ABSTRACT
The oocyte cytoplasmic lattices (CPLs) have long been predicted to function as a storage form for the maternal contribution of ribosomes to the early embryo. Our previous studies have demonstrated that ribosomal component S6 is stored in the oocyte CPLs and peptidylarginine deiminase 6 (PADI6) is critical for CPLs formation. Additionally, we found that depletion of PADI6 reduced de novo protein synthesis prior to the maternal-to-embryonic transition, therefore causing embryos to arrest at the 2-cell stage. Here, we present evidence further supporting the association of ribosomes with the CPLs by demonstrating that rRNAs are dramatically decreased in Padi6 KO oocytes. We also show that the abundance and localization of mRNAs is affected upon PADI6 depletion, suggesting that mRNAs are very possibly associated with CPLs. Consistent with this observation, the amount of the major RNA binding protein, MSY2, that is associated with the insoluble fraction of the oocytes after Triton X-100 extraction is also markedly decreased in the Padi6 KO oocytes. Furthermore, treatment of the oocytes with RNase A followed by Triton X-100 extraction severely impairs the localization of PADI6 and MSY2 in oocytes. These results indicate that mRNAs, possibly in a complex with MSY2 and PADI6, are bound in the CPLs and may play a role in securing the mRNA-MSY2 complex to the CPLs.
Introduction
During oocyte growth, many essential maternal factors are produced and stored for later use. Although the exact identity of most of these factors remains a mystery, they are nevertheless instrumental in preparing the oocyte for the egg to embryo transition.Citation1-5 The presence and correct storage of these maternal factors is crucial for embryonic genome activation (EGA) which needs to occur for proper embryonic development.Citation6,7 During this transition, mRNAs are released, translated, and the resulting proteins, along with other stored maternal factors, are targeted to the nucleus in a regulated fashion where they then orchestrate the nuclear reprogramming events leading to the EGA.Citation2,8-11
It has been reported that 75–80% of ribosomes in the mouse oocytes are not incorporated into polysomes at ovulation and do not engage in protein synthesis in vitro.Citation12,13 Interestingly, previous studies have suggested that these inactive ribosomes are embedded in the oocyte cytoplasmic lattices (CPLs), a highly abundant structure that has been well-described in the literature but remains poorly understood.Citation14 Electron microscopy studies have demonstrated that the lattices are absent from non-growing oocytes but increase in number throughout oocyte growth, eventually becoming the dominant feature of the fully grown oocyte.Citation15-18 Regarding the molecular composition of the lattices, older studies suggested that the lattices were largely comprised of intermediate filaments, however more recent studies suggest that the lattices are also associated with microtubules.Citation19 In addition to their cytoskeletal composition, older and more recent studies have suggested that one function of the lattices is to store a pool of ribosomes that is needed for early cleavage divisions. Perhaps most interestingly, the CPLs undergo extensive spatial reorganization at fertilization, compaction, and blastocyst formation.Citation20 This extensive reorganization suggests a role for the CPLs in embryonic reprogramming.
The peptidylarginine deiminase 6 (PADI6), is a member of PADI family and was originally identified from the murine egg proteome.Citation21 Padi6 knockout (KO) oocytes, while competent to mature and be fertilized, do not develop past the 2-cell stage, thus indicating that Padi6 represents a novel maternal effect gene.Citation22 Our previous studies have shown that PADI6 localizes to the CPLs and that the CPLs are absent from Padi6 KO oocytes/embryos,Citation21 therefore, it seems likely that the developmental defect for the Padi6 KO embryos is due to loss of the CPLs and that this defect may initiate in oocytes, when the CPLs begin to nucleate. Thus, the Padi6 KO oocytes provide a unique tool to study the composition and function of the CPLs. It was later found that transcriptional activity in Padi6 KO 2-cell embryos is severely compromised, suggesting that the mechanism of the Padi6 KO defect arises due to failure of embryonic transcription activation.Citation23 Given the essential nature of PADI6 in embryonic development with respect to the CPLs, unmasking the exact function of this protein and how it aids the assembly of the CPLs may provide important insight to human infertility issues.
