ABSTRACT
Members of the E2F family of transcription factors have been reported to regulate the expression of genes involved in cell cycle control, DNA replication, and DNA repair in multicellular eukaryotes. Here, E2FL1, a meiosis-specific E2F transcription factor gene, was identified in the model ciliate Tetrahymena thermophila. Loss of this gene resulted in meiotic arrest prior to anaphase I. The cytological experiments revealed that the meiotic homologous pairing was not affected in the absence of E2FL1, but the paired homologous chromosomes did not separate and assumed a peculiar tandem arrangement. This is the first time that an E2F family member has been shown to regulate meiotic events. Moreover, BrdU incorporation showed that DSB processing during meiosis was abnormal upon the deletion of E2FL1. Transcriptome sequencing analysis revealed that E2FL1 knockout decreased the expression of genes involved in DNA replication and DNA repair in T. thermophila, suggesting that the function of E2F is highly conserved in eukaryotes. In addition, E2FL1 deletion inhibited the expression of related homologous chromosome segregation genes in T. thermophila. The result may explain the meiotic arrest phenotype at anaphase I. Finally, by searching for E2F DNA-binding motifs in the entire T. thermophila genome, we identified 714 genes containing at least one E2F DNA-binding motif; of these, 235 downregulated represent putative E2FL1 target genes.
Introduction
Meiosis is a key step in the eukaryotic sexual reproduction. By duplicating chromosomes only once during 2 cell divisions, it provides a mechanism for halving the chromosome number in progeny germ cells. Meiotic events include DNA replication, the formation of DNA double-strand breaks (DSBs), recombinational repair of DSBs, homologous chromosome pairing, homologous chromosome segregation, and sister chromatid resolution. Progression of meiosis is regulated by numerous factors, such as cyclins, cyclin-dependent kinases (CDKs), and transcription factors.
It is well established that E2F transcription factor family members play essential roles in the early stages of the cell cycle.Citation1-3 E2F family members have been identified in a wide range of eukaryotes, including mammals, Arabidopsis thaliana, and Caenorhabditis elegans.Citation3,4 E2F family members have E2F proteins and its dimerization partner (DP) proteins. They formed heterodimers and controlled the expression of genes related to cell cycle, DNA replication, and DNA repair.Citation5 And, E2F family member has a conserved DNA binding domain in eukaryotes, which stimulates E2F-dependent transcription.Citation5,6 At least 8 different E2F proteins (E2f1–E2f8) and 3 DP proteins (DP1, DP2, and DP3) have been identified in mammals.Citation3 In A. thaliana cells, there are 6 E2F genes and 2 DP genes.Citation4 Two E2F genes and one DP gene have been identified in C. elegans.Citation3 Eukaryotic E2F family members can act as transcriptional activators or repressors. In mammalian cells, E2f1, E2f2, and E2f3a act primarily as transcriptional activators, while E2f3b, E2f4, E2f5, E2f6, E2f7, and E2f8 are transcriptional repressors for a subset of E2F-regulated genes.Citation3,7 The E2f4 and E2f5 transcriptional repressors interact with the retinoblastoma protein (RB) to inhibit target gene transcription in quiescent cells and during early G1 phase of the cell cycle.Citation8,9 At G1 or S phase entry, transcriptional activators with free RB replace the repressors, leading to the increase in G1/S phase gene expression and promote DNA replication at S phase.Citation10 During progression from quiescent to S phase, RB phosphorylation by cyclin–CDK complexes activates E2F activity. E2F pathway is highly conserved in higher eukaryotes.Citation11,12
E2F target genes have been identified in mammals and plants.Citation5,13 Large-scale systematic approaches have shown that many E2F target genes function in cell cycle control, DNA replication, DNA damage repair, and many other processes.Citation5,14 Characterization of these genes revealed that E2F DNA-binding motifs are largely conserved during evolution.Citation15 Thus, E2F DNA-binding motifs in different multicellular eukaryotes can serve as a reference for studying E2F target genes in unicellular eukaryotes. The E2F family members are conserved in most eukaryotic lineages.Citation16 While E2F proteins and their function in the cell cycle are extensively investigated among multicellular eukaryotes, little is known about their roles in the cell cycle control of unicellular eukaryotes. Here, we report homologs of E2F family proteins in the model ciliate Tetrahymena thermophila and study the role of a meiosis-specific member of the E2F family.
T. thermophila is a unicellular ciliated protist capable of vegetative and sexual reproduction. All cells contain 2 nuclei. The diploid (2n) germline micronucleus (MIC) contains 5 pairs of metacentric chromosomes. It is transcriptionally inactive in vegetative propagation and undergoes mitotic divisions and meiosis. The polyploid (∼50n) somatic macronucleus (MAC) divides by amitotic splitting and is not transmitted to sexual progeny. Genes from the MAC are transcribed during cell proliferation and determine the phenotype of the cell. When starved cells of 2 different mating types are mixed together, they will conjugate (i.e. form pairs) and both will initiate meiosis. All meiotic nuclear divisions in Tetrahymena (and other ciliates) are closed, i.e., the nuclear envelope is not transiently disassembled.Citation17
Meiosis has been reasonably well characterized at the cytological and molecular levels in T. thermophila. The earliest detectable event is the formation of DNA double-strand breaks (DSBs) by the conserved endonuclease Spo11.Citation18 DSBs trigger formation of the phosphorylated histone H2A.X (γ-H2A.X) and elongation of the MIC to about twice the length of the cell via an ataxia-telangiectasia and Rad3-related protein (ATR)-dependent response.Citation19,20 Within the elongated nucleus, chromosome arms are arranged in parallel, with all centromeres clustered at one end and all telomeres clustered at the opposite end. This arrangement promotes homologous pairing.Citation19 At the same time, DSBs are processed in a manner that is widely conserved in eukaryotes: DSB ends are resected and single-stranded 3′ overhangs become decorated by the recombination protein Dmc1 and Rad51. These single strands can invade DNA tracts on the homologous chromosome.Citation21 The heteroduplexes created by this strand exchange are extended by DNA repair synthesis, which can be monitored by the incorporation of 5-bromo-2′-deoxyuridine (BrdU; in Tetrahymena, the pre-meiotic DNA replication is completed prior to mixing the starved cells and thus before pair formation).Citation22 Then, subsets of these DNA joint molecules become crossovers. After, 5 condensed bivalents are appeared.Citation23 Ultimately, owing to the role of separase Esp1, homologous chromosomes and sister chromatids are segregated.Citation24
Little is known about developmental programs controlled by transcription factors that initiate and regulate the progression of meiosis in T. thermophila. In this study, screening E2F family members by gene prediction analysis and bioinformatic analysis, we found an E2F transcription factor homolog E2FL1 (TTHERM_00695710), was specifically expressed during conjugation. Based on the cytological experiments and transcriptome sequencing data, we found that disruption of this gene resulted in arrested meiosis and decreased expression of genes involved in DNA replication, DNA repair, and the associated homologous chromosome segregation. Finally, using a bioinformatics approach, we have identified many putative E2FL1 target genes in T. thermophila.
Results
E2FL1 is a meiosis-specific transcription factor in T. thermophila
Gene prediction analysis identified 51 conjugation-related transcription factors in T. thermophila.Citation25 Among them, 7 E2F transcription factor family members had been identified: these were named as E2FL1, E2FL2, E2FL3, E2FL4 and DPL1, DPL2, DPL3 (Table S1). Domain prediction showed that all E2F family members in T. thermophila have a conserved E2F/DP family winged-helix DNA-binding domain (), which has demonstrated that all E2F family members are transcription factor in T. thermophila. It also can be seen that the T. thermophila E2fl1 showed the greatest similarity to C. elegans Efl-2 (), which is a candidate transcriptional activator.
