ABSTRACT
Recent loss-of-function studies in tissue-specific as well as global Tspo (Translocator Protein 18 kDa) knockout mice have not confirmed its long assumed indispensability for the translocation of cholesterol across the mitochondrial inter-membrane space, a rate-limiting step in steroid biosynthesis. Instead, recent studies in global Tspo knockout mice indicate that TSPO may play a more fundamental role in cellular bioenergetics, which may include the indirect down-stream regulation of transport or metabolic functions. To examine whether overexpression of the TSPO protein alters the cellular bioenergetic profile, Jurkat cells with low to absent endogenous expression were transfected with a TSPO construct to create a stable cell line with de novo expression of exogenous TSPO protein. Expression of TSPO was confirmed by RT-qPCR, radioligand binding with [3H]PK11195 and immunocytochemistry with a TSPO antibody. We demonstrate that TSPO gene insertion causes increased transcription of genes involved in the mitochondrial electron transport chain. Furthermore, TSPO insertion increased mitochondrial ATP production as well as cell excitability, reflected in a decrease in patch clamp recorded rectified K channel currents. These functional changes were accompanied by an increase in cell proliferation and motility, which were inhibited by PK11195, a selective ligand for TSPO. We suggest that TSPO may serve a range of functions that can be viewed as downstream regulatory effects of its primary, evolutionary conserved role in cell metabolism and energy production.
Introduction
The translocator protein 18 kDa (TSPO) or peripheral ben-zodiazepine receptor (PBR) is a highly conserved,Citation1-3 multi-functional,Citation4 mitochondrial, 5 transmembrane-domain protein.Citation5-8 It is thought to interact with the voltage-dependent anion channel (VDAC; 32 kDa),Citation4,9-11 the adenine nucleotide transporter (ANT; 30 kDa).Citation4,9-11 and the mitochondrial permeability transition pore (mPTP).Citation4,9-11 The protein structure of the bacterial homolog RsTspOCitation8 and Bacillus cereus (BcTspO)Citation6 has been crystalized. Based on in vivo and in vitro observations, the ‘translocation’ of cholesterol across the mitochondrial inter-membrane space has been identified as a rate-determining step in steroid biosynthesis and as the most prominent, and essential for life, function of the TSPO,Citation4,12 thus providing the rationale for its renaming as the Translocator Protein 18 kDa (TSPO). The conceptual history of the ‘translocator protein’ and scientific publication trends (as reviewed byCitation13) illustrate the emerging role of this protein as a diagnostic biomarker of active disease in the nervous system, and a potential therapeutic target for a broad range of inflammatory, neurodegenerative, neoplastic, metabolic and behavioral diseases.Citation13
Recent observations in mice with global and conditional deletions of Tspo with apparently normal phenotypes in independent laboratoriesCitation14-17 unexpectedly failed to confirm an essential role of TSPO/PBR in cholesterol import into mitochondria and steroid synthesis. Additionally, the role of TSPO in the regulation of mPTP also failed to be confirmed.Citation16 Our observations over 24 months in 700 animals of the GuwiyangWurra (‘Fire Mouse’) strain of Tspo knockout mice revealed no differences in growth rate, fertility, cholesterol transport and steroid biosynthesis, or blood levels of the endogenous TSPO ligand, protoporphyrin IX (PPIX) compared with littermate wild-type animals. However, a decreased level of ATP production by mitochondria in microglia extracted from the knockout animals indicates the potential existence of a latent phenotype that may come to the fore under disease rather than normal physiological conditions.Citation14 As a complement to the loss-of-function phenotypic data in mitochondrial energy production from the GuwiyangWurra Tspo−/− animals,Citation13,14,18,19 we re-examined the role of the TSPO/PBR in energy production and cell metabolism in an in vitro model.
To this end, we stably transfected a T-cell line, Jurkat cells, with the human TSPO gene. Jurkat cells have low or absent expression of TSPO due to a high degree of promoter methylation as revealed in our previous study.Citation20 Confirming the stable expression of the inserted exogenous human TSPO gene by PCR, RT-qPCR, membrane receptor binding with TSPO-specific ligand [3H]PK11195, and immunohistochemistry with a TSPO antibody, we describe significant gain-of-function effects in the mitochondrial electron transport chain, cell membrane excitability, as well as marked changes in the highly energy-demanding functions of cell proliferation and motility.
Results
Confirmation of stable TSPO transfection
Jurkat cell lines, derived from human leukemia cells, have previously been reported to have very low or absent TSPO expression.Citation21 Wild-type Jurkat cells used in this study were 82% identical to Jurkat Clone E6–1 (ATCC: TIB-152), determined using short tandem repeat (STR) profiling (Table S1). The promoter of TSPO in our Jurkat cell line is highly methylated in the region surrounding the transcription start site,Citation20 resulting in the expression of TSPO mRNA and TSPO protein at barely detectable levels in the wild-type Jurkat cells ().
Figure 1. Confirmation of TSPO expression in Jurkat cells after stable transfection of TSPO expressing plasmids. (A) Relative abundance of exogenous TSPO mRNA expression in genetically modified Jurkat cell lines. Bar graph shows exogenous TSPO mRNA levels normalized to GAPDH and β-actin mRNAs. Expression of endogenous TSPO mRNA in MDA-MB-231 cells was set to 1.0. Jurkat cells transfected with TSPO expressed a comparable level of TSPO mRNA (exogenous) with that of the MDA-MB-231 cells (endogenous). No exogenous TSPO mRNA could be detected in wild-type and empty plasmid Jurkat cells. The data are presented as means ± SD (n = 4). (B) The level of TSPO protein expression in Jurkat cells was measured by radioligand binding using the TSPO specific ligand [3H]PK11195. The absence of specific binding in the wild-type and empty plasmid Jurkat cells confirms the negiligible level of TSPO protein. TSPO-Jurkat cells exhibited specific [3H]PK11195 binding though its Bmax is about one-third of that obtained from the positive control MDA-MB-231 cells. (C) Endogenous and exogenous expression of TSPO in Jurkat cells was determined by immunostaining with a TSPO specific antibody (green). There was no positive immunostaining observed in both wild-type and empty plasmid transfected Jurkat cells. However, strong staining with localized intracellular distribution along with mitochondria (red) was observed in TSPO-Jurkat cells. The blue color indicates the staining of nuclei. The yellow fluorescence in the right image demonstrates the overlap between TSPO and mitochondria. Scale bars: 10 μm.