Results
rRNA may constitute part of the CPLs
Analyzing the solubility of rRNA and mRNA in relation to the CPLs will allow a better understanding of how these RNAs are associated with these structures.Citation18,24 The rationale of studying solubility in the case of the CPLs is as follows: Triton X-100 extraction allows removal of soluble material from the oocyte cytoplasm, leaving behind insoluble structures such as the cytoskeleton and in particular the CPLs. Using the Padi6 KO oocytes will be crucial for this aspect. In the case of Padi6 KO oocytes, any factor that would normally be attached to or associated with the CPLs thus becomes soluble since the CPLs are absent in Padi6 KO oocytes. Therefore, comparison of Triton-extracted Padi6 WT and KO oocytes by subtracting what is identical in both WT and KO oocytes leaves behind the differences due solely to the presence or absence of the CPLs. This provides a unique tool to examine what happens to CPL-associated factors without their putative storage compartment.
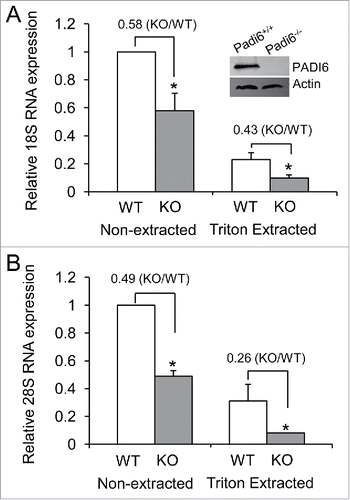
We first started working with PADI6 and the CPLs to investigate whether functional ribosomes are part of the CPLs. Our previous studies have showed that S6, a protein found in the small ribosomal subunit of ribosomes, was affected by the absence of the CPLs in Padi6 KO oocytes, suggesting that ribosomes are probably stored in the CPLs.Citation23 If ribosomes are indeed stored in the CPLs, then one would expect to find rRNAs there as well, being an integral part of functional ribosomes. We therefore decided to determine the fate of the rRNAs in Padi6 WT and KO oocytes. PADI6 knock out efficiency in the Padi6KO mouse oocytes was first confirmed by immuboblot (). We compared the relative quantities of 2 major rRNA components, the 18S and 28S rRNAs by quantitative real time PCR. Results showed a 42 ± 12.3% of 18S RNA (), and 51 ± 3.8% of 28S RNA () reduction (p < 0.05, respectively) in Padi6 KO oocytes when compare with Padi6 WT oocytes under the non-extracted condition. Triton X-100 extraction of the oocytes prior to qRT-PCR analysis showed that the levels were further reduced, with a 57 ± 4.1% of 18S RNA (), and 74 ± 11.6% of 28S RNA () reduction (p < 0.05, respectively) in Padi6 KO oocytes when compare with Padi6 WT oocytes under the Triton-extracted condition. These results indicate that part of the rRNAs is soluble, but the remainder is likely associated with the insoluble CPLs. Furthermore, the total loss of the 18S and 28S rRNAs is greater in both cases in the extracted samples, underlying the possibility that these rRNAs are indeed part of the CPLs. The localization of rRNAs to the CPLs, in addition to the fact that S6 ribosomal protein has already been demonstrated to be associated with the CPLs,Citation23 strongly suggests that functional ribosomes are indeed stored in the these structures.
Figure 1. rRNA quantities are lower and more soluble in Padi6 KO oocytes. (A) Quantitative real time PCR analysis of the 18S rRNA expression in Padi6 WT or KO GV stage oocytes before or after Triton X-100 extraction. The insert shows the depletion of PADI6 in Padi6 knock out oocytes by western blot. Actin was used as a protein loading control. (B) Quantitative real time PCR analysis of the 28S rRNA expression in Padi6 WT or KO GV stage oocytes before or after Triton X-100 extraction. The expression data were normalized to Gapdh. The graph represents the mean ± SD (n = 3). *p < 0.05.