Figure 1. T. thermophila E2fl1 is a meiosis-specific transcription factor. (A) Phylogenetic tree shows the E2F and DP transcription factors protein in H. sapiens, A. thaliana, C. elegans, and T. thermophila. The red box indicates T. thermophila E2fl1. H, A, C, T stand for H. sapiens, A. thaliana, C. elegans, and T. thermophila, respectively. Every protein has different domains. (B) Expression profiles of 7 E2F homologs in T. thermophila based on microarray data from the TetraFGD. The blue box indicates the expression of E2FL1 during vegetative growth and starvation in T. thermophila. (C) The T. thermophila E2FL1 expression profile based on microarray data from the TetraFGD. G and S in (B) and (C) indicate vegetative growth and starvation. “0”, “2”, “4”, “6”, “8”, “10”, “12”, “14”, “16”, ,” “18” in (B) and (C) indicate the time post mixing. (D) E2fl1-HA (green) localizes to the MAC during meiosis but not during vegetative growth (both mating partners express HA-tagged E2fl1). Scale bar: 10 μm.
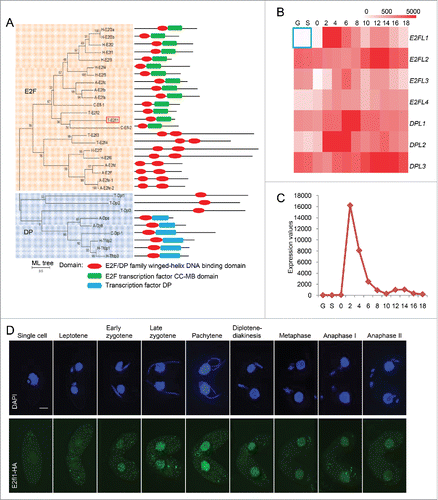
The majority of the T. thermophila meiosis genes were only transcribed during conjugation and the genes expression profiles were peaked at early stage of conjugation (around 2 h post mixing). We assumed that the transcription factor which specifically activated the meiotic genes transcription would exhibit the similar expression profile. So, the expression profiles of all E2F family homologs were investigated using microarray data from the Tetrahymena Functional Genomics Database (TetraFGD, http://tfgd.ihb.ac.cn/). E2FL1 (TTHERM_00695710) was not expressed under vegetative growth or starvation conditions, but it was specifically expressed during conjugation (). Transcriptome sequencing analysis showed that E2FL1 gene expression was particularly high at 2 h post mixing, which was consistent with previous microarray data (Fig. S1). Therefore, we hypothesized that E2fl1 is a meiosis-specific transcription factor and may play an important role at an early stage of meiosis.
Our conjecture that E2fl1 acted as a meiosis-specific transcription factor in T. thermophila was supported by the subcellular localization of a C-terminal hemagglutinin (HA)-tagged E2fl1 fusion protein during conjugation. Granular HA signals were not detected under vegetative growth. However, we observed HA signals in MACs from the beginning of the meiosis until anaphase II (), and in the crescent stage at 4 h post mixing, HA signals was very highlight, which indicated that the expression of E2fl1 protein was highest at this time.
E2fl1 is essential for sexual reproduction
To study E2FL1 gene function, we knocked out its open reading frame (ORF) in the Tetrahymena macronuclear (MAC) genome. Transcriptome sequencing analysis confirmed that the E2FL1 gene was completely knocked out in both mating types (Fig. S1). Consistent with the previous result that E2FL1 is a meiosis specific gene, thus E2FL1 gene knockout had no effect on vegetative growth (Fig. S2). To understand whether E2fl1 is required for the sexual reproduction, the viability of sexual progeny of e2fl1△ is evaluated (see material and methods for details). Strikingly, none of sexual progeny of e2fl1△ cell matings was survived, whereas 71.2% of the progeny of WT matings was viable. The results indicated that E2fl1 is essential for sexual reproduction in T. thermophila.
Bivalents are arranged abnormally at a meiotic stage corresponding to anaphase I in e2fl1△ cells
During T. thermophila meiosis, prophase has 5 stages that are classified by the degree of MIC elongation: leptene, zygotene, pachytene, diplotene and diakinesis.Citation26 At stages roughly corresponding to leptotene, zygotene, and pachytene of meiotic prophase, chromosomes are arranged in parallel bundles within the elongated MIC. Once chromosomes are paired, the nucleus shortens and 5 condensed bivalents become visible; they assemble at the nuclear equator at metaphase I. Finally, the MIC undergoes 2 meiotic divisions to separate the homologous chromosomes and sister chromatids (). Early meiotic MIC elongation and shortening had similar temporal properties (initiation and duration) in e2fl1△ cells to those of WT cells, which seemed normal at meiotic prophase by the disruption of E2FL1. However, meiosis progression was temporarily halted at a metaphase and then until 6 h post mixing arrested at a stage prior to anaphase I with 5 dense chromatin masses arranged in tandem. Finally, only at 7 h post mixing did the MIC begin to form a single nucleus (). Because one of the most critical events at meiotic prophase is homologous pairing, fluorescence in situ hybridization (FISH) confirmed that homologous chromosomes were paired normally at pachytene in e2fl1△ cells (), suggesting that homologous chromosomes pairing was not affected by deletion of E2FL1. However, FISH also confirmed that the 5 dense chromatin masses in e2fl1△ cells were equivalent to bivalents (). Their tandem arrangement was remarkable because in the WT such an arrangement did not occur, even transiently. In a word, the results indicated that in e2fl1△ cells bivalents are arranged abnormally at a meiotic stage corresponding to anaphase I.
Figure 2. The e2fl1△ phenotype is arrested a meiotic stage prior to anaphase I. (A) The e2fl1△ phenotype and meiotic events in T. thermophila. DAPI staining shows abnormal meiosis in e2fl1△: cells are arrested a stage prior anaphase I. At anaphase I, homologous chromosome segregation can be seen in WT cells, whereas e2fl1△ cells have 5 dense chromatin masses arranged in tandem. Scale bars: 10 μm. (B) The meiotic time course, classified according to the morphological criteria,Citation26 is similar in WT and e2fl1△ MICs before the metaphase stage. The red asterisk at 6 h post mixing (which corresponds to WT anaphase I) indicates that e2fl1△ cells have formed 5 dense chromatin masses arranged in tandem after metaphase stage. The blue asterisk at 7 h post mixing (which corresponds to WT anaphase II) indicates that some MICs in e2fl1△ cells have formed a single nucleus. At least 100 MICs are evaluated for each time point.
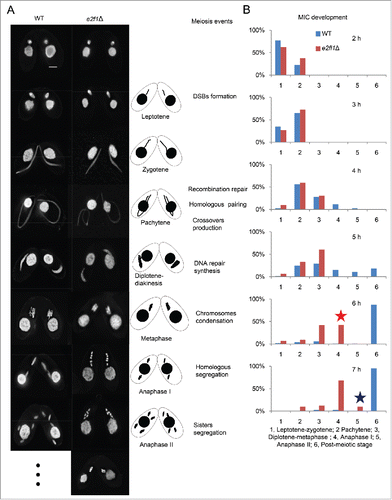
Figure 3. Bivalents are arranged abnormally in e2fl1△ cells at a stage corresponding to anaphase I. (A) FISH (red) analysis shows that homologous chromosome pairing is normal in e2fl1△ cells. ‘d’ shows the distance between 2 dots in elongated MICs at meiotic prophase. The statistical analysis used Mann-Whitney test. (B) FISH (red) analysis shows that MICs formed bivalents at metaphase in WT cells and an abnormal structure in e2fl1△ cells. (C) α-tubulin immunostaining (green) shows abnormal microtubule stretching in e2fl1△ cells from meiotic pachytene to anaphase. Scale bar: 10 μm.