![Figure 1. Confirmation of TSPO expression in Jurkat cells after stable transfection of TSPO expressing plasmids. (A) Relative abundance of exogenous TSPO mRNA expression in genetically modified Jurkat cell lines. Bar graph shows exogenous TSPO mRNA levels normalized to GAPDH and β-actin mRNAs. Expression of endogenous TSPO mRNA in MDA-MB-231 cells was set to 1.0. Jurkat cells transfected with TSPO expressed a comparable level of TSPO mRNA (exogenous) with that of the MDA-MB-231 cells (endogenous). No exogenous TSPO mRNA could be detected in wild-type and empty plasmid Jurkat cells. The data are presented as means ± SD (n = 4). (B) The level of TSPO protein expression in Jurkat cells was measured by radioligand binding using the TSPO specific ligand [3H]PK11195. The absence of specific binding in the wild-type and empty plasmid Jurkat cells confirms the negiligible level of TSPO protein. TSPO-Jurkat cells exhibited specific [3H]PK11195 binding though its Bmax is about one-third of that obtained from the positive control MDA-MB-231 cells. (C) Endogenous and exogenous expression of TSPO in Jurkat cells was determined by immunostaining with a TSPO specific antibody (green). There was no positive immunostaining observed in both wild-type and empty plasmid transfected Jurkat cells. However, strong staining with localized intracellular distribution along with mitochondria (red) was observed in TSPO-Jurkat cells. The blue color indicates the staining of nuclei. The yellow fluorescence in the right image demonstrates the overlap between TSPO and mitochondria. Scale bars: 10 μm.](/cms/asset/7c4cd746-8b95-4ed6-b842-5eaba8bd1353/kccy_a_1281477_f0001_oc.gif)
Stable TSPO transfection of Jurkat cells (TSPO-Jurkat cells) was initially confirmed by RT-qPCR using primers specifically designed for exogenous TSPO mRNA. Since the exogenous TSPO mRNA was codon-optimised, primers could be designed to allow discrimination between exogenous and endogenous TSPO variants (Table S2). Insertion of TSPO into wild-type Jurkat cells and selection to create a stable cell line did not change the Jurkat cell profile that was still 82% identical to Jurkat, Clone E6–1 (ATCC: TIB-152) (Table S3). As a positive control, the level of TSPO mRNA () was compared with the MDA-MB-231 cell line, a breast cancer cell line with high TSPO mRNA and protein expression.Citation22,23
The level of TSPO protein in wild-type, empty plasmid-transfected and exogenous TSPO-transfected Jurkat cells, and positive control MDA-MB-231 cells was measured by membrane receptor binding using the TSPO ligand [3H]PK11195.Citation24 No difference between total and non-specific binding of [3H]PK11195 was found in the wild-type Jurkat cells, confirming the barely detectable level of endogenous TSPO protein in these cells (). This was also the case for Jurkat cells transfected with the empty plasmid (). In contrast, Jurkat cells transfected with TSPO showed specific [3H]PK11195 binding (). Scatchard analysis revealed a maximal number of binding sites (Bmax) of 4.8 ± 0.3 pmol/mg protein with a dissociation constant (KD) of 17.8 ± 2.2 nM in the TSPO-Jurkat cells, which is approximately a third of the expression levels seen in the highly expressing tumor cell line MDA-MB-231 (Bmax of 15.4 ± 0.8 pmol/mg protein and a Kd of 8.9 ± 1.1 nM), similar to previous reports.Citation23,25 Scatchard analysis of wild-type and empty plasmid-containing Jurkat cells was not possible due to the absence of specific binding.
Expression of TSPO in the wild-type, TSPO-transfected, and empty plasmid-transfected Jurkat cells was examined by immunostaining using a TSPO specific antibody.Citation14,17 Undetectable to faint non-localized background staining was observed in wild-type () and empty plasmid Jurkat cells (). In contrast, staining was clearly visible in the TSPO-Jurkat cells (). The intracellular distribution of TSPO expression closely matched that of mitochondria, indicating that the exogenous TSPO co-localized with mitochondria in the transfected Jurkat cells (). The morphology of mitochondria in the wild-type and empty plasmid Jurkat cells did not show a discernible difference compared with TSPO-Jurkat cells (). Thus, TSPO overexpression in Jurkat cells did not appear to induce mitochondrial stress, which might cause non-specific differences from the control cells.
Genes involved in the mitochondrial electron transport chain are upregulated following TSPO transfection
Gene set enrichment analysis (GSEA) and Enrichment Map were used to identify functional signatures within the expression data. As shown in , TSPO transfection resulted in an enrichment of clusters related to translation, transcription, protein folding, nucleotide excision repair, the electron transport chain and the proteasome. The role of TSPO in energy production and cell metabolism, as indicated by the upregulation of genes associated with these functions, was subsequently examined.
Figure 2. Changes in mRNA abundance following TSPO transfection. (A) Microarray analysis was used to detect differences in gene expression between the 3 Jurkat cell types. GSEA results comparing empty plasmid and TSPO-Jurkat cells were visualised using Enrichment Map Cytoscape Plug-in. Gene sets are represented by circular nodes that are proportional to the number of genes present in the gene set. The thickness of the lines between nodes is proportional to the overlap between gene sets. Nodes with highly similar gene sets are placed close to each other to form clusters. Red nodes are upregulated and blue nodes are downregulated. Expanded clusters in high resolution are shown in Figs. S1–S6. (B) Heat map of the leading edge genes from the mitochondrial electron transport chain cluster. The top 60 genes are shown with high expression represented in red. (C) Gene expression of mitochondrial respiratory chain complex IV (COX IV, subunit 2) in the TSPO-Jurkat cells was decreased by the TSPO ligand PK11195 (100 nM) when compared with control (without PK11195 but containing 0.1% DMSO). The same concentration of PK11195 had no effect on the wild-type and empty plasmid Jurkat cells.
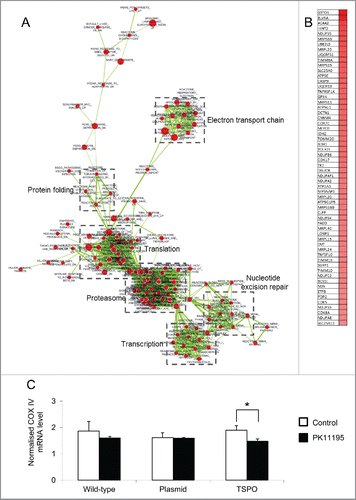
To further examine changes in the mitochondrial respiratory chain following TSPO gene insertion, RT-qPCR was used to detect mRNA of complex IV of the mitochondrial respiratory chain (cytochrome c oxidase). The mRNA level of the COX IV subunit 2 in TSPO overexpressed Jurkat cells was higher, but not significantly, than those of the wild-type and empty plasmid Jurkat cells (). The mRNA level of the COX IV subunit 2 was significant decreased by the TSPO ligand PK11195 at 100 nM (p < 0.05, n = 4), while the mRNA levels of the COX IV subunit 2 in wild-type and empty plasmid Jurkat cells were not significantly altered by PK11195 at the same concentration ().
Increased mitochondrial ATP production in Jurkat cells with TSPO transfection
The role of TSPO in ATP production was examined using permeabilised Jurkat cells with intact mitochondria. Ap5A (25 μM) was used to inhibit adenylate kinase which prevents the inter-conversion of ATP to ADP and AMP. Iodoacetate (25 μM) was used to inhibit glyceraldyde-3-phosphate dehydrogenase to block glycolysis-induced ATP production. Bioluminescent luciferin-luciferase was then used to detect ATP by generating photons correlating with the number of ATP molecules present.
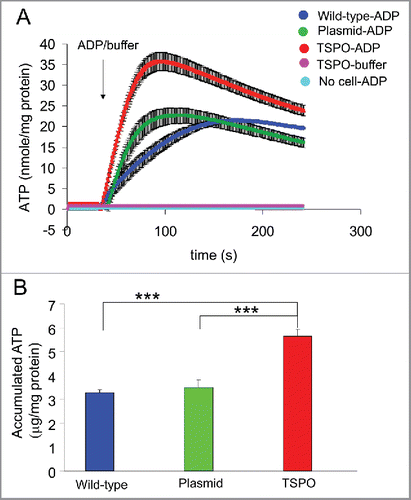
ATP production increased markedly after the addition of 2 mM ADP () in all 3 cell types (wild-type, empty plasmid and TSPO-Jurkat cells). However, the ATP production in TSPO-Jurkat cells rose significantly higher (accumulated ATP: 5.6 ± 0.28 μg/mg protein, n = 6 wells from 3 batches of cultures) than in wild-type (3.3 ± 0.14 μg/mg protein, n = 3 wells, p < 0.001) and empty plasmid Jurkat cells (3.5 ± 0.30 μg/mg protein, n = 6 wells, p < 0.001) (). No apparent ATP production was observed in either TSPO transfected, wild-type or empty plasmid Jurkat cells after the addition of the buffer solution used for dissolving ADP and luciferin-luciferase, or in cell-free luciferin-luciferase solution after addition of ADP (). Moreover, there was no observable bioluminescence in the Jurkat cells in the absence of luciferin-luciferase, nor any observable ATP contamination in the pure ADP preparation. The results suggest that TSPO transfection increased mitochondrial ATP production.