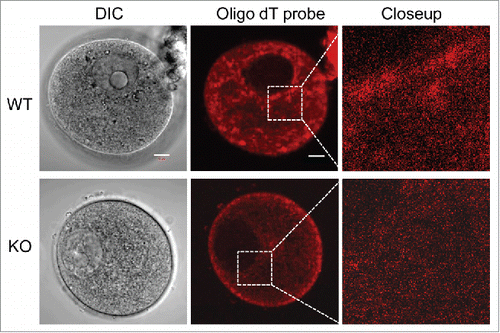
mRNA is also an integral part of the CPLs
Storing mRNAs during oocyte growth is an essential part of ensuring that the oocyte, once fertilized, will be capable of using these mRNAs as needed, thus allowing for the proper staging of development. Using pelleted insoluble oocytes that had been extracted with Triton X-100, our previous in vitro translation experiments have showed that the remaining mRNA in the insoluble fraction was competent for protein translation, and possibly in the CPLs.Citation23 To further confirm the possibility that mRNA is associated with the CPLs, we performed fluorescent in situ hybridization (FISH) experiments using an oligo dT probe that hybridizes with the poly A tail of mRNA, which reveals the localization of these mRNAs. As shown in , the cytoplasmic localization of the mRNAs in Padi6 WT oocytes can be readily detected. The mRNAs were found throughout the cytoplasm and granular concentrations were abundant around the perinuclear region as well as on the inside of the cytoplasmic membrane. However, this particular staining pattern was not observed in the Padi6 KO oocytes. Though the mRNA was diffusely distributed in the cytoplasm, the granular component was absent. These results suggest that these granules are likely to be associated with the CPLs. The absence of the granules in the Padi6 KO oocyte likely reflects the absence of the CPLs, thus suggesting that mRNAs are very possibly associated with these structures.
MSY2 localization is altered in the absence of PADI6 and the CPLs
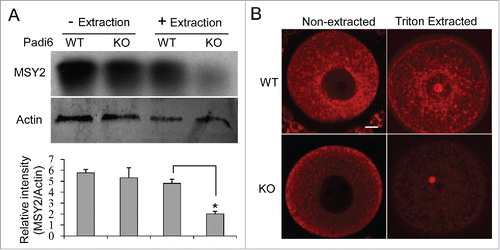
Two oocyte-restricted and potentially interrelated mechanisms are thought to regulate the storage of maternal mRNAs in oocytes. First, in fully grown oocytes, as opposed to somatic cells, the bulk of maternal mRNA partitions into the Triton X-100 insoluble cytoskeletal fraction, thus segregating the transcripts from the translational machinery.Citation24 Second, these maternal mRNAs are thought to be further stabilized by “RNA masking” which is likely mediated via interaction with MSY2, the major RNA binding protein in mammalian oocytes.Citation25,26 If mRNAs are possibly associated with the CPLs, then MSY2 is also likely to be related to the same structure. Interestingly, in oocytes, MSY2 has previously been shown to be Triton X-100 insoluble.Citation27 Given the fact that the CPLs are also insoluble in Triton X-100, it is possible that MSY2 plays a critical role in protecting maternal mRNAs from degradation in the oocyte by sequestering the transcripts on the CPLs. If this hypothesis is correct, then MSY2 should be found to partition in the Triton X-100 soluble fraction in Padi6 KO oocytes which lack CPLs. We therefore performed the experiments to test this hypothesis. Oocytes from Padi6 WT and KO mice were either extracted or not with Triton X-100, and the proteins from the insoluble fractions were analyzed by western blot (). Results showed that MSY2 levels were comparable between non-extracted Padi6 WT and KO oocytes. However, following extraction, there was a dramatic decrease in the amount of MSY2 that remains associated with the insoluble fraction in the Padi6 KO oocytes. Thus, our results further confirm that MSY2 may localize to CPLs.
Figure 3. Effect of PADI6 depletion on MSY2 localization in the insoluble fraction of the Padi6 KO oocytes. (A) Western blot showing that Triton X-100 extraction released MSY2 from the insoluble fraction in Padi6 KO oocytes, with actin as a loading control. Histograms represent normalized band densitometry readings averaged from 3 independent experiments. *p < 0.05. (B) Representative confocal images showing the mislocalization of MSY2 in Padi6 KO oocytes after triton extraction. Scale bar, 10 μm.