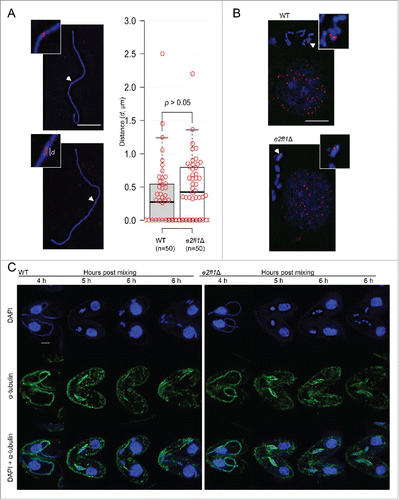
Immunostaining revealed that the arrangement of α-tubulin in e2fl1△ cells is normal before anaphase I. At pachytene, α-tubulin and MICs became stretched to form a so-called full crescent shape at 4 h post mixing. At metaphase (at 5 h post mixing), MICs were rearranged and located in the middle of spindle-like bundles in both WT and e2fl1△ cells. However, due to the meiotic abnormalities caused by E2FL1 deletion, α-tubulin in e2fl1△ cells formed aberrant structures at anaphase (at 6 h post mixing). Instead of forming the bundle-like structure seen in WT cells, α-tubulin in e2fl1△ cells formed a spindle-like bundle (). Therefore, α-tubulin arrangement in the bivalents of e2fl1△ cells appears abnormal.
The DNA repair synthesis stage of DSB processing is abnormal in e2fl1△ cells
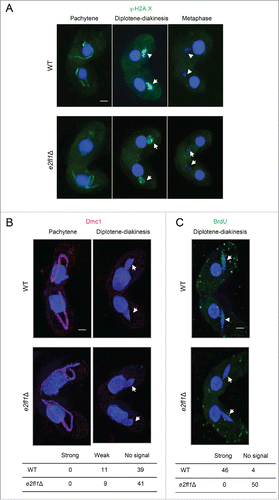
The appearance at pachytene and diplotene–diakinesis and disappearance at metaphase of γ-H2A.X (a marker of DSB formation) was similar in both WT and e2fl1△ cells (). The appearance at pachytene and disappearance at diplotene-diakinesis of Dmc1 (a marker of DSB processing) indicated that DSB repair was normal in e2fl1△ cells (). At diplotene–diakinesis, 22% of WT cells (n = 50) had a weak Dmc1 signal and 78% had no signal. In e2fl1△ cells, 18% (n = 50) had a weak Dmc1 signal and the remainder had no signal (). This suggests that DSBs are repaired with a similar efficiency in WT and e2fl1△ cells. However, we unexpectedly found that DNA repair synthesis differed in e2fl1△ and WT cells. When BrdU was added at 3.5 h post mixing, it became incorporated in WT MICs during the shortening phase, when repair-related DNA synthesis is known to take place.Citation22 Consequently, we observed foci using an anti-BrdU antibody in 46 out of 50 WT MICs at this or a later stage. In contrast, 0 out of 50 MICs at corresponding stages in e2fl1△ cells had BrdU signals (). This may indicate that DSB repair is arrested in e2fl1△ cells at an as yet unknown intermediate stage.
Figure 4. The DNA repair synthesis stage of DSB processing is abnormal in e2fl1△ cells. (A) γ-H2A.X (green) foci indicate the same meiotic prophase in MICs of both e2fl1△ and WT cells in a mating pair. The foci also appear at diplotene or diakinesis; however, they are absent at metaphase in WT and e2fl1△ cells. (B) Intense Dmc1 immunostaining (red) is present in WT MICs at pachytene but completely absent at diplotene or diakinesis, similar to in e2fl1△ cells. (C) BrdU immunostaining (green) shows numerous foci at diplotene–diakinesis in WT meiotic MICs, but none in e2fl1△ cells. Scale bars: 10 μm.
Genes with roles in DNA metabolic and homologous chromosome segregation are downregulated in e2fl1△ cell
We next investigated the effect of E2FL1 knockout on gene expression using a twofold cutoff for differentially expressed genes (DEGs; see Material and methods for details). At 2 h post mixing, the expression of only 914 DEGs differed from WT. The proportion of DEGs increased with time up to 5589 at 6 h post mixing (Fig. S3). This is consistent with a function of E2fl1 from about 6 h post mixing, which matches with the onset of a cytologically observable defect in the mutant (). Gene Ontology (GO) enrichment analysis of downregulated or upregulated DEGs at 2 h, 3 h, 4 h, 5 h, 6 h, and 7 h post mixing was performed. Then, we calculated the number of genes belongs to the enriched terms at each time point. It is obvious that the numbers of downregulated DEGs in enriched terms are far more than upregulated DEGs (). Therefore, the function of downregulated genes seem more concentrated, and perhaps is more important for us to understand the regulation of E2fl1. A significant proportion of the 5179 DEGs that were downregulated at least at one time point post mixing are classified into GO terms related to translation, DNA metabolism, cell cycle, and organelle organization (). The most important GO term was DNA metabolism which was involved in DNA replication and DNA repair. The expression of DEGs to DNA replication and DNA repair which were found by GO analysis was completely downregulated at 2–7 h post mixing (Table S2; ). This may indicated that genes of DNA replication and DNA repair were regulated by E2fl1.
Figure 5. Differentially expressed genes (DEGs) are analyzed in WT and e2fl1△ cells. (A) Distribution of upregulated and downregulated DEGs after GO enrichment at 2–7 h post mixing. The histogram shows that the number of downregulated DEGs in enriched terms is far more than upregulated DEGs. (B) GO enrichment analysis of DEGs that are downregulated at least at one time point post mixing. The GO terms include translation, DNA metabolism, cell cycle, and organelle organization. (C) Heat map shows fold change in the expression of genes related to DNA replication and DNA repair in the e2fl1△ cells. (D) Heat map shows fold changes in meiotic gene expression, including gene related to homologous chromosome segregation in T. thermophila. (E) Heat map shows fold change in the expression of putative E2FL1 target genes in the e2fl1△ cells. (F) GO enrichment analysis of putative E2FL1 target genes. The GO terms include DNA metabolism and organelle organization.
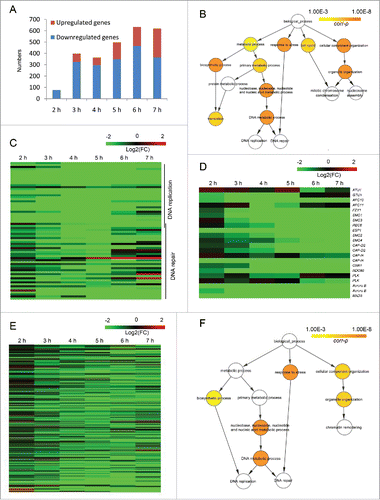
Microtubules are crucial for homologous chromosome segregation at anaphase I of meiosis.Citation27 Therefore, E2FL1 knockout may lead to a microtubule defect. Transcriptome sequencing analysis showed that α-tubulin gene (ATU1) expression was normal in e2fl1△ cells, which may explain that α-tubulin localization is normal before anaphase and appears abnormal at anaphase in e2fl1△ cells. However, expression of the γ-tubulin gene (GTU1), another microtubule component, was strongly inhibited in e2fl1△ cells at 2–5 h post mixing (). Genes encoding anaphase-promoting complex (APC) components, APC activator Fzy1/Cdc20, the cohesion complex (Smc1, Smc3 and Rec8), the Esp1 separase, the condensation complex (Smc2, Smc4, Cap-D, and Cap-H), the kinetochore (CenH3 (Cna1 in T. thermophila) and Ndc80), the microtubule–kinetochore attachment (polo-like kinases, aurora B kinases, and the Mad3 spindle checkpoint protein) (Table S3) are important regulators of homologous chromosome segregation. The strong downregulation of all these genes in e2fl1△ cells at 2–7 h post mixing (), may explain the abnormal homologous chromosome segregation seen during meiosis.
Cytological experiments and differential expression analysis of genes related to DNA replication and DNA repair indicated that incomplete DNA repair synthesis may be responsible for the e2fl1△ phenotype. In addition, a sharp reduction in the expression of genes encoding proteins related to homologous chromosome segregation at 2–7 h post mixing in e2fl1△ cells, is expected to weaken microtubule–chromosome attachment and microtubule–kinetochore attachment, resulting in abnormal or absent homologous chromosome segregation, which may explain the meiotic arrest phenotype.