Figure 3. TSPO increased mitochondrial ATP production. (A) Averaged time-course of ATP production from permeabilised Jurkat cells was measured with luciferin-luciferase and a microplate reader. ATP production after injection of 2 mM ADP was markedly increased in all 3 cell types (wild-type, empty plasmid and TSPO-Jurkat cells). ATP production in TSPO-Jurkat cells increased at a greater rate than that in wild-type and empty plasmid Jurkat cells. (B) Accumulated ATP following injection of ADP in TSPO-Jurkat cells, calculated by subtracting the baseline (before ADP injection), was significantly greater than that observed in wild-type and empty plasmid Jurkat cells.
Increased cell proliferation in TSPO transfected Jurkat cells
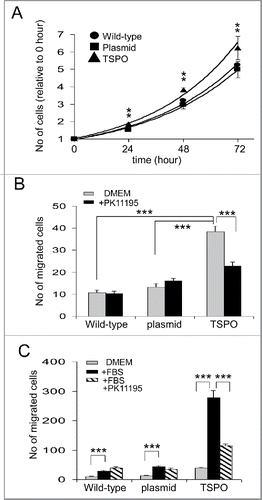
Observed over 3 days, all 3 cell types proliferated exponentially (R2 = 0.986, y = 0.9342e0.282x for wild-type; R2 = 0.987, y = 0.955e0.253x for empty plasmid Jurkat cells and R2 = 0.993, y = 0.9637e0.282x for TSPO-Jurkat cells). However, the TSPO-Jurkat cells proliferated significantly faster than wild-type and empty plasmid Jurkat cells across all measurements (i.e. 24, 48 and 72 hours, p < 0.01) (). There was no difference in proliferation rate between wild-type and empty plasmid Jurkat cells ().
Figure 4. TSPO increased cell proliferation and motility. (A) Time course of Jurkat cell proliferation normalized to day 0, i.e., 1 hour after plating each of the cell types. TSPO-Jurkat cells proliferated significantly faster than wild-type and empty plasmid Jurkat cells at all time points measured (i.e. days 1–3). There was no difference in proliferation rate between wild-type and empty plasmid Jurkat cells. (B) TSPO-Jurkat cells had significantly higher spontaneous movement/migration compared with wild-type and empty plasmid Jurkat cells measured with the Boyden chamber (Transwell). The inhibitory effect of TSPO specific ligand PK11195 on spontaneous movement/migration was only seen in the TSPO-Jurkat cells, indicating the effect is TSPO-specific. (C) Foetal bovine serum (FBS, 0.1%) in DMEM, a chemo-attractant, non-specifically attracted all 3 types of Jurkat cells. However, PK11195 at 100 nM significantly decreased FBS-induced cell migration in TSPO-Jurkat cells only.
Increased cell motility in TSPO transfected Jurkat cells
The motility of TSPO-Jurkat cells was examined by measuring spontaneous activity and induced cell migration through Transwell (Boyden) chambers. TSPO-Jurkat cells (38 ± 2.6 cells, n = 13 Transwells) had significantly higher spontaneous movement/migration than wild-type (10 ± 1.2 cells, n = 11 Transwells, p < 0.001) and empty plasmid Jurkat cells (13 ± 2.5 cells, n = 11, p < 0.001) (), as determined by the number of cells passing through the Transwell chamber. There was no significant difference in spontaneous movement/migration between wild-type and empty plasmid Jurkat cells (). The spontaneous activity of TSPO-Jurkat cells, but not wild-type and empty plasmid Jurkat cells, was significantly attenuated by the TSPO specific ligand PK11195 at 100 nM (23 ± 1.8 cells, n = 9 for TSPO + PK11195, p < 0.001; ).
The addition of 0.1% foetal bovine serum (FBS), which acts as a chemo-attractant, significantly increased the number of cells that migrated through the Transwell membrane for all 3 cell types – wild-type, empty plasmid and TSPO-Jurkat cells, compared with cells in DMEM only (). However, the FBS-induced increase in the migration of TSPO-Jurkat cells was significantly attenuated by the addition of 100 nM PK11195, which had no effect on the wild-type and empty plasmid Jurkat cells (277 ± 24 cells, n = 6 for FBS + TSPO-Jurkat cells compared with 115 ± 5 cells, n = 9 for PK11195 + FBS + TSPO-Jurkat cells, p < 0.001; ). This suggests that the effect of PK11195 on motility is TSPO-specific.
Decreased outward rectified K+ currents in TSPO transfected Jurkat cells
Changes in cell excitability as a result of altered membrane potential were determined by patch clamp measurements of the electrophysiological cell membrane properties in a whole-cell mode configuration. Episodic traces of voltage-activation were applied from −100 mV to +80 mV, increasing by 20 mV increments. The membrane potential was held at −60 mV. Outwardly rectified whole-cell currents were then recorded in all 3 cell types (i.e., wild-type, empty plasmid and TSPO-Jurkat cells; ). The shape and amplitude of voltage-induced outward currents in wild-type (220 ± 41 pA at 0 mV, n = 4) and empty plasmid (197 ± 20 pA at 0 mV, n = 10) Jurkat cells are comparable, while the amplitude in TSPO-Jurkat cells was markedly smaller (78 ± 17 pA at 0 mV, n = 12, p < 0.001 when compared with wild-type or empty plasmid Jurkat cells). It should be noted that outward currents in empty plasmid Jurkat cells peaked at −20 mV (261 ± 51 pA, n = 10), while in wild-type and TSPO-Jurkat cells it peaked at 0 mV (). The amplitude of voltage-gated outward currents in TSPO-Jurkat cells was significantly smaller (particularly at −20 mV to 20 mV) than both wild-type and empty plasmid Jurkat cells (). This suggests that TSPO transfection increased membrane excitability by decreasing the voltage-activated outwardly rectified current.
Figure 5. TSPO decreased whole-cell outward rectified K+ currents. (A) Episodic current traces of voltage-activated (from −100 mV to +80 mV with 20 mV increment for A, C) outward currents recorded in wild-type, empty plasmid, and TSPO-Jurkat cells. The membrane potential was held at −60 mV. The shape and amplitude of voltage induced outward currents in wild-type and empty plasmid Jurkat cells were comparable, while the amplitude in TSPO-Jurkat cells was smaller. (B) Current-voltage (I-V) curves of voltage-activated currents shown in (A). The amplitude of voltage-gated outward currents in TSPO-Jurkat cells was significantly smaller at −20 mV to 20 mV than either wild-type or empty plasmid Jurkat cells. (C) Current traces of empty plasmid Jurkat cells in normal bath solution (containing NaCl) and normal pipette solution (containing KCl), in normal bath solution with addition of 20 mM TEA-Cl, in normal pipette solution with addition of 20 mM TEA-Cl, and in a pipette solution with CsCl replacing KCl. TEA in the bath solution partly inhibited the voltage-gated outward current, but a greater effect was seen when TEA was in the pipette solution. Cs+ replacement of K+ in the pipette solution abolished the outward currents. (D) The I-V curves of corresponding traces shown in (C).