We also compared MSY2 localization in Padi6 WT and KO oocytes under both non-extracted and extracted conditions by immunofluorescence analysis (). Results showed that MSY2 staining reveals a fibrillar and granular structure that surrounds the nucleus and branches out to the cytoplasm in the Padi6 WT oocyte. However, in the absence of PADI6 and the CPLs, the staining pattern is much more diffuse with a loss of this structure. This mislocalization was even more evident once the oocytes have been extracted with Triton X-100, with a dramatic loss of the staining signals. Taken together, these results support the hypothesis that MSY2 is indeed associated with the CPLs. Also, it is possible that maternal mRNAs associate with the CPLs via MSY2 interaction, and therefore the Padi6 KO developmental defect is likely due to the premature release of stored mRNAs from the CPLs.
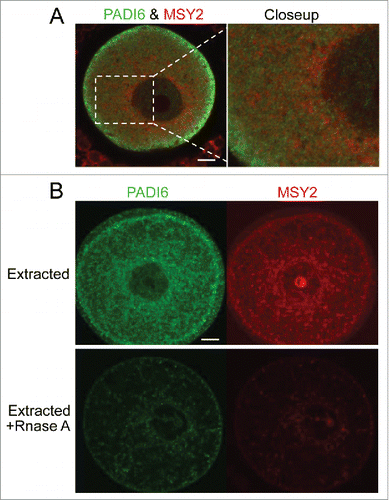
PADI6 and MSY2 proteins are retained in the CPLs
If MSY2 is associated with the CPLs, then it can be expected that MSY2 and PADI6 are found in close proximity to each other. To explore this possibility, Padi6 WT oocytes were stained with both PADI6 and MSY2 antibodies and analyzed via immunofluorescent confocal microscopy. The data showed that though PADI6 and MSY2 did not perfectly co-localize, they were found adjacent to one another (). While these images would tend to indicate that the 2 proteins are not found exactly in the same location, it does not however eliminate the possibility that MSY2 is still part of the CPLs, perhaps in a distinct subdomain of the CPLs compare with PADI6. This result suggests that the CPLs may be more variable in composition and structure than initially thought. When the localization of PADI6 and MSY2 was examined in Padi6 WT oocytes after Triton X-100 extraction, the relationship between them became clearer. In extracted oocytes, the staining of both proteins remained present, as demonstrated in . However, if the oocytes were treated with RNase A and followed by Triton X-100 extraction, the quantities of signals for both PADI6 and MSY2 were dramatically reduced, suggesting that digestion of the RNA element of the CPLs releases most of the PADI6 and MSY2 proteins, thus rendering them soluble.
Figure 4. PADI6 and MSY2 localize adjacent to each other in the normal oocytes and Triton extraction followed by RNase A treatment induces the loss of PADI6 and MSY2 from the oocytes. (A) Representative confocal images of PADI6 (green) and MSY2 (red) in the normal oocytes. (B) Representative confocal images of Padi6 WT oocytes stained with PADI6 (green) and MSY2 (red). All images parameters are the same in both conditions. Scale bar, 10 μm.
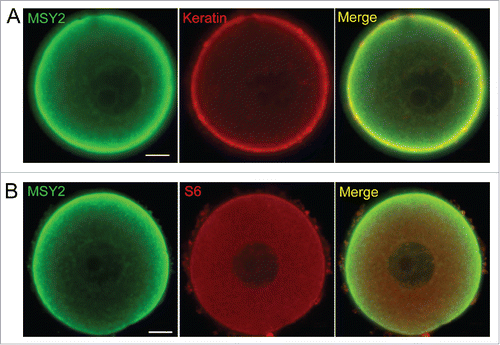
Given the facts that CPLs are largely comprised of intermediate filaments, and our recent study showing that one function of CPLs is to store a pool of ribosomes that is needed for early cleavage divisions,Citation23 we further utilized double immunofluoresence labeling procedures and confocal microscopy to test whether cytokeratin intermediate filaments or S6, a protein found in the small ribosomal subunit of ribosomes, are co-localized with MSY2 in oocytes. Results showed that both Keratin and S6 co-localized throughout much of the cytoplasm and the cortex of mouse oocytes, suggesting that MSY2 is associated with CPLs (). Taken together, these results suggest that both PADI6 and MSY2-mRNA complexes are retained in an insoluble structure, likely the CPLs.