Many genes containing E2F DNA-binding motifs are downregulated in e2fl1△ cells
Identifying the complete pool of target genes of E2F transcription factor family members in the Tetrahymena genome would provide a valuable resource for investigating the roles of E2F proteins in the Tetrahymena life cycle. All E2F family members have conserved DNA-binding motifs that are necessary and sufficient for binding to DNA-binding motifs in target gene promoters. Therefore, to predict E2F DNA-binding motifs in the Tetrahymena genome, we analyzed the E2F DNA-binding motifs of several well-studied model eukaryotes, including humans, Mus musculus, A. thaliana, C. elegans, and Drosophila melanogaster (). Using the approach (see material and methods for details), we identified E2F family target genes containing these sequences within 1 kb upstream of the translation start site (TSS) in the Tetrehymena genome. A total of 714 Tetrahymena genes have at least one E2F DNA-binding motif, representing putative target genes of the 7 E2F family members in T. thermophila. Of these, 316 DEGs were identified at 2 h, 3 h, 4 h, 5 h, 6 h, and 7 h post mixing in e2fl1△ cells, of which 235 genes (74.4%) were downregulated for at least at one time point post mixing (, Table. S4). Many genes containing E2F DNA-binding motifs were downregulated in e2fl1△ cells, thus supporting their classification as putative E2FL1 target genes.
Table 1. The number of genes with E2F DNA-binding motifs (identified in several well-studied eukaryotes) in Tetrahymena.
GO enrichment analysis of these downregulated genes showed over-representation of terms related to DNA metabolism, and organellar organization (, which is similar to ). Genes related to DNA metabolism in overlap with those related to DNA replication and DNA repair that we listed previously. Some enriched genes, such as RFC2, RFC3, RFC4, MCM5, MSH2, and MSH3, are reported to be E2F target genes in other eukaryotes, and are therefore likely to also be E2FL1 target genes in Tetrehymena. Downregulation of these genes in e2fl1△ cells may be responsible for the abnormal DNA repair synthesis, and possibly for the meiotic arrest.
Discussion
E2fl1 is a meiosis-specific putative transcriptional activator
Gene prediction analysis, domain prediction analysis, and transcriptome sequencing analysis suggest that T. thermophila E2fl1 may be a meiosis-specific transcription factor. Then, according to the phylogenetic analysis, the T. thermophila E2fl1 is a close neighbor of the C. elegans Efl-2, which is a candidate transcriptional activator and had a role in DNA damage-induced germ cell apoptosis.Citation3 In addition, mammalian E2F family members regulate the expression of genes involved in DNA replication, DNA repair, nucleosome assembly, and multiple cell cycle control points.Citation28 However, genes related to DNA replication and DNA repair were mostly downregulated in e2fl1△ cells. Many genes containing E2F DNA-binding motifs are also downregulated in e2fl1△ cells, including several such as RFC2, RFC3, RFC4, MCM5, MSH2, and MSH3 that are reported to be E2F target genes in other eukaryotes.Citation5 Therefore, E2fl1 may be a meiosis-specific transcriptional activator.
This study provides evidence from transcriptome sequencing analysis and cytological experiments of E2FL1 knockout cells that E2fl1 may encode a meiosis-specific transcriptional activator involved in regulating meiotic DNA repair synthesis and homologous chromosome segregation.
E2fl1 is important for DNA repair synthesis
On the one hand, the e2fl1△ phenotype in T. thermophila is unusual. The appearance of Dmc1 in MICs after the elongation stage and the disappearance of well-condensed, intact bivalents suggest that DSBs are repaired. In contrast, no evidence of DNA repair synthesis (by BrdU incorporation) was found. An alternative method of meiotic DSB repair by synthesis-independent non-homologous end joining in T. thermophila is unlikely,Citation29 but remains a possible explanation. It is also conceivable that strand invasion and heteroduplex formation occurs, but the subsequent elongation of invading strands fails. In this case, we hypothesized that mature chiasmata were not formed and that bivalents were held together by crossover intermediates in the absence of E2FL1. Therefore, homologous chromosomes would be stably connected by these molecular recombination intermediates.
On the other hand, in response to DSB signals, the repair protein Rad51 functions in the attachment and strand invasion. RAD51 gene expression is also reported to be regulated by E2F family members.Citation5 In T. thermophila, RAD51 gene disruption resulted in the fragmented chromatin at metaphase I during meiosis.Citation21 RAD51 gene expression was also downregulated after E2FL1 inhibition (). Therefore, the level of RAD51 expression may affect which method is used for strand invasion in T. thermophila. In addition, DNA replication genes such as those encoding DNA replication licensing factor mini-chromosome maintenance (MCM) proteins and replication factor C (RFC) proteins are regulated by E2F family.Citation5 Moreover, MCM complex proteins that colocalize with γ-H2A.X are involved in chromatin remodeling in response to DNA damage.Citation30 Expression of these genes was sharply downregulated in e2fl1△ cells, which might have impaired the process of DNA repair synthesis. However, this phenomenon has not been previously reported and further investigation is therefore necessary.
E2fl1 is also important for homologous chromosome segregation
Microtubules are important for homologous chromosome segregation in many eukaryotes,Citation27 although little is known about their role in this process in T. thermophila. γ-tubulin has conserved roles in microtubule nucleation, centrosome duplication, and spindle formation in many eukaryotes.Citation27,31,32 It also interacts with Rad51 in DNA recombination repair,Citation33 and modulates the transcriptional activity of E2F family members during S phase entry.Citation34 However, GTU1 expression was strongly downregulated at meiotic prophase in the e2fl1△ cells, suggesting that E2FL1 might cooperate with γ-tubulin to regulate downstream genes.
The APC activator Fzy1/Cdc20, the cohesin complex (Smc1, Smc3, and Rec8), and the Esp1 separase are essential for homologous chromosome segregation.Citation27 The APC activator Fzy1/Cdc20 promotes endocycle onset and progression.Citation35 Atypical E2F proteins control gene expression during the cell cycle.Citation36 The cohesin complex is loaded onto chromosomes during premeiotic S phase. It is involved in DNA damage repair and promotes homologous recombination repair.Citation37 In T. thermophila, loss of the cohesin complex genes lead to abnormal meiosis, causing cells to be arrested at the metaphase-anaphase transition along with chromosome missegregation and a failure in DSB repair. In addition, loss of ESP1 prevented homologous chromosome separation at meiosis I.Citation24
Then, the condensation complex (Smc2, Smc4, Cap-D, and Cap-H) has an important role in chromosome condensation and segregation in many eukaryotes.Citation27 E2F members are reported to directly regulate the expression of mammalian Smc2 and Smc4.Citation14 In T. thermophila, the expression of the condensation complex genes is sharply downregulated at 2–7 h post mixing in e2fl1△ cells, indicating that E2fl1 may control the condensation complex genes expression, leading to a chromosome condensation defect that alters homologous chromosome segregation in e2fl1△ cells.
In summary, although DSB formation, DSB repair, and homologous chromosome pairing occur normally during meiosis in e2fl1△ cells, DNA repair synthesis is incomplete and homologous chromosomes do not segregate. Therefore, the 5 dense chromatin masses may represent 5 pairs of homologous chromosomes in the e2fl1△ MIC. However, further investigation is needed to determine what eventually happens to the T. thermophila MIC in the absence of E2FL1.