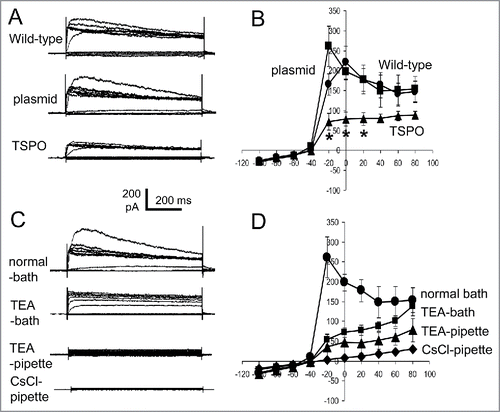
To examine whether the outward rectified currents were due to the efflux of K+, current traces from empty plasmid Jurkat cells with the addition of K+ channel inhibitors, TEA-Cl and CsCl were studied. Adding TEA-Cl (20 mM to replace equal molar amounts of NaCl) to the bath solution partly inhibited the voltage-gated outward current, but a greater effect was seen when TEA-Cl (20 mM) was in the pipette solution (). CsCl replacement of KCl in the pipette solution abolished the outward currents (). These results suggest that the outwardly rectified whole-cell currents were due to the efflux of K+.
Discussion
TSPO/PBR-dependent regulation has been reported for a number of biological processes, including steroid biosynthesis,Citation4,26 cell division,Citation23,27 energy metabolism,Citation9,13,14,28,29 production of reactive oxygen species,Citation9,11,30 cell apoptosis,Citation9,31-33 immune responses,Citation34,35 and hematopoiesis.Citation36 It is well established that the expression of TSPO increases significantly after injury and under neuropathological conditions, such as multiple sclerosis, or neurodegenerative conditions such as Alzheimer's disease.Citation13,37-46 Recently, the global and conditional Tspo knockout mouse models created in different laboratories have not confirmed the previously identified putative function of TSPO in cholesterol transport, biosynthesis, and the mitochondrial permeability transmission pore (mPTP).Citation14-17,47 There were also no abnormalities in these mice in terms of reproduction and life span up to 24 months. Our study using microglia isolated from global Tspo knockout mice had demonstrated that mitochondrial respiration and ATP production may be different from wild-type littermates.Citation14 This finding has prompted us to further investigate the functions of TSPO in mitochondrial energy/ATP production and associated high energy intensive cell functions. In contrast to the Tspo knockout method, a loss-of-function approach, we opted for a gain-of-function approach by inserting an additional TSPO gene into Jurkat cells, a human T-lymphocyte leukemia cell line, which we had observed to have a barely detectable TSPO protein expression.
The wild-type Jurkat cells used for transfection of the human TSPO gene in this study have shown TSPO expression at barely detetctable levels, confirmed by radioligand receptor membrane binding with [3H]PK11195 and immunocytochemistry with the most specific TSPO antibody (Abcam) that we have used in our laboratory. A high degree of TSPO promoter methylation in wild-type Jurkat cells contributes to the low TSPO expression observed in these cells.Citation28 Our barely detectable TSPO expression is consistent with the finding of Bertomeu et al.Citation21 who demonstrated negligible TSPO expression in the clone E6.1 Jurkat cells and also overexpressed TSPO for studying the role of TLN-4601, a farnesylated dibenzodiazepinone. The affinity constant (Kd) for receptor membrane binding with [3H]PK11195 in the TSPO overexpressed Jurkat cells are in the low nano-molar range in both studies. However, our study does not support the findings by Costa et al,Citation48 who have shown TSPO expression in Jurkat cells. The differences are (i) they used Jurkat cells sourced from the Interlab Cell Line Collection without providing the clone name; (ii) unusual binding of [3H]PK11195 with Kd values in the micro-molar range; (iii) all their studies on TSPO protein expression (Western blot, immunocytochemistry and electron microscopy) were based on the use of their TSPO antibody, which showed diffused staining throughout whole cells (even though they claimed mitochondrial and nuclear membrane co-localization). This diffused staining is in stark contrast to our results in TSPO overexpressed Jurkat cells which demonstrated mitochondrial co-localization of TSPO using a known highly specific TSPO antibody.
The successful transfection of TSPO was confirmed by PCR, RT-qPCR, membrane receptor binding with the TSPO-specific ligand [3H]PK11195, and immunohistochemistry with a TSPO antibody. We confirmed that the TSPO protein in transfected cells was localized to the mitochondria. Using microarray gene expression profiling and gene set enrichment analysis (GSEA), we found that a set of genes involved in mitochondrial energy production via the mitochondrial electron transport chain were upregulated in the TSPO-transfected Jurkat cells. This upregulation was associated with increased mitochondrial ATP synthesis and an increase in cell excitability by decreased rectified K+channel currents. These contributed to the increase in cell proliferation and motility.
The finding that the expression of TSPO correlates with ATP production and the upregulation of genes involved in mitochondrial respiration are consistent with our previous report using global Tspo knockout GuwiyangWurra (‘Fire Mouse’) animals.Citation14 In the microglia from Tspo knockout mice, we observed lower ATP production and basal oxygen consumption.Citation14 Similar findings on the role of TSPO in ATP production have been previously reported.Citation9,11,49-54 The observed TSPO–dependent modulations in energy metabolism have been suggested to occur through its interaction with VDAC on the outer mitochondrial membrane and ANT on the inner mitochondrial membrane. Krestinina et al.Citation51 demonstrated that TSPO ligands can modulate energy production in mitochondria by controlling phosphorylation of the FoF1-ATPase subunit c. TSPO may modulate ATP synthase via a direct interaction between the ATP ‘synthasome’ (composed of ATP synthase, phosphate carrier, and ANT) and the PBR complex composed of TSPO, VDAC, and ANT.Citation55,56 Veenman et al.Citation9,10 and Zeno et al.Citation11 have suggested that reactive oxygen species (ROS) generation is modulated by TSPO through regulation of the Fo subunit of FoF1 ATP(synth)ase.
Both AMP-activated protein kinase (AMPK) and sirtuins (NAD+-dependent histone/protein deacetylases) modulate energy balance in cells.Citation57 They are activated by reduced cellular energy levels (e.g. increased AMP/ATP ratio).Citation57 It would be of great interest to conduct future studies to examine the relationship between TSPO and AMPK/sirtuins, which has not yet been documented. Based on the reported functions, it can be speculated that under normal physiological conditions, overexpression of TSPO along with positive cell metabolism/energy production negatively correlates with the level of expression of AMPK/sirtuins. This also means that high expression levels of AMPK and sirtuins in reduced energy expenditure and halted cell cycles should correlate with lower TSPO expression. To validate this conjecture, the levels of AMPK/sirtuins in TSPO over-expressing Jurkat cells and in TSPO knockout animals needs to be examined.
The increased cell proliferation and motility observed in TSPO-Jurkat cells is consistent with observations in other cancer cell lines.Citation23,49,58-60 TSPO overexpression in C6 rat glioblastoma cells increases cell motility and proliferation.Citation61 Overexpression of TSPO in the breast cancer cell line MCF7 also increases cell proliferation/migration and inhibits Mammary Epithelial morphogenesis.Citation62 However, these results should be interpreted carefully as the cells already had high endogenous TSPO expression before they were manipulated, possibly resulting in different functional changes compared with cell lines with little to no endogenous TSPO.
Outward rectified K+ channel currents were recorded in whole Jurkat cells, and were found to be significantly decreased in amplitude in TSPO-Jurkat cells. It is possible that increased ATP production could cause increased cell excitability by decreasing whole-cell outward rectified K+ channel currents. It is also possible that the currents might be ATP sensitive K+ channel (KATP) currents as they have the same outward rectified shape,Citation63,64 although this needs to be confirmed using the KATP channel blocker glibenclimade.Citation63 Mitochondria also possess KATP channel currentsCitation65,66 that may contribute to whole cell KATP currents, which could directly influence other cellular functions (i.e. ATP secretion, cell proliferation and motility).
