptor α, β, and γ in rat central nervous system. J Neurochem 1998; 70:1366-75; PMID:9523552; http://dx.doi.org/10.1046/j.1471-4159.1998.70041366.x
- Farioli-Vecchioli S, Moreno S, Cerù MP. Immunocytochemical localization of acyl-CoA oxidase in the rat central nervous system. J Neurocytol 2001; 30:21-33; PMID:11577243; http://dx.doi.org/10.1023/A:1011913223541
- Roth AD, Leisewitz AV, Jung JE, Cassina P, Barbeito L, Inestrosa NC, Bronfman M. PPAR gamma activators induce growth arrest and process extension in B12 oligodendrocyte-like cells and terminal differentiation of cultured oligodendrocytes. J Neurosci Res 2003; 72:425-35; PMID:12704804; http://dx.doi.org/10.1002/jnr.10596
- Di Loreto S, D'Angelo B, D'Amico MA, Cristiano L, Benedetti E, Cinque B, Cifone MG, Cerù MP, Cimini A. PPARβ agonists trigger neuronal differentiation in the human neuroblastoma cell line SH-SY5Y. J Cell Physiol 2007; 211:837-47; PMID:17390299; http://dx.doi.org/10.1002/jcp.20996
- D'Angelo B, Benedetti E, Di Loreto S, Cristiano L, Laurenti G, Cerù MP, Cimini A. Signal transduction pathways involved in PPARβ/δ-induced neuronal differentiation. J Cell Physiol 2011; 226:2170-80; PMID:21520069; http://dx.doi.org/10.1002/jcp.22552
- Wada K, Nakajima A, Katayama K, Kudo C, Shibuya A, Kubota N, Terauchi Y, Tachibana M, Miyoshi H, Kamisaki Y, et al. Peroxisome proliferator-activated receptor γ-mediated regulation of neural stem cell proliferation and differentiation. J Biol Chem 2006; 81:12673-81; http://dx.doi.org/10.1074/jbc.M513786200
- Mulholland DJ, Dedhar S, Coetzee GA, Nelson CC. Interaction of nuclear receptors with the Wnt/β-catenin/Tcf signaling axis: Wnt you like to know? Endocrine Rev 2008; 26:898-915; http://dx.doi.org/10.1210/er.2003-0034
- Jung Y, Song S, Choi C. Peroxisome proliferator activated receptor γ agonists suppress TNFα-induced ICAM-1 expression by endothelial cells in a manner potentially dependent on inhibition of reactive oxygen species. Immunol Lett 2008; 117:63-9; PMID:18206249; http://dx.doi.org/10.1016/j.imlet.2007.12.002
- Cimini A, Cristiano L, Benedetti E, D'Angelo B, Cerù MP. PPAR expression in adult mouse neural stem cells (NSC). Modulation of PPARs during astroglial differentiation. PPAR Res 2007; 2007:48242
- Cimini A, Cerù MP. Emerging roles of peroxisome proliferator-activated receptors (PPARs) in the regulation of neural stem cells proliferation and differentiation. Stem Cell Rev 2008; 4:293-303; PMID:18561036; http://dx.doi.org/10.1007/s12015-008-9024-2
- Hämmerle B, Yañez Y, Palanca S, Cañete A, Burks DJ, Castel V, Font de Mora J. Targeting neuroblastoma stem cells with retinoic acid and proteasome inhibitor. PLoS One 2013; 8:e76761; http://dx.doi.org/10.1371/journal.pone.0076761
- Foreman JE, Sorg JM, McGinnis KS, Rigas B, Williams JL, Clapper ML, Gonzalez FJ, Peters JM. Regulation of peroxisome proliferator-activated receptor-beta/delta by the APC/beta-CATENIN pathway and nonsteroidal antiinflammatory drugs. Mol Carcinog 2009; 48:942-52; PMID:19415698; http://dx.doi.org/10.1002/mc.20546
- Walton NM, Shin R, Tajinda K, Heusner CL, Kogan JH, Miyake S, Chen Q, Tamura K, Matsumoto M. Adult neurogenesis transiently generates oxidative stress. PLoS One 2012; 7:e35264; PMID:22558133; http://dx.doi.org/10.1371/journal.pone.0035264
- Doetsch F, Caille I, Lim DA, Garcia-Verdugo JM, Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell 1999; 97:703-16; PMID:10380923; http://dx.doi.org/10.1016/S0092-8674(00)80783-7
- Hermann A, Suess C, Fauser M, Kanzler S, Witt M, Fabel K, Schwarz J, Höglinger GU, Storch A. Rostro-caudal gradual loss of cellular diversity within the periventricular regions of the ventricular system. Stem Cells 2009; 27:928-41; PMID:19353521; http://dx.doi.org/10.1002/stem.21
- Fuentealba LC, Obernier K, Alvarez-Buylla A. Adult neural stem cells bridge their niche. Cell Stem Cell 2012; 10:698-708; PMID:22704510; http://dx.doi.org/10.1016/j.stem.2012.05.012
- Kazanis I. Neurogenesis in the adult mammalian brain: how much do we need, how much do we have? Curr Top Behav Neurosci 2013; 15:3-29; PMID:22976273; http://dx.doi.org/10.1007/7854_2012_227
- Denham M, Dottori M. Signals involved in neural differentiation of human embryonic stem cells. Neurosignals 2009; 17:234-41; PMID:19816060; http://dx.doi.org/10.1159/000231890
- Alvarez Z, Hyroššová P, Perales JC, Alcántara S. Neuronal progenitor maintenance requires lactate metabolism and PEPCK-M-directed cataplerosis. Cereb Cortex 2014; 26:1046-58; PMID:25452568; http://dx.doi.org/10.1093/cercor/bhu281
- Saez I, Duran J, Sinadinos C, Beltran A, Yanes O, Tevy MF, Martínez-Pons C, Milán M, Guinovart JJ. Neurons have an active glycogen metabolism that contributes to tolerance to hypoxia. J Cereb Blood Flow Metab 2014; 34:945-55; PMID:24569689; http://dx.doi.org/10.1038/jcbfm.2014.33
- Magistretti PJ, Sorg O, Naichen Y, Pellerin L, de Rham S, Martin JL. Regulation of astrocyte energy metabolism by neurotransmitters. Ren Physiol Biochem 1994 May-Aug; 17(3-4):168-71.
- Brown AM, Ransom BR. Astrocyte glycogen and brain energy metabolism. Glia 2007; 55:1263-71; PMID:17659525; http://dx.doi.org/10.1002/glia.20557
- Shi Y, Hon M, Evans RM. The peroxisome proliferator-activated receptor delta, an integrator of transcriptional repression and nuclear receptor signaling. Proc Natl Acad Sci U S A 2001; 99:2613-8; http://dx.doi.org/10.1073/pnas.052707099
- Bacci S, Pieri L, Buccoliero AM, Bonelli A, Taddei G, Romagnoli P. Smooth muscle cells, dendritic cells and mast cells are sources of TNFalpha and nitric oxide in human carotid artery atherosclerosis. Thromb Res 2008; 122:657-67; PMID:18561985; http://dx.doi.org/10.1016/j.thromres.2008.04.013