The E2F family has a conserved function in eukaryotes
Studies in model eukaryotes, such as Mus musculus, A. thaliana, Drosophila melanogaster, and C. elegans, have revealed the conserved function of various E2F family members.Citation3 In these eukaryotes, E2f transcriptional activators regulate cell cycle progression and cell proliferation, and control the expression of genes related to cell cycle and DNA replication.Citation12,38 When these genes are mutated, DNA replication and cell cycle progression are inhibited, even resulting in ectopic cell division; and, the expression of genes related to cell cycle and DNA replication is decreased.Citation39,40 However, E2f transcriptional suppressors are also important for cell cycle progression and cell differentiation.Citation39 Mutation these genes increase the expression of cell cycle genes and decrease DNA endoreplication; cell proliferation and cell division are also affected.Citation41-43 Some atypical E2Fs have diverse functions, for example, A. thaliana E2Ff represses cell wall biosynthesis genes.Citation44
So far, no E2F family member with a meiosis-specific function has been reported. T. thermophila contains homologs of several E2F family members, including a novel example of a meiosis-specific E2F family gene, E2FL1. When E2FL1 was mutated, the expression of DNA metabolic genes was strongly downregulated and the progression of cell cycle was inhibited. Furthermore, most putative E2FL1 target genes are downregulated in e2fl1△ cells and some are conserved between T. thermophila and other eukaryotes. This study therefore showed that E2F family members have highly conserved functions in model eukaryotes (such as regulating DNA replication gene expression and controlling cell cycle progression) and thus have conserved target genes. The other T. thermophila E2F homologs will be further investigated.
Materials and methods
Strains and cell culture
T. thermophila strains CU427 (VI) and CU428 (VII) were served as WT controls which were obtained from the Tetrahymena Stock Center, Cornell University (http://tetrahymena.vet.cornell.edu/). Cells were propagated in super proteose peptone (SPP) medium (1% proteose peptone, 0.2% glucose, 0.1% yeast extract, 0.003% Sequestrene) on a rotary shaker at 135 rpm at 30°C.Citation45 For meiosis experiments, cells were grown to a density of 3 × 105 cells ml−1 and then starved in 10 mM Tris-HCl (pH 7.4) for at least 20 h. Conjugation was induced by mixing 2 populations of starved cells of different mating types at equal densities (∼2 × 105 cells ml−1). Samples of conjugating cells were collected at the indicated time points and prepared for cytological analysis.
Macronuclear gene knockout
To construct E2FL1 knockout strains, ∼1 kb fragments upstream and downstream of the of E2FL1 gene ORF were amplified using primers from Table S5: E2FL1KO-5FW, E2FL1KO-5RV, E2FL1KO-3FW, and E2FL1KO-3RV. Using fusion polymerase chain reaction (PCR), the 2 flanking fragments were joined to the ends of the neo4 cassette, comprising the neo4 resistance gene under the control of the Cd2+-inducible MTT1 metallothionein promoter.Citation46 Then the product was then cloned into the pBlueScript SK (+) vector to construct the pE2FL1KO plasmid. Finally, the knockout construct was introduced into WT cells of CU427 and CU428 by biolistic transformation as described previously.Citation47 The transformants were selected in SPP media containing decreasing concentrations of CdCl2 (from 1 mg ml−1 to 0.1 mg ml−1) and increasing paromomycin (from 150 μg ml−1 to 64 mg ml−1) until all MAC copies of the WT allele were replaced by the knockout construct completely owing to phenotypic assortment.Citation47
Protein tagging
E2fl1-HA-expressing cells were created using a knock-in method to fuse the HA tag to the 3’ of the gene at its target genomic locus. To create the tagged construct, the last 500 bp of the ORF and 2 regions of downstream of the E2FL1 gene were amplified by fusion PCR to connect to the neo4 cassette. The gene fragments were amplified using the primers from Table S5: E2FL1HA-CDSFW, E2FL1HA-CDSRV, E2FL1HA-3UPFW, E2FL1HA-3UPRV, E2FL1HA-3DOWNFW, and E2FL1HA-3DOWNRV. The three PCR products were cloned into the pBlueScript SK (+) vector to construct the pE2FL1HA plasmid. Finally, transformations and selections were performed as described for the knockout strains.
Cytological methods
For 4′, 6′-diamidino-2-phenylindole (DAPI) staining and HA immunostainings, cells were fixed and prepared by the methods of Loidl and Scherthan (2004). In short, 5 ml conjugating cells suspension, 250 μl of 10% Triton X-100 and 500 μl of 37% formaldehyde were added and incubated for 30 min at room temperature (RT). The fixed cell suspension was then centrifuged and resuspended in 500 μl fixative solution (4% paraformaldehyde and 3.4% sucrose in water). Cell suspension (80 μl) was then spread onto a slide and air-dried in a fume hood.Citation22 For Dmc1 immunostaining need a cold condition with a higher concentration of 10% Triton X-100, which will remove free proteins from nucleus.Citation48
For γ-H2A.X immunostaining, cells were prepared by the methods of Mochizuki et al. (2008). Conjugating cell suspension (5 ml) was fixed with 20 μl partial Schaudinn's fixative (saturated HgCl2, absolute ethanol 2:1). After incubation for 5 min at RT, cells were washed twice with 500 μl methanol, and then resuspended in 500 μl methanol. A few drops of this suspension were applied to a slide from a height of at least 50 cm and then air-dried. For FISH, 495 μl of Schaudinn fixative was supplemented with 5 μl 100% acetic acid to cell pellet prepared from 5 ml conjugating cell suspension. Incubating 1 h at RT, the cells suspension was centrifuged and resuspended in 300 μl M/A solution (methanol, acetic acid 2:1). Several drops were applied to the slides and then air-dried.
For 5-Bromo-2’-deoxyuridine (BrdU) immunostaining, BrdU reagent (from a 0.2 M stock in dimethyl sulfoxide(DMSO)) was added to conjugating cells at 3.5 h post to a final concentration of 2 × 10−4 M and incubated in the dark for 1 h. Slides were prepared as DAPI staining. Incubating the slides using 1 M potassium thiocyanate at 90°C for 15 min, rinsing twice with 2 × sodium chloride-sodium citrate (SSC) solution, and denaturing in 70% formamide containing 2 × SSC solution for 2 min at 68°C expose the labeled nucleotides to the antibody.Citation22 Then, the slides were placed in ice-cold water to stop denaturation.
For α-tubulin immunostaining, 5 ml conjugating cell suspension was centrifuged and incubated with 5 ml PEM buffer for 1-2 min at RT, and then with 243 µl of 37% formaldehyde for 30 min at RT. The cell suspension was re-centrifuged and resuspended in 0.5-1 ml of PEM buffer containing 0.1 M glycine. Cell suspension (150 μl) was spread onto the coated poly-L-lysine and air-dried.
For immunostaining, the slides were washed twice with 1 × phosphate buffered saline (PBS) and once with 1 × PBS containing 0.05% Triton (5 min each). Primary and FITC- or Cy3-labeled secondary antibodies were applied as described previously and mounted with anti-fading agent (Vector Laboratories Inc., Burlingame, CA, USA) supplemented with 0.5 mg ml−1 DAPI.Citation49 Primary antibodies: HA immunostaining (1:100 rabbit anti-HA monoclonal antibody, Sigma, St. Louis, MO, USA), Dmc1 immunostaining (1:100 mouse anti-DMC1 monoclonal antibody, NeoMarkers, Fremont, CA, USA), γ-H2A.X immunostaining (1:100 mouse anti-γ-H2A.X monoclonal antibody, BioLegend, San Diego, CA), BrdU immunostaining (1:50 rat anti-BrdU antibody, Abcam, Cambridge, UK), a-tubulin immunostaining (1:150, mouse anti-a-tubulin monoclonal antibody, Lab Vision, Fremont, CA, USA). For FISH, a probe corresponding to a 22.1 kb intercalary chromosomal region was produced by PCR and Cy3 labeled by nick translation.Citation50 The probe and chromosomal DNA were denatured with hot formamide and hybridized for 36-48 h at 37°C.Citation22 Then the slides were washed with 1 × PBS and mounted with DAPI.
Cell staining was observed by fluorescence microscopy, Z-stacks of images were recorded using MetaViewsoftware (Universal Imaging, Downingtown, PA, USA), deconvoluted using AutoDeblur (AutoQuant Imaging, Watervliet, NY, USA), and projected with ImageJ (Wayne Rasband, N.I.H.; http://rsb.info.nih.gov/ij/) software.