The TSPO/PBR antagonist PK11195 has been confirmed as a highly selective TSPO ligand using the global Tspo knockout GuwiyangWurra mouse.Citation14 However, it has been reported that PK11195 may exert functions independently from TSPO,Citation47 particularly at higher concentrations. Using Langmuir monolayers, quartz crystal microbalance with dissipation monitoring and neutron reflectometry, we have previously investigated the behavior of PK11195 in lipid bilayers with and without integrated TSPO,Citation67 indicating that there might be a synergistic effect resulting from the oriented insertion of PK11195 into the bi-lipid layer and its high affinity binding to TSPO. In the present study, we show that PK11195 at concentrations less than 10 μM has a significant antagonistic effect on the mRNA level of COX IV subunit 2 and cell motility. It would be of great interest to conduct further studies to determine whether other TSPO ligands, which do not bind to the PK11195 binding site or CRAC domain of TSPO protein, have similar effects. These ligands include endogenous ligands, such as cholesterol and protoporphyrin IX, and synthetic ligands, such as benzodiazepine Ro5–4864. The binding sites of Ro5–4864 and PK11195 on TSPO have distinct but overlapping binding sites that involve amino acid residues on the cytoplasmic loop and the C-terminal.Citation13
In summary and outlook, the overexpression of TSPO in Jurkat cells with barely detectable TSPO expression was successfully achieved by stable transfection with a functional human TSPO gene. This resulted in a gain-of-function compared with the non-transfected wild-type Jurkat cells, characterized by increased cell metabolism, energy production, cell cycle, motility and cell membrane excitability. We speculate that the broad range of reported regulatory effects associated with the mitochondrial TSPO may be driven by its known evolutionary role in energy -the current study describes a range of cellular effects due to the insertion of a TSPO gene, it is limited in regard to the precise effect of TSPO on the cell's bioenergetics profile. Extensive studies, including the probing of the electron transport chain under different substrate conditions, pharmacological dose range studies, and different compounds across a range of cells types, are required to determine which functions of the TSPO are additional rather than merely resultant from its potential primary role in bioenergetics.
Methods and materials
Cell culture
The wild-type Jurkat cells were donated by Professor Ian Campbell at the University of Sydney, Australia. Jurkat cells transfected with pcDNA3.1(+) empty vector, pcDNA3.1(+)-TSPO (TSPO), and wild-type Jurkat cells were maintained in Dulbecco's Modified Eagle's Medium (DMEM), supplemented with 10% v/v fetal calf serum (FCS) (Invitrogen, Carlsbad, CA, USA) and 2 mM glutamine in 5% CO2 at 37°C before and during functional studies. The TSPO positive control cells MDA-MB-231 (a human breast cancer cell line) were cultured under the same conditions.
Plasmid construction
Optimisation and synthesis of the human TSPO coding sequence, and subsequent cloning into the pMA vector, was performed by GeneArt (Life Technologies). The TSPO sequence in pMA was then subcloned into pcDNA3.1(+) (Invitrogen). DNA sequencing was used to verify all of the constructs (AGRF, Sydney, Australia). The plasmids were linearized before stable transfection.
Cell transfection and selection
Jurkat cells were transfected using the Amaxa Nucleofection system according to instructions provided by the manufacturer (Lonza, Basel, Switzerland). Briefly, 1 × 106 cells were resuspended in Nucleofector Solution V, mixed with 2 µg of plasmid DNA and transfected using program X-001. To create stable cell lines, G418 was added to the culture media 2 days after transfection. G418 was added at 1 mg/mL to the culture media of cells transfected with the TSPO containing plasmid or the empty vector pcDNA3.1(+). Once stable cell lines had been produced, cells were maintained in media supplemented with a lower concentration of G418 (0.5 mg/mL).
RNA isolation and reverse transcription
Wild-type and stable transfected Jurkat cells were plated at 1.6 × 105 cells per well in 24-well plates and grown overnight. Total RNA was extracted from each of the Jurkat cell lines using the PureLink RNA mini kit (Invitrogen) following the manufacturer's protocol. The isolated RNA was further treated with DNase using the PureLink DNase Set (Invitrogen), to ensure the removal of contaminating DNA before RT-qPCR. The concentration of the RNA was determined using a NanoDrop 2000c Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). The purity of the extracted RNA was assessed spectrophotometrically using the A260/A280 ratio, while RNA integrity was assessed using the 28s/18s ratio after agarose gel electrophoresis.
First-strand cDNA was synthesized from 100 ng of total RNA by first incubating for 5 minutes at 65°C with 0.5 mM dNTP mix and 2.5 µM of oligo(dT)20 primer, this mix was then placed on ice for at least 1 minute. After adding 1 x RT buffer, 5 mM MgCl2, 10 mM DTT, 40 units RNaseOUT and 200 units of Superscript III (Invitrogen), the reaction (total volume 20 µl) was incubated for 50 minutes at 50°C. The reverse transcription was terminated by incubating the samples at 85°C for 5 minutes. Samples without the addition of Superscript III were included as control for genomic DNA contamination. The freshly prepared cDNA was further diluted by DEPC-treated water so that approximately 1/20 of the original stock was used for the subsequent RT-qPCR.
Real-time PCR
TSPO and COX IV subunit 2 mRNA expression in each of the Jurkat cell lines was assessed using a CFX 384 Real-Time PCR Detection System (BioRad, Hercules, CA, USA). Primer-BLAST was used to design primers that detect the exogenous and endogenous TSPO mRNA (Table S2). The primers for COX IV subunit 2 PCR were used according to a previous publicationCitation68 (Table S2). Diluted cDNA (1 µl) was added to 4 µl of reaction mixture containing 2.5 µl of SsoFast supermix (BioRad) and forward and reverse primers at a final concentration of 0.5 µM each. PCR conditions were empirically optimised and the efficiency was further assessed at the selected annealing temperature. The PCR conditions were 98°C for 30 seconds, followed by 45 cycles at 98°C for 5 seconds and 63°C for 10 seconds. At the end of the 45th cycle, the temperature was raised to 72°C for 10 minutes to ensure complete extension of the PCR products. A melt curve analysis was subsequently performed to confirm the specificity of the results. The PCR was repeated without melt curve analysis for resolving the PCR product on a 1.5% agarose gel.
β-actin (ACTB)Citation68 as well as glyceraldehyde 3-phosphate dehydrogenase (GAPDH)Citation69 were used as internal standards (i.e. housekeeping genes (Table S2)). Samples were run in triplicates. The relative expression of target genes was quantified using comparative Ct analysis incorporated into the CFX Manager Software (version 1.5) (BioRad).
Microarray analysis
Total RNA was extracted from wild-type, empty plasmid and TSPO-Jurkat cells using TRIzol and the PureLink RNA mini kit (Invitrogen) with on-column DNase treatment following the manufacturer's protocol. The quality of the RNA was assessed using an Agilent 2100 Bioanalyzer. Sample labeling, hybridization to Affymetrix Human Gene 1.0 ST Arrays, washing and scanning was performed by the Ramaciotti Centre for Gene Function Analysis at the University of New South Wales. Two biological replicates for each sample were processed. The GenePattern platform (Broad Institute, MIT) was used to analyze the microarray data. Normalization was carried out using RMA via the NormalizeAffymetrixST module followed by the identification of differentially expressed genes using the LimmaGP module (http://pwbc.garvan.unsw.edu.au/gp). Using the t-statistic from LimmaGP, gene set enrichment analysis (GSEA) was performed in pre-ranked mode with 1000 permutations. The GSEA results were visualized using the Cytoscape plug-in Enrichment Map. Only gene sets with a FDR value of less than 0.05 are shown.