Discussion
Accumulating evidence suggests that mRNAs encoding part of the maternal factors are actually sequestered into an insoluble cytoplasmic compartment during oocyte growth, but recent findings support the hypothesis that they may be stored in the CPLs which are found in the cytoplasm of oocytes.Citation12-14 Padi6 KO oocytes, while capable of correct maturation to the metaphase II stage and being fertilized, do not develop past the 2-cell stage and arrest in development at the time when the EGA should occur.Citation22 It is now known that the CPLs contain the PADI6 protein which has been shown to be essential for their formation.Citation21 Furthermore, the CPLs are absent from Padi6 KO oocytes, and therefore are likely to play a role in the developmental arrest.
Recent data have demonstrated that the S6 protein of ribosomes is also a part of the CPLs, suggesting that intact mature ribosomes are likely part of the CPLs.Citation23 Our results also demonstrated that mRNAs are very possibly associated with CPLs (). To further explore the molecular mechanisms, we showed that mRNAs, possibly in a complex with MSY2 and PADI6, are bound in the CPLs and may play a role in securing the mRNA-MSY2 complexes to the CPLs (). If the mRNAs and ribosomes necessary for the EGA are prematurely translated or degraded because of incorrect storage, this might explain the developmental arrest observed in Padi6 KO 2-cell staged embryos. Given PADI6s essential nature for development, mutations or misregulation of this protein may be responsible for female infertility. Actually, a recent report has just identified such a homozygous premature nonsense and compound-heterozygous PADI6 mutations in infertile individuals with early embryonic development arrest.Citation28 They found a lack of PADI6 in affected individuals' oocytes, and both the amount of phosphorylated RNA polymerase II and expression levels of several genes involved in embyonic genome activation were reduced in the affected individuals' embryos.Citation28
In summary, our results suggest that MSY2, mRNA and rRNA (probably in the form of functional ribosomes) are associated with the CPLs. If our hypothesis is correct, then this would provide the early embryo with an extremely powerful tool for regulating the timely translation of stored maternal mRNAs. Indeed, during the first stages of development following fertilization, the CPLs could be disassembled at the appropriate time allowing the translation of proteins necessary for the transition from maternally dependent transcription to maternally independent transcription in the embryo. Keeping the ribosomes and mRNAs stored within the same structure in an inactive form until they are needed provides a simple but robust mechanism for ensuring that both mRNA and ribosomes are released and available at the same time, eventually culminating in successful activation of the embryonic genome.
Materials and methods
All chemicals and reagents were obtained from Sigma unless otherwise stated. Animal care and experimental procedures were approved by the Animal Care and Use Committee of Nanjing Medical University. The study protocol was performed in accordance with institutional guidelines
Collection and preparation of oocytes
All germinal vesicle (GV) stage oocytes were collected from 3- to 5-week-old Padi6 wild type (WT) and Padi6 KO female mice. GV stage oocytes were isolated from ovarian follicles ∼46 hours after 10 international units (IU) of pregnant mare serum gonadotropin (PMSG) stimulation. The mice were then sacrificed and the ovaries were isolated and extensively punctured with 2 30 gauge needles. Immature oocytes displaying germinal vesicles were collected from ovaries in M2 medium.
Quantitative reverse transcription PCR (qRT-PCR)
Oocytes collected from female Padi6 WT and KO mice were either extracted with 0.5% Triton X-100 for 30 minutes or untreated serving as a control. Total RNA from both groups of oocytes were extracted using Trizol and chloroform. The RNA contained within the aqueous fraction was then purified using RNeasy Mini Kit (Qiagen), reverse transcribed, and subjected to real-time PCR using sequence-specific 18S rRNA and 28S rRNA primers for TaqMan Gene Expression Assays (Applied Biosystems). PCR was performed using the TaqMan PCR Master Mix using the following parameters: 50°C for 2 minutes and 95°C for 10 minutes, followed by 40 cycles at 95°C for 15 seconds and 60°C for 1 minute. 18S and 28S rRNA expression levels were normalized to Gapdh.