T. thermophila growth analysis
Cells of different strains at same density (3250∼5000 cells ml−1) were propagated in 30 ml SPP medium on a rotary shaker at 135 rpm at 30°C until they reached log phase (about after 12 h), and were then counted every 2 h using a Z Series Coulter Counter until they reached the plateau stage (at about 48 h). The doubling time was then calculated using the formula: t = ln (2) (growth rate)−1.Citation51 The experiment had 3 replications to calculate the doubling time.
Testing the viability of sexual progeny
For testing the viability of sexual progeny, cells were prepared by the method from Mochizuki et al. (2008). In short, conjugating-cell pairs after 6-10 h post mixing were selected into single droplets of SPP medium and kept 2 days at 30°C. After this time colonies that had formed were transferred to 96-well plates. One day later, aliquots were transffered to SPP medium containing 100 ug ml−1 paromomycin and 1 ug ml−1 CdCl2. After meiosis, the old MAC carried the neo4 paromomycin resistance gene, whereas the new MAC did not carry. Colonies that survived paromomycin treatment for 2 days were scored as parental; otherwise, they were scored as progeny because only those that had successfully undergone meiosis and produced paromomycin-sensitive colonies were counted.
Gene identification, phylogenetic and heat map analysis
E2F family members were identified based on the new gene predictions (version 2014: http://ciliate.org/index.php/home/downloads).
To identify homologous genes involved in microtubule–kinetochore attachment and spindle checkpoint in Tetrehymena, BLASTP analysis was performed to compare gene expression in model eukaryotes and T. thermophila. The gene expression profiles were retrieved from TetraFGD database.Citation25,52,53
To identify putative Tetrehymena E2FL1 putative target genes, E2F DNA-binding motifs in several eukaryotes, including humans, Mus musculus, A. thaliana, Oryza sativa, Nicotiana benthamiana, C. elegans, and D. melanogaster, were analyzed to determine the E2F DNA-binding motifs. These E2F DNA-binding motifs were used to search 1 kb regions upstream of the translation start site in all Tetrehymena genes. Genes containing the E2F DNA-binding motifs were putative E2F family members target genes. The number of target genes associated with the DNA-binding motif for each E2F family member is listed in .Citation13,14,54-58
For phylogenetic analysis, multiple sequence alignment was performed using clustalW.Citation59 Phylogenetic tree was constructed using MEGA6 with the Maximum Likelihood method and 1000 bootstrap replicates.Citation60 The protein domain prediction was based on NCBI's conserved domain database.Citation61 A heat map was generated using the MeV program (version 4.9.0).Citation62
Transcriptome sequencing analysis
Total RNA was extracted from WT and e2fl1△ mating cells at 2 h, 3 h, 4 h, 5 h, 6 h and 7 h post mixing using the RNeasy Protect Cell Mini Kit (Qiagen, Germany), as previously described (TetraFGD: http://tfgd.ihb.ac.cn/index/smphelp).Citation63 The poly-A tailed mRNA as enriched using the Sera‐Mag magnetic oligo (dT) beads (GE), and Illumina sequencing libraries were constructed according to the manufacturer's protocols. Paired-end (150 bp × 2) sequencing was performed for all the samples using Illumina Hiseq4000 sequencer. All RNA sequence data have been submitted to GenBank databases under accession number GSE81856. Raw reads adaptors were trimmed with Trim-Galore (version 0.4.0)Citation64 and then mapped to the T. thermophila Macronuclear genome assembly (version 2014: http://ciliate.org/index.php/home/downloads) using TopHat (version 2.0.9).Citation65 Expression levels were normalized to fragments per kilobase of exon per million fragments mapped (FPKM) using the Cuffdiff program (version 2.1.1).Citation66
Genes with very low expression (FPKM < 10) were excluded from the DEGs analysis. DEGs between WT and e2fl1△ cells were identified using a twofold cutoff. GO enrichment analysis was carried out using BiNGO (version 3.0.2).Citation67,68 False discovery rate correction was used in multiple testing to control for false positives, and significantly enriched GO terms were defined by a corrected P-value of <0.001.
Abbreviations
| APC | = | anaphase-promoting complex |
| ATR | = | ataxia-telangiectasia and Rad3-related protein |
| BrdU | = | 5-Bromo-2′-deoxyUridine |
| CDKs | = | cyclin-dependent kinases |
| e2fl1Δ | = | E2FL1 knockout strains |
| DEGs | = | differentially expressed genes |
| DP | = | dimerization partner |
| DSBs | = | double-strand breaks |
| FPKM | = | fragments per kilobase of exon per million fragments mapped |
| GO | = | gene ontology |
| MAC | = | macronucleus |
| MIC | = | micronucleus |
| RB | = | retinoblastoma protein |
| TSS | = | translation start site |
| WT | = | wild-type |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
KCCY_A_1259779_Supplementary_material.zip
Download Zip (141.8 KB)Acknowledgment
We are grateful to Prof. Josef Loidl from University of Vienna for his critical comments and English improvement.
Funding
This work was supported by grants from the Projects of International Cooperation and Exchanges Ministry of Science and Technology of China (2013DFG32390) and the Natural Science Foundation of China (No. 31525021) to Wei Miao, and grants from the OEAD (CN10/2013) and the Austrian Science Fund (P27313-B20 and W1238-B20) to Josef Loidl.
References
- Lathangue NB. Drtf1/E2f: an expanding family of heterodimeric transcription factors implicated in cell cycle control. Trends Biochem Sci 1994; 19:108-14; PMID:8203017; http://dx.doi.org/10.1016/0968-0004(94)90202-X
- Weinberg RA. The retinoblastoma protein and cell cycle control. Cell 1995; 81:323-30; PMID:7736585; http://dx.doi.org/10.1016/0092-8674(95)90385-2
- van den Heuvel S, Dyson NJ. Conserved functions of the pRB and E2F families. Nat Rev Mol Cell Bio 2008; 9:713-24; http://dx.doi.org/10.1038/nrm2469
- Mariconti L, Pellegrini B, Cantoni R, Stevens R, Bergounioux C, Cella R, Albani D. The E2F family of transcription factors from Arabidopsis thaliana: novel and conserved components of the retinoblastoma/E2F pathway in plants. J Biol Chem 2002; 277:9911-9; PMID:11786543; http://dx.doi.org/10.1074/jbc.M110616200
- Bracken AP, Ciro M, Cocito A, Helin K. E2F target genes: unraveling the biology. Trends Biochem Sci 2004; 29:409-17; PMID:15362224; http://dx.doi.org/10.1016/j.tibs.2004.06.006
- Dynlacht BD, Brook A, Dembski M, Yenush L, Dyson N. DNA-binding and transactivation properties of Drosophila E2f and Dp proteins. P Natl Acad Sci USA 1994; 91:6359-63; http://dx.doi.org/10.1073/pnas.91.14.6359
- Blais A, Dynlacht BD. Hitting their targets: an emerging picture of E2F and cell cycle control. Curr Opin Genet Dev 2004; 14:527-32; PMID:15380244; http://dx.doi.org/10.1016/j.gde.2004.07.003
- Lindeman GJ, Gaubatz S, Livingston DM, Ginsberg D. The subcellular localization of E2f-4 is cell-cycle dependent. P Natl Acad Sci USA 1997; 94:5095-100; http://dx.doi.org/10.1073/pnas.94.10.5095