Cell membrane preparation and radioligand binding
Membranes of wild-type, empty plasmid and TSPO-Jurkat cells for radioligand binding experiments were prepared as follows. Cells were lysed in 20 volumes of ice-cold 50 mM Tris-HCl pH 7.4 using 50 strokes of a Potter-Elvehjem homogenizer. Cell lysate was centrifuged at 48 000 × g for 20 minutes at 4°C and supernatant discarded. The membrane fraction collected was homogenized in fresh buffer, centrifuged, and the final membrane preparation resuspended in 50 mM Tris-HCl pH 7.4. The protein concentration of membrane preparations was measured using the BCA protein assay (Thermo Fisher Scientific). The saturation binding of [3H] PK11195 in MDA-MB-231 cells and wild-type, empty plasmid, and TSPO-Jurkat cells was performed according to Banati et al.Citation14
Determination of TSPO expression and subcellular distribution
Immunocytochemistry was performed according to Liu et al.Citation70 Briefly, cells from each of the Jurkat cell lines were allowed to settle-down onto glass coverslips and were then fixed with 3.7% paraformaldehyde for 10 minutes. After 3 washes with phosphate buffered saline (PBS, 5 minutes per wash), the cell membranes were permeabilised with 0.1% Triton X-100 for 1 minute. The cells were then incubated with 2% bovine serum albumin (BSA) for 30 minutes before being incubated with primary antibodies (rabbit anti-TSPO monoclonal antibody (Abcam, Cambridge, UK) and mouse anti-human/mouse mitochondrial electron transport chain complex IV antibody (Abcam) overnight at 4°C. After 3 washes, the cells were incubated with the secondary antibodies Alexa Fluor (AF) 488-conjugated goat anti-rabbit antibody (Life Technologies) and AF594-conjugated goat anti-mouse antibody (Life Technologies) for 1 hour. After 3 washes, the cells on coverslips were mounted with ProLong Gold antifade reagent containing DAPI (Life Technologies) and viewed under a BX61WI Olympus microscope. Images were acquired with a digital camera (CoolSNAP, Photometrics, Tucson, AZ, USA) and the Image InVivo program (Photometrics). Deconvolution of images was performed with the Auto Deblur program (Photometrics) and further processed with ImageJ (NIH, Baltimore, MD, USA).
Cell permeabilisation
Jurkat cells used for measuring mitochondrial ATP production were permeabilised according to Milakovic and Johnson (2005).Citation71 Briefly, Jurkat cells were pelleted and resuspended in media A (20 mM HEPES, 10 mM MgCl2, 250 mM sucrose, pH 7.3). The cells (1×107 cells for each cell line) were then incubated in media A containing digitonin (Sigma-Aldrich, final concentration 100 μg/ml) and mixed for 1 minute at room temperature. After pelleting the permeablised Jurkat cells, the digitonin-containing media A was removed, and the permeabilised cells washed with 15 ml of media A for 3 times to remove any remaining digitonin. The permeablised Jurkat cells were finally resuspended in respiratory buffer (media A plus 0.72 mM KH2PO4, 1.28 mM K2HPO4, pH 7–7.1, plus 1% BSA) in preparation for detecting mitochondrial ATP production.
Detection of mitochondrial ATP production
ATP production was determined in Jurkat cells that were permeabilised but had intact mitochondria. Other possible ATP production pathways, besides aerobic metabolism, were blocked with Ap5A (25 μM) to inhibit adenylate kinase, which prevents the inter-conversion of ATP, ADP and AMP, and with iodoacetate (25 μM) to inhibit glyceraldyde-3-phosphate dehydrogenase to block glycolysis-induced ATP production. ATP production was detected with the luciferin-luciferase bioluminescence assay, which generates photons correlating to the number of ATP molecules present.Citation71
The permeabilised Jurkat cells were re-suspended in the respiratory buffer containing Ap5A and iodoacetate and respiratory substrates including 10 mM glutamate (for complex I of the respiratory chain), 10 mM succinate (for complex II), 10 mM ascorbate and 25 μΜ TMPD (for complex IV). Aliquots of 50 μl (5×106 cells/ml density) were placed into an optical Nunk 96-well plate (Thermo Scientific) and 50 μl ATP assay mix (luciferin-luciferase, Sigma-Aldrich) was added. After a 30 minute incubation (to allow luciferase to consume the free ATP in the aliquots), the plate was placed in the BMG plate reader. After an initial recording, 200 μM of ultra-pure ADP (> 99.99%, Cell Technology, Mountain View, CA, USA) was injected, and the recording continued for 2 more minutes. At least 3 repeats for each cell line were performed. The amount of protein from permeabilised cells was also quantified using a NanoDrop (Thermo Scientific) with the Pierce BCA protein assay kit (Thermo Scientific). The photon emission was normalized against the amount of protein in each sample.
Cell proliferation
Jurkat cell proliferation was measured using a Scepter cell counter (Millipore, Billerica, MA, USA), which is based on Coulter Count-impedance measurement technology. Wild-type, empty plasmid and TSPO-Jurkat cells were all seeded at the same density of 3 × 105 cells/ml before the experiment. Three days later, cells from each Jurkat cell line were counted with the Scepter cell counter and plated at 3 × 105 cells/ml into 24-well culture plates (0.5 ml/well, 4 repeats for each cell line and for each time point). One hour after plating, the cells from each cell line were counted with the Scepter cell counter and recorded as day zero. The cells were then counted at 24, 48 and 72 hours after plating. The number of cells in each cell line was normalized to the average number of cells at day 0. The cell proliferation for each cell line was then plotted with an exponential function and compared across groups.
Chemotaxis
Chemotaxis of Jurkat cells was investigated using the Transwell migration assay (also called Boyden chamber) which consisted of 24-well polyethylene terephthalate membranes with 5−μm pores (Falcon, Franklin Lakes, NJ, USA). One hundred thousand cells in 0.4 ml DMEM were added to the upper reservoir and 0.7 mL DMEM was placed in the lower reservoir with either a TSPO specific ligand or 0.1% FBS as a general chemoattractant. To investigate the role of TSPO in FBS-induced chemotaxis, 100 nM was added to both the upper and lower reservoirs while 0.1% FBS was only placed in the lower reservoir. After 4 hours of incubation at 37°C and 5% CO2, the plate containing the Transwell chambers was shaken 5 times to free the cells that had migrated but were still attached to the Transwell membrane in the lower reservoir. The migrated cells on the bottom of each well of the plate were viewed and images were captured using an Olympus microscope (BX61WI, Olympus) mounted with 4x objective and a CCD camera (CoolSNAP HQ2). Migrated cells were counted from at least 3 Transwell chambers (3 fields per chamber) with Image J.
Patch clamp electrophysiology
Whole-cell patch-clamp recordings from potassium channels in Jurkat cells were made using a Multiclamp 700B amplifier (Molecular Devices, MDS Analytical Technologies, Toronto, Canada) at room temperature (22–24°C) as described previously.Citation63 Briefly, resistances of glass pipettes were 4–8 Mohm when a patch pipette was filled with pipette solution containing 140 mM KCl, 1 mM MgCl2, 10 mM EGTA, 10 mM HEPES and pH 7.3; the external bath solution contained 135 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 10 mM glucose and 10 mM HEPES (pH 7.4). To verify K+ channels, general 20 mM K+ blocker TEA was added to the bath solution or in the normal pipette solution, or CsCl replaced KCl in the pipette solution. Episodic voltage-activated current traces (from −100 mV to +100 mV with 20 mV increment) were recorded in wild-type, empty plasmid and TSPO-Jurkat cells. The membrane potential was held at −60 mV. Whole-cell currents were also recorded in gap-free mode at a holding membrane potential of −60 mV. The currents were sampled online using a Digidata 1400 interface and pClamp10 program (Molecular Devices).
Statistics
Data presented are the averages of at least 4 repeat measurements. Results are expressed as means ± standard deviation. The statistical analysis of significance of results was carried out with one-way analysis of variance (ANOVA) and Post-hoc comparisons were made using Tukey's HSD test. Two-tailed unpaired Student's t-tests were also used to evaluate statistical significance. Results with p < 0.05 were considered significant.