Fluorescent in situ hybridization (FISH)
Oocytes were collected from female Padi6 WT and KO mice. They were fixed for 1 hour with 4% paraformaldehyde in Dulbecco's PBS (DPBS) followed by permeabilization with 0.5% Triton X-100 in PBS for 30 minutes. All of the oocytes were incubated in hybridization buffer containing 50% formamide, 10% dextran sulfate, 2X SSC, 35ug/ml tRNA, 2mM Ribonucleoside Vanadyl Complex and biotinylated oligo dT probes (Promega) as a marker of mRNA for 24 hours at 37°C. Following overnight hybridization, the oocytes were washed in 2X SSC/50% formamide twice for 30 minutes and finally in 1X PBS. For single color detection, the slides were incubated with fluorescein isothiocyanate (FTTC)-labeled avidin for 15 min at 37°C. After the final washing step, the oocytes were mounted in Slowfade Gold antifade reagent (Molecular Probes, Eugene, OR) and examined for the presence of positive fluorescent signals by confocal laser scanning microscope (LSM 700, Zeiss).
Western blotting
For western blotting, Padi6 WT and Padi6 KO oocytes at the GV stage were isolated, boiled for 10 minutes in Laemmli buffer, and directly loaded onto a 10% SDS-PAGE gel. Proteins were separated at 150 V for 60 minutes and then transferred to a nitrocellulose membrane by applying a current of 90 V for 120 minutes. All blots were blocked with 5% nonfat dry milk in TBS containing 0.5% Tween-20 (TBS-T), washed and incubated with anti-MSY2 (ab105336, Abcam) or anti-Actin (ab8227, Abcam) in blocking buffer overnight at 4°C. The blots were then washed 3 times for 10 minutes in TBS-T and incubated with peroxidase-conjugated goat anti-rabbit IgG secondary antibody (Jackson ImmunoResearch) for 1 hour. Following incubation in secondary antibody, the membranes were washed 3 times for 10 minutes in TBS-T, and Immobilon Western HRP Chemiluminescent Substrate (Millipore, Temecula, CA) was applied for 5 minutes and developed. ImageJ software was used to determine the band intensity values.
Scanning confocal microscopy
Oocytes were either immediately fixed in 4% paraformaldehyde for 1 hour at room temperature or incubated first for 10 minutes in extraction buffer containing 0.1 M KCl, 20 mM MgCl2, 3 mM EGTA, 20 mM HEPES (pH 6.8), 0.1% Triton X-100, Complete Protease Inhibitor Cocktail (Roche, Mannheim, Germany), then rinsed quickly 3 times in PBS. The oocytes were then washed 5 times in 1% BSA, permeabilized with 0.5% Triton X-100 in PBS for 30 minutes, and incubated with guinea pig anti-PADI6 (1:500),Citation21 anti-MSY2 (ab105336, Abcam), anti-S6 (#2217, CST), or anti-AE1/AE3 (MAB3412, Chemicon) antibody in 1% BSA overnight at 4°C. Oocytes were washed again and then incubated for 2 hours at room temperature with appropriate secondary antibodies. Oocytes were mounted on slides in Slowfade Gold antifade reagent (Molecular Probes, Eugene, OR), and imaged using confocal laser scanning microscope (LSM 700, Zeiss).
RNase A treatment
To check whether RNA integrity was required for the retention of MSY2 following Triton permeabilization, oocytes were incubated with 50 μg/ml RNaseA for 30 min at 37°C following permeabilization with 0.1% Triton X-100 as described above.
Statistical analysis
All experiments used for quantification were repeated at least 3 times. Values are given as mean ± STD and were analyzed using student's t-test. Differences of p < 0.05 were defined as significant.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the National Natural Science Foundation of China (81372850), the Key University Natural Science Research Project of Jiangsu Province (15KJA320003), and Jiangsu Entrepreneurship & Innovation Award. Coonrod lab support was provided by NIH grant NICHD-38353.