- Muller H, Moroni MC, Vigo E, Petersen BO, Bartek J, Helin K. Induction of S-phase entry by E2F transcription factors depends on their nuclear localization. Mol Cell Biol 1997; 17:5508-20; PMID:9271426; http://dx.doi.org/10.1128/MCB.17.9.5508
- DeGregori J, Leone G, Miron A, Jakoi L, Nevins JR. Distinct roles for E2F proteins in cell growth control and apoptosis. P Natl Acad Sci USA 1997; 94:7245-50; http://dx.doi.org/10.1073/pnas.94.14.7245
- Cobrinik D. Regulatory interactions among E2Fs and cell cycle control proteins. Curr Top Microbiol Immunol 1996; 208:31-61; PMID:8575212
- Dyson N. The regulation of E2F by pRB-family proteins. Gene Dev 1998; 12:2245-62; PMID:9694791; http://dx.doi.org/10.1101/gad.12.15.2245
- Vandepoele K, Vlieghe K, Florquin K, Hennig L, Beemster GTS, Gruissem W, Van De Peer Y, Inze D, De Veylder L. Genome-wide identification of potential plant E2F target genes. Plant Physiol 2005; 139:316-28; PMID:16126853; http://dx.doi.org/10.1104/pp.105.066290
- Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, Dynlacht BD. E2F integrates cell cycle progression with DNA repair, replication, and G2/M checkpoints. Gene Dev 2002; 16:245-56; PMID:11799067; http://dx.doi.org/10.1101/gad.949802
- Rabinovich A, Jin VX, Rabinovich R, Xu XQ, Farnham PJ. E2F in vivo binding specificity: comparison of consensus versus nonconsensus binding sites. Genome Res 2008; 18:1763-77; PMID:18836037; http://dx.doi.org/10.1101/gr.080622.108
- Cao LH, Peng B, Yao L, Zhang XM, Sun KA, Yang XM, Yu L. The ancient function of RB-E2F pathway: insights from its evolutionary history. Biol Direct 2010; 5:55; PMID:20849664; http://dx.doi.org/10.1186/1745-6150-5-55
- Orias E, Cervantes MD, Hamilton EP. Tetrahymena thermophila, a unicellular eukaryote with separate germline and somatic genomes. Res Microbiol 2011; 162:578-86; PMID:21624459; http://dx.doi.org/10.1016/j.resmic.2011.05.001
- Mochizuki K, Novatchkova M, Loidl J. DNA double-strand breaks, but not crossovers, are required for the reorganization of meiotic nuclei in Tetrahymena. J Cell Sci 2008; 121:2148-58; PMID:18522989; http://dx.doi.org/10.1242/jcs.031799
- Loidl J, Lukaszewicz A, Howard-Till RA, Koestler T. The Tetrahymena meiotic chromosome bouquet is organized by centromeres and promotes interhomolog recombination. J Cell Sci 2012; 125:5873-80; PMID:22976299; http://dx.doi.org/10.1242/jcs.112664
- Song XY, Gjoneska E, Ren QH, Taverna SD, Allis CD, Gorovsky MA. Phosphorylation of the SQ H2A.X motif is required for proper meiosis and mitosis in Tetrahymena thermophila. Mol Cell Biol 2007; 27:2648-60.
- Howard-Till RA, Lukaszewicz A, Loidl J. The recombinases Rad51 and Dmc1 play distinct roles in DNA break repair and recombination partner choice in the meiosis of Tetrahymena. Plos Genet 2011; 7:e1001359; PMID:21483758; http://dx.doi.org/10.1371/journal.pgen.1001359
- Loidl J, Scherthan H. Organization and pairing of meiotic chromosomes in the ciliate Tetrahymena thermophila. J Cell Sci 2004; 117:5791-801; PMID:15522890; http://dx.doi.org/10.1242/jcs.01504
- Shodhan A, Lukaszewicz A, Novatchkova M, Loidl J. Msh4 and Msh5 function in SC-independent chiasma formation during the streamlined meiosis of Tetrahymena. Genetics 2014; 198:983-93; PMID:25217051; http://dx.doi.org/10.1534/genetics.114.169698
- Howard-Till RA, Lukaszewicz A, Novatchkova M, Loidl J. A single cohesin complex performs mitotic and meiotic functions in the protist Tetrahymena. Plos Genet 2013; 9:e1003418; PMID:23555314; http://dx.doi.org/10.1371/journal.pgen.1003418
- Miao W, Xiong J, Bowen J, Wang W, Liu YF, Braguinets O, Grigull J, Pearlman RE, Orias E, Gorovsky MA. Microarray analyses of gene expression during the Tetrahymena thermophila life cycle. Plos One 2009; 4:e4429; PMID:19204800; http://dx.doi.org/10.1371/journal.pone.0004429
- Sugai T, Hiwatashi K. Cytologic and autoradiographic studies of the micronucleus at meiotic prophase in Tetrahymena pyriformis. J Protozool 1974; 21:542-8; PMID:4214068; http://dx.doi.org/10.1111/j.1550-7408.1974.tb03695.x
- Morgan DO. The cell cycle: principles of control. London: New Science Press Ltd 2007.
- Polager S, Kalma Y, Berkovich E, Ginsberg D. E2Fs up-regulate expression of genes involved in DNA replication, DNA repair and mitosis. Oncogene 2002; 21:437-46; PMID:11821956; http://dx.doi.org/10.1038/sj.onc.1205102
- Loidl J, Lorenz A. DNA double-strand break formation and repair in Tetrahymena meiosis. Semin Cell Dev Biol 2016; 54:126-34; PMID:26899715; http://dx.doi.org/10.1016/j.semcdb.2016.02.021
- Drissi R, Dubois ML, Douziech M, Boisvert FM. Quantitative proteomics reveals dynamic interactions of the minichromosome maintenance complex (MCM) in the cellular response to etoposide induced DNA damage. Mol Cell Proteomics 2015; 14:2002-13; PMID:25963833; http://dx.doi.org/10.1074/mcp.M115.048991
- Shang YH, Li B, Gorovsky MA. Tetrahymena thermophila contains a conventional γ-tubulin that is differentially required for the maintenance of different microtubule-organizing centers. J Cell Biol 2002; 158:1195-206; PMID:12356864; http://dx.doi.org/10.1083/jcb.200205101
- Kushida Y, Takaine M, Nakano K, Sugai T, Numata O. Dynamic change of cellular localization of microtubule-organizing center during conjugation of ciliate Tetrahymena thermophila. Zool Sci 2015; 32:25-32; PMID:25660693; http://dx.doi.org/10.2108/zs140149
- Lesca C, Germanier M, Raynaud-Messina B, Pichereaux C, Etievant C, Emond S, Burlet-Schiltz O, Monsarrat B, Wright M, Defais M. DNA damage induce γ-tubulin-Rad51 nuclear complexes in mammalian cells. Oncogene 2005; 24:5165-72; PMID:15897881; http://dx.doi.org/10.1038/sj.onc.1208723
- Hoog G, Zarrizi R, von Stedingk K, Jonsson K, Alvarado-Kristensson M. Nuclear localization of γ-tubulin affects E2F transcriptional activity and S-phase progression. Faseb J 2011; 25:3815-27; PMID:21788450; http://dx.doi.org/10.1096/fj.11-187484
- Narbonne-Reveau K, Senger S, Pal M, Herr A, Richardson HE, Asano M, Deak P, Lilly MA. APC/CFzr/Cdh1 promotes cell cycle progression during the Drosophila endocycle. Development 2008; 135:1451-61; PMID:18321983; http://dx.doi.org/10.1242/dev.016295
- Lammens T, Boudolf V, Kheibarshekan L, Zalmas LP, Gaamouche T, Maes S, Vanstraelen M, Kondorosi E, La Thangue NB, Govaerts W, et al. Atypical E2F activity restrains APC/CCCS52A2 function obligatory for endocycle onset. P Natl Acad Sci USA 2008; 105:14721-6; http://dx.doi.org/10.1073/pnas.0806510105
- Watrin E, Peters JM. Cohesin and DNA damage repair. Exp Cell Res 2006; 312:2687-93; PMID:16876157; http://dx.doi.org/10.1016/j.yexcr.2006.06.024
- Sozzani R, Maggio C, Varotto S, Canova S, Bergounioux C, Albani D, Cella R. Interplay between Arabidopsis activating factors E2fb and E2fa in cell cycle progression and development. Plant Physiol 2006; 140:1355-66; PMID:16514015; http://dx.doi.org/10.1104/pp.106.077990
- DeGregori J. The genetics of the E2F family of transcription factors: shared functions and unique roles. Bba-Rev Cancer 2002; 1602:131-50.