Disclosure of Potential Conflicts of Interest
We have no competing interests.
Authors' contributions
G.-J.L. and R.B.B. devised and supervised the project. G.-J.L. performed most of the experiments. R.J.M. created the TSPO-Jurkat cells, performed cell culture and analyzed microarray data. D.Y.C. designed and created the TSPO plasmid. W.W.-Y.K. performed RT-qPCR. C.R.H. and RC contributed to radioligand membrane receptor binding studies. G.-J.L. wrote the manuscript with assistance by R.B.B. and R.J.M.
KCCY_A_1281477_supplemental_material.pdf
Download PDF (452 KB)Acknowledgments
The authors would like to thank their colleagues at the Australian Nuclear Science and Technology Organisation (ANSTO): C. Betlazar, M. Harrison-Brown, G. Dhand, E. Dodson, for technical laboratory support and assistance; RBB holds an ANSTO Distinguished Research Fellowship.
References
- Eshleman AJ, Murray TF. Differential binding properties of the peripheral-type benzodiazepine ligands [3H]PK 11195 and [3H]Ro 5-4864 in trout and mouse brain membranes. J Neurochem 1989; 53:494-502; PMID:2746235; http://dx.doi.org/10.1111/j.1471-4159.1989.tb07361.x
- Fan J, Lindemann P, Feuilloley MG, Papadopoulos V. Structural and functional evolution of the translocator protein (18 kDa). Curr Mol Med 2012; 12:369-86; PMID:22364126
- Snyder MJ, Van Antwerpen R. Evidence for a diazepam-binding inhibitor (DBI) benzodiazepine receptor-like mechanism in ecdysteroidogenesis by the insect prothoracic gland. Cell Tissue Res 1998; 294:161-8; PMID:9724466; http://dx.doi.org/10.1007/s004410051166
- Papadopoulos V, Baraldi M, Guilarte TR, Knudsen TB, Lacapere JJ, Lindemann P, Norenberg MD, Nutt D, Weizman A, Zhang MR, et al. Translocator protein (18kDa): new nomenclature for the peripheral-type benzodiazepine receptor based on its structure and molecular function. Trends Pharmacol Sci 2006; 27:402-9; PMID:16822554; http://dx.doi.org/10.1016/j.tips.2006.06.005
- Antkiewicz-Michaluk L, Guidotti A, Krueger KE. Molecular characterization and mitochondrial density of a recognition site for peripheral-type benzodiazepine ligands. Mol Pharmacol 1988; 34:272-8. PMID: 2843747
- Guo Y, Kalathur RC, Liu Q, Kloss B, Bruni R, Ginter C, Kloppmann E, Rost B, Hendrickson WA. Structure and activity of tryptophan-rich TSPO proteins. Science 2015; 347:551-5; PMID:25635100; http://dx.doi.org/10.1126/science.aaa1534
- Jaremko L, Jaremko M, Giller K, Becker S, Zweckstetter M. Structure of the mitochondrial translocator protein in complex with a diagnostic ligand. Science 2014; 343:1363-6; PMID:24653034; http://dx.doi.org/10.1126/science.1248725
- Li F, Liu J, Zheng Y, Garavito RM, Ferguson-Miller S. Crystal structures of translocator protein (TSPO) and mutant mimic of a human polymorphism. Science 2015; 347:555-8; PMID:25635101; http://dx.doi.org/10.1126/science.1260590
- Veenman L, Alten J, Linnemannstons K, Shandalov Y, Zeno S, Lakomek M, Gavish M, Kugler W. Potential involvement of F0F1-ATP(synth)ase and reactive oxygen species in apoptosis induction by the antineoplastic agent erucylphosphohomocholine in glioblastoma cell lines : a mechanism for induction of apoptosis via the 18 kDa mitochondrial translocator protein. Apoptosis 2010; 15:753-68; PMID:20107899; http://dx.doi.org/10.1007/s10495-010-0460-5
- Veenman L, Gavish M. The role of 18 kDa mitochondrial translocator protein (TSPO) in programmed cell death, and effects of steroids on TSPO expression. Curr Mol Med 2012; 12:398-412; PMID:22348610
- Zeno S, Veenman L, Katz Y, Bode J, Gavish M, Zaaroor M. The 18 kDa mitochondrial translocator protein (TSPO) prevents accumulation of protoporphyrin IX. Involvement of reactive oxygen species (ROS). Curr Mol Med 2012; 12:494-501; PMID:22376065
- Papadopoulos V, Amri H, Boujrad N, Cascio C, Culty M, Garnier M, Hardwick M, Li H, Vidic B, Brown AS, et al. Peripheral benzodiazepine receptor in cholesterol transport and steroidogenesis. Steroids 1997; 62:21-8; PMID:9029710; http://dx.doi.org/10.1016/S0039-128X(96)00154-7
- Liu GJ, Middleton RJ, Hatty CR, Kam WW, Chan R, Pham T, Harrison-Brown M, Dodson E, Veale K, Banati RB. The 18 kDa translocator protein, microglia and neuroinflammation. Brain Pathol 2014; 24:631-53; PMID:25345894; http://dx.doi.org/10.1111/bpa.12196
- Banati RB, Middleton RB, Chan R, Hatty CR, Kam WWY, Quin C, Graeber MB, Parmar A, Zahra D, Callaghan P, et al. Positron emission tomography and functional characterization of a complete PBR/TSPO-knockout. Nat Commun 2014; 5:5452; PMID:25406832; http://dx.doi.org/10.1038/ncomms6452
- Morohaku K, Pelton SH, Daugherty DJ, Butler WR, Deng W, Selvaraj V. Translocator protein/peripheral benzodiazepine receptor is not required for steroid hormone biosynthesis. Endocrinology 2014; 155:89-97; PMID:24174323; http://dx.doi.org/10.1210/en.2013-1556
- Sileikyte J, Blachly-Dyson E, Sewell R, Carpi A, Menabo R, Di Lisa F, Ricchelli F, Bernardi P, Forte M. Regulation of the mitochondrial permeability transition pore by the outer membrane does not involve the Peripheral Benzodiazepine Receptor (TSPO). J Biol Chem 2014; 289:13769-81; PMID:24692541; http://dx.doi.org/10.1074/jbc.M114.549634
- Tu LN, Morohaku K, Manna PR, Pelton SH, Butler WR, Stocco DM, Selvaraj V. Peripheral Benzodiazepine Receptor/Translocator protein global knockout mice are viable with no effects on steroid hormone biosynthesis. J Biol Chem 2014; 289:27444-54; PMID:24936060; http://dx.doi.org/10.1074/jbc.M114.578286
- Gut P, Zweckstetter M, Banati RB. Lost in translocation: the functions of the 18-kD translocator protein. Trends Endocrinol Metab 2015; 26:349-56; PMID:26026242; http://dx.doi.org/10.1016/j.tem.2015.04.001
- Middleton RJ, Liu G-J, Banati RB. Guwiyang Wurra–‘Fire Mouse’: a global gene knockout model for TSPO/PBR drug development, loss-of-function and mechanisms of compensation studies. Biochem Soc Trans 2015; 43:553-8; PMID:26551692; http://dx.doi.org/10.1042/BST20150039
- Middleton RJ, Liu GJ, Banati RB. Epigenetic silencing of the human 18 kDa translocator protein (TSPO) in a T-cell leukemia cell line. DNA Cell Biol 2017; 36:103-8; PMID:28004979; http://dx.doi.org/10.1089/dna.2016.3385