References
- Kim KH, Lee KA. Maternal effect genes: Findings and effects on mouse embryo development. Clin Exp Reprod Med 2014; 41(2):47-61; PMID:25045628; http://dx.doi.org/10.5653/cerm.2014.41.2.47
- Li L, Lu X, Dean J. The maternal to zygotic transition in mammals. Mol Aspects Med 2013; 34(5):919-38; PMID:23352575; http://dx.doi.org/10.1016/j.mam.2013.01.003
- De Leon V, Johnson A, Bachvarova R. Half-lives and relative amounts of stored and polysomal ribosomes and poly(A) + RNA in mouse oocytes. Dev Biol 1983; 98(2):400-8; PMID:6683687; http://dx.doi.org/10.1016/0012-1606(83)90369-X
- Mermillod P, Dalbiès-Tran R, Uzbekova S, Thélie A, Traverso JM, Perreau C, Papillier P, Monget P. Factors affecting oocyte quality: who is driving the follicle? Reprod Domest Anim 2008; 43 Suppl 2:393-400; http://dx.doi.org/10.1111/j.1439-0531.2008.01190.x
- Bettegowda A, Smith GW. Mechanisms of maternal mRNA regulation: implications for mammalian early embryonic development. Front Biosci 2007; 12:3713-26; PMID:17485333; http://dx.doi.org/10.2741/2346
- Bianchi E, Sette C. Post-transcriptional control of gene expression in mouse early embryo development: a view from the tip of the iceberg. Genes (Basel) 2011; 2(2):345-59; PMID:24710195; http://dx.doi.org/10.3390/genes2020345
- Lim CY, Knowles BB, Solter D, Messerschmidt DM. Epigenetic Control of Early Mouse Development. Curr Top Dev Biol 2016; 120:311-60; PMID:27475856; http://dx.doi.org/10.1016/bs.ctdb.2016.05.002
- Oh B, Hwang S, McLaughlin J, Solter D, Knowles BB. Timely translation during the mouse oocyte-to-embryo transition. Development 2000; 127(17):3795-803; PMID:10934024
- Conti M. Phosphodiesterases and regulation of female reproductive function. Curr Opin Pharmacol 2011; 11(6):665-9; PMID:22019564; http://dx.doi.org/10.1016/j.coph.2011.10.004
- Downs SM. Regulation of the G2/M transition in rodent oocytes. Mol Reprod Dev 2010; 77(7):566-85; PMID:20578061; http://dx.doi.org/10.1002/mrd.21175
- Yu XJ, Yi Z, Gao Z, Qin D, Zhai Y, Chen X, Ou-Yang Y, Wang ZB, Zheng P, Zhu MS, et al. The subcortical maternal complex controls symmetric division of mouse zygotes by regulating F-actin dynamics. Nat Commun 2014; 5:4887; PMID:25208553; http://dx.doi.org/10.1038/ncomms5887
- Bachvarova R, De Leon V. Stored and polysomal ribosomes of mouse ova. Dev Biol 1977; 58(2):248-54; PMID:560325; http://dx.doi.org/10.1016/0012-1606(77)90090-2
- Gallicano GI, McGaughey RW, Capco DG. Cytoskeletal sheets appear as universal components of mammalian eggs. J Exp Zool 1992; 263(2):194-203; PMID:1500884; http://dx.doi.org/10.1002/jez.1402630209
- Sternlicht AL, Schultz RM. Biochemical studies of mammalian oogenesis: kinetics of accumulation of total and poly(A)-containing RNA during growth of the mouse oocyte. J Exp Zool 1981; 215(2):191-200; PMID:6168731; http://dx.doi.org/10.1002/jez.1402150209
- Capco DG, Gallicano GI, McGaughey RW, Downing KH, Larabell CA. Cytoskeletal sheets of mammalian eggs and embryos: a lattice-like network of intermediate filaments. Cell Motil Cytoskeleton 1993; 24(2):85-99; PMID:8440027; http://dx.doi.org/10.1002/cm.970240202