- Royzman I, Whittaker AJ, OrrWeaver TL. Mutations in Drosophila DP and E2F distinguish G1-S progression from an associated transcriptional program. Gene Dev 1997; 11:1999-2011; PMID:9271122; http://dx.doi.org/10.1101/gad.11.15.1999
- del Pozo JC, Diaz-Trivino S, Cisneros N, Gutierrez C. The balance between cell division and endoreplication depends on E2Fc-DPb, transcription factors regulated by the ubiquitin-SCFSKP2A pathway in Arabidopsis. Plant Cell 2006; 18:2224-35; PMID:16920782; http://dx.doi.org/10.1105/tpc.105.039651
- Frolov MV, Huen DS, Stevaux O, Dimova D, Balczarek-Strang K, Elsdon M, Dyson NJ. Functional antagonism between E2F family members. Gene Dev 2001; 15:2146-60; PMID:11511545; http://dx.doi.org/10.1101/gad.903901
- Rempel RE, Saenz-Robles MT, Storms R, Morham S, Ishida S, Engel A, Jakoi L, Melhem MF, Pipas JM, Smith C, et al. Loss of E2F4 activity leads to abnormal development of multiple cellular lineages. Mol Cell 2000; 6:293-306; PMID:10983977; http://dx.doi.org/10.1016/S1097-2765(00)00030-7
- Lammens T, Li J, Leone G, De Veylder L. Atypical E2Fs: new players in the E2F transcription factor family. Trends Cell Biol 2009; 19:111-8; PMID:19201609; http://dx.doi.org/10.1016/j.tcb.2009.01.002
- Orias E, Hamilton EP, Orias JD. Tetrahymena as a laboratory organism: useful strains, cell culture, and cell line maintenance. Method Cell Biol 2000; 62:189-211; http://dx.doi.org/10.1016/S0091-679X(08)61530-7
- Mochizuki K. High efficiency transformation of Tetrahymena using a codon-optimized neomycin resistance gene. Gene 2008; 425:79-83; PMID:18775482; http://dx.doi.org/10.1016/j.gene.2008.08.007
- CassidyHanley D, Bowen J, Lee JH, Cole E, VerPlank LA, Gaertig J, Gorovsky MA, Bruns PJ. Germline and somatic transformation of mating Tetrahymena thermophila by particle bombardment. Genetics 1997; 146:135-47; PMID:9136007
- Lukaszewicz A, Howard-Till RA, Novatchkova M, Mochizuki K, Loidl J. Mre11 and Com1/Sae2 are required for double-strand break repair and efficient chromosome pairing during meiosis of the protist Tetrahymena. Chromosoma 2010; 119:505-18; PMID:20422424; http://dx.doi.org/10.1007/s00412-010-0274-9
- Howard-Till RA, Lukaszewicz A, Loidl J. The recombinases Rad51 and Dmc1 play distinct roles in DNA break repair and recombination partner choice in the meiosis of Tetrahymena. Plos Genet 2011; 7:e1001359; PMID:21483758; http://dx.doi.org/10.1371/journal.pgen.1001359
- Loidl J, Mochizuki K. Tetrahymena meiotic nuclear reorganization is induced by a checkpoint kinase-dependent response to DNA damage. Mol Biol Cell 2009; 20:2428-37; PMID:19297526; http://dx.doi.org/10.1091/mbc.E08-10-1058
- Gao S, Xiong J, Zhang CC, Berquist BR, Yang RD, Zhao M, Molascon AJ, Kwiatkowski SY, Yuan DX, Qin ZH, et al. Impaired replication elongation in Tetrahymena mutants deficient in histone H3 Lys 27 monomethylation. Gene Dev 2013; 27:1662-79; PMID:23884606; http://dx.doi.org/10.1101/gad.218966.113
- Xiong J, Lu XY, Lu YM, Zeng HH, Yuan DX, Feng LF, Chang Y, Bowen J, Gorovsky M, Fu CJ, et al. Tetrahymena Gene Expression Database (TGED): a resource of microarray data and co-expression analyses for Tetrahymena. Sci China Life Sci 2011; 54:65-7; PMID:21253873; http://dx.doi.org/10.1007/s11427-010-4114-1
- Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997; 25:3389-402; PMID:9254694; http://dx.doi.org/10.1093/nar/25.17.3389
- Ramirez-Parra E, Frundt C, Gutierrez C. A genome-wide identification of E2F-regulated genes in Arabidopsis. Plant J 2003; 33:801-11; PMID:12609051; http://dx.doi.org/10.1046/j.1365-313X.2003.01662.x
- Kirienko NV, Fay DS. Transcriptome profiling of the C. elegans Rb ortholog reveals diverse developmental roles. Dev Biol 2007; 305:674-84; PMID:17368442; http://dx.doi.org/10.1016/j.ydbio.2007.02.021
- Kehoe SM, Oka M, Hankowski KE, Reichert N, Garcia S, McCarrey JR, Gaubatz S, Terada N. A conserved E2F6-binding element in murine meiosis-specific gene promoters. Biol Reprod 2008; 79:921-30; PMID:18667754; http://dx.doi.org/10.1095/biolreprod.108.067645
- Lincker F, Messmer M, Houlne G, Devic M, Chaboute ME. E2F factors rate controls the dual role of CDE/E2F composite element: a model of E2F-regulated gene expression in plant development. Febs Lett 2006; 580:5167-71; PMID:16962586; http://dx.doi.org/10.1016/j.febslet.2006.08.067
- Korenjak M, Anderssen E, Ramaswamy S, Whetstine JR, Dyson NJ. RBF binding to both canonical E2F targets and noncanonical targets depends on functional dE2F/dDP complexes. Mol Cell Biol 2012; 32:4375-87; PMID:22927638; http://dx.doi.org/10.1128/MCB.00536-12
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and clustal X version 2.0. Bioinformatics 2007; 23:2947-8; PMID:17846036; http://dx.doi.org/10.1093/bioinformatics/btm404
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 2013; 30:2725-9; PMID:24132122; http://dx.doi.org/10.1093/molbev/mst197
- Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu SN, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, et al. CDD: NCBI's conserved domain database. Nucleic Acids Res 2015; 43:D222-6; PMID:25414356; http://dx.doi.org/10.1093/nar/gku1221
- Mar JC, Wells CA, Quackenbush J. Defining an informativeness metric for clustering gene expression data. Bioinformatics 2011; 27:1094-100; PMID:21330289; http://dx.doi.org/10.1093/bioinformatics/btr074
- Xiong J, Lu XY, Zhou ZM, Chang Y, Yuan DX, Tian M, Zhou ZG, Wang L, Fu CJ, Orias E, et al. Transcriptome analysis of the model protozoan, Tetrahymena thermophila, using deep RNA sequencing. Plos One 2012; 7:e30630; PMID:22347391; http://dx.doi.org/10.1371/journal.pone.0030630
- Wu ZP, Wang X, Zhang XG. Using non-uniform read distribution models to improve isoform expression inference in RNA-Seq. Bioinformatics 2011; 27:502-8; PMID:21169371; http://dx.doi.org/10.1093/bioinformatics/btq696
- Kim D, Salzberg SL. TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol 2011; 12:R72; PMID:21835007; http://dx.doi.org/10.1186/gb-2011-12-8-r72
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 2012; 7:562-78; PMID:22383036; http://dx.doi.org/10.1038/nprot.2012.016
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene Ontology: tool for the unification of biology. Nat Genet 2000; 25:25-9; PMID:10802651; http://dx.doi.org/10.1038/75556
- Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 2005; 21:3448-9; PMID:15972284; http://dx.doi.org/10.1093/bioinformatics/bti551