- Gallicano GI, Larabell CA, McGaughey RW, Capco DG. Novel cytoskeletal elements in mammalian eggs are composed of a unique arrangement of intermediate filaments. Mech Dev 1994; 45(3):211-26; PMID:8011554; http://dx.doi.org/10.1016/0925-4773(94)90009-4
- García RB, Pereyra-Alfonso S, Sotelo JR. Protein-synthesizing machinery in the growing oocyte of the cyclic mouse. A quantitative electron microscopic study. Differentiation 1979; 14(1-2):101-6; PMID:573223
- Bachvarova R, De Leon V, Spiegelman I. Mouse egg ribosomes: evidence for storage in lattices. J Embryol Exp Morphol 1981; 62:153-64; PMID:7196940
- Kan R, Yurttas P, Kim B, Jin M, Wo L, Lee B, Gosden R, Coonrod SA. Regulation of mouse oocyte microtubule and organelle dynamics by PADI6 and the cytoplasmic lattices. Dev Biol 2011; 350(2):311-22; PMID:21147087; http://dx.doi.org/10.1016/j.ydbio.2010.11.033
- Gallicano GI, Capco DG. Remodeling of the specialized intermediate filament network in mammalian eggs and embryos during development: regulation by protein kinase C and protein kinase M. Curr Top Dev Biol 1995; 31:277-320; PMID:8746668; http://dx.doi.org/10.1016/S0070-2153(08)60231-8
- Wright PW, Bolling LC, Calvert ME, Sarmento OF, Berkeley EV, Shea MC, Hao Z, Jayes FC, Bush LA, Shetty J, et al. ePAD, an oocyte and early embryo-abundant peptidylarginine deiminase-like protein that localizes to egg cytoplasmic sheets. Dev Biol 2003; 256(1):73-88; PMID:12654293; http://dx.doi.org/10.1016/S0012-1606(02)00126-4
- Esposito G, Vitale AM, Leijten FP, Strik AM, Koonen-Reemst AM, Yurttas P, Robben TJ, Coonrod S, Gossen JA. Peptidylarginine deiminase (PAD) 6 is essential for oocyte cytoskeletal sheet formation and female fertility. Mol Cell Endocrinol 2007; 273(1-2):25-31; PMID:17587491; http://dx.doi.org/10.1016/j.mce.2007.05.005
- Yurttas P, Vitale AM, Fitzhenry RJ, Cohen-Gould L, Wu W, Gossen JA, Coonrod SA. Role for PADI6 and the cytoplasmic lattices in ribosomal storage in oocytes and translational control in the early mouse embryo. Development 2008; 135(15):2627-36; PMID:18599511; http://dx.doi.org/10.1242/dev.016329
- Brower PT, Schultz RM. Biochemical studies of mammalian oogenesis: possible existence of a ribosomal and poly(A)-containing RNA-protein supramolecular complex in mouse oocytes. J Exp Zool 1982; 220(2):251-60; PMID:6176672; http://dx.doi.org/10.1002/jez.1402200214
- Yu J, Hecht NB, Schultz RM. Expression of MSY2 in mouse oocytes and preimplantation embryos. Biol Reprod 2001; 65(4):1260-70; PMID:11566752; http://dx.doi.org/10.1095/biolreprod65.4.1260
- Yu J, Hecht NB, Schultz RM. RNA-binding properties and translation repression in vitro by germ cell-specific MSY2 protein. Biol Reprod 2002; 67(4):1093-8; PMID:12297523; http://dx.doi.org/10.1095/biolreprod67.4.1093
- Yu J, Hecht NB, Schultz RM. Requirement for RNA-binding activity of MSY2 for cytoplasmic localization and retention in mouse oocytes. Dev Biol 2003; 255(2):249-62; PMID:12648488; http://dx.doi.org/10.1016/S0012-1606(02)00094-5
- Xu Y, Shi Y, Fu J, Yu M, Feng R, Sang Q, Liang B, Chen B, Qu R, Li B, et al. Mutations in PADI6 cause female infertility characterized by early embryonic arrest. Am J Hum Genet 2016; 99(3):744-52; PMID:27545678; http://dx.doi.org/10.1016/j.ajhg.2016.06.024