- Bertomeu T, Zvereff V, Ibrahim A, Zehntner S, Aliaga A, Rosa-Neto P, Bedell B, Falardeau P, Gourdeau H. TLN-4601 peripheral benzodiazepine receptor (PBR/TSPO) binding properties do not mediate apoptosis but confer tumor-specific accumulation. Biochem pharmacol 2010; 80:1572-9; PMID:20655882; http://dx.doi.org/10.1016/j.bcp.2010.07.018
- Bribes E, Carriere D, Goubet C, Galiegue S, Casellas P, Simony-Lafontaine J. Immunohistochemical assessment of the peripheral benzodiazepine receptor in human tissues. J Histochem Cytochem 2004; 52:19-28; PMID:14688214; http://dx.doi.org/10.1177/002215540405200103
- Hardwick M, Fertikh D, Culty M, Li H, Vidic B, Papadopoulos V. Peripheral-type benzodiazepine receptor (PBR) in human breast cancer: correlation of breast cancer cell aggressive phenotype with PBR expression, nuclear localization, and PBR-mediated cell proliferation and nuclear transport of cholesterol. Cancer Res 1999; 59:831-42; PMID:10029072
- Le Fur G, Vaucher N, Perrier M, Flamier A, Benavides J, Renault C, Dubroeucq M, Gueremy C, Uzan A. Differentiation between two ligands for peripheral benzodiazepine binding sites,[3 H] R05-4864 and [3 H] PK 11195, by thermodynamic studies. Life Sci 1983; 33:449-57; PMID:6308375; http://dx.doi.org/10.1016/0024-3205(83)90794-4
- Hardwick M, Cavalli LR, Barlow KD, Haddad BR, Papadopoulos V. Peripheral-type benzodiazepine receptor (PBR) gene amplification in MDA-MB-231 aggressive breast cancer cells. Cancer Genet Cytogenet 2002; 139:48-51; PMID:12547158; http://dx.doi.org/10.1016/S0165-4608(02)00604-0
- Lacapere JJ, Papadopoulos V. Peripheral-type benzodiazepine receptor: structure and function of a cholesterol-binding protein in steroid and bile acid biosynthesis. Steroids 2003; 68:569-85; PMID:12957662; http://dx.doi.org/10.1016/S0039-128X(03)00101-6
- Brown RC, Degenhardt B, Kotoula M, Papadopoulous V. Location-dependent role of the human glioma cell peripheral-type benzodiazepine receptor in proliferation and steroid biosynthesis. Cancer Lett 2000; 156:125-32; PMID:10880761; http://dx.doi.org/10.1016/S0304-3835(00)00451-1
- Alvarez-Maubecin V, Garcia-Hernandez F, Williams JT, Van Bockstaele EJ. Functional coupling between neurons and glia. J Neurosci 2000; 20:4091-8; PMID:10818144
- Gut P, Baeza-Raja B, Andersson O, Hasenkamp L, Hsiao J, Hesselson D, Akassoglou K, Verdin E, Hirschey MD, Stainier DYR. Whole-organism screening for gluconeogenesis identifies activators of fasting metabolism. Nat Chem Biol 2013; 9:97-104; PMID:23201900; http://dx.doi.org/10.1038/nchembio.1136
- Xiao J, Liang D, Zhang H, Liu Y, Li F, Chen YH. 4′-Chlorodiazepam, a translocator protein (18 kDa) antagonist, improves cardiac functional recovery during postischemia reperfusion in rats. Exp Biol Med (Maywood) 2010; 235:478-86; PMID:20407080; http://dx.doi.org/10.1258/ebm.2009.009291
- Kugler W, Veenman L, Shandalov Y, Leschiner S, Spanier I, Lakomek M, Gavish M. Ligands of the mitochondrial 18 kDa translocator protein attenuate apoptosis of human glioblastoma cells exposed to erucylphosphohomocholine. Cell Oncol 2008; 30:435-50; PMID:18791274
- Veenman L, Papadopoulos V, Gavish M. Channel-like functions of the 18-kDa translocator protein (TSPO): regulation of apoptosis and steroidogenesis as part of the host-defense response. Curr Pharm Des 2007; 13:2385-405; PMID:17692008; http://dx.doi.org/10.2174/138161207781368710
- Zeno S, Zaaroor M, Leschiner S, Veenman L, Gavish M. CoCl(2) induces apoptosis via the 18 kDa translocator protein in U118MG human glioblastoma cells. Biochemistry 2009; 48:4652-61; PMID:19358520; http://dx.doi.org/10.1021/bi900064t
- Gavish M, Bachman I, Shoukrun R, Katz Y, Veenman L, Weisinger G, Weizman A. Enigma of the peripheral benzodiazepine receptor. Pharmacol Rev 1999; 51:629-50; PMID:10581326
- Veenman L, Gavish M. The peripheral-type benzodiazepine receptor and the cardiovascular system. Implications for drug development. Pharmacol Ther 2006; 110:503-24; PMID:16337685; http://dx.doi.org/10.1016/j.pharmthera.2005.09.007
- Carayon P, Portier M, Dussossoy D, Bord A, Petitpretre G, Canat X, Le Fur G, Casellas P. Involvement of peripheral benzodiazepine receptors in the protection of hematopoietic cells against oxygen radical damage. Blood 1996; 87:3170-8; PMID:8605331
- Banati RB. Visualising microglial activation in vivo. Glia 2002; 40:206-17; PMID:12379908; http://dx.doi.org/10.1002/glia.10144
- Banati RB. Neuropathological imaging: in vivo detection of glial activation as a measure of disease and adaptive change in the brain. Br Med Bull 2003; 65:121-31; PMID:12697620; http://dx.doi.org/10.1093/bmb/65.1.121
- Banati RB, Cagnin A, Brooks DJ, Gunn RN, Myers R, Jones T, Birch R, Anand P. Long-term trans-synaptic glial responses in the human thalamus after peripheral nerve injury. Neuroreport 2001; 12:3439-42; PMID:11733686; http://dx.doi.org/10.1097/00001756-200111160-00012
- Banati RB, Goerres GW, Tjoa C, Aggleton JP, Grasby P. The functional anatomy of visual-tactile integration in man: a study using positron emission tomography. Neuropsychologia 2000; 38:115-24; PMID:10660224; http://dx.doi.org/10.1016/S0028-3932(99)00074-3
- Cagnin A, Brooks DJ, Kennedy AM, Gunn RN, Myers R, Turkheimer FE, Jones T, Banati RB. In-vivo measurement of activated microglia in dementia. Lancet 2001; 358:461-7; PMID:11513911; http://dx.doi.org/10.1016/S0140-6736(01)05625-2
- Cagnin A, Myers R, Gunn RN, Lawrence AD, Stevens T, Kreutzberg GW, Jones T, Banati RB. In vivo visualization of activated glia by [11C] (R)-PK11195-PET following herpes encephalitis reveals projected neuronal damage beyond the primary focal lesion. Brain 2001; 124:2014-27; PMID:11571219; http://dx.doi.org/10.1093/brain/124.10.2014
- Kuhlmann AC, Guilarte TR. Cellular and subcellular localization of peripheral benzodiazepine receptors after trimethyltin neurotoxicity. J Neurochem 2000; 74:1694-704; PMID:10737628; http://dx.doi.org/10.1046/j.1471-4159.2000.0741694.x
- Pedersen MD, Minuzzi L, Wirenfeldt M, Meldgaard M, Slidsborg C, Cumming P, Finsen B. Up-regulation of PK11195 binding in areas of axonal degeneration coincides with early microglial activation in mouse brain. Eur J Neurosci 2006; 24:991-1000; PMID:16930426; http://dx.doi.org/10.1111/j.1460-9568.2006.04975.x
- Venneti S, Lopresti BJ, Wiley CA. The peripheral benzodiazepine receptor (Translocator protein 18kDa) in microglia: from pathology to imaging. Prog Neurobiol 2006; 80:308-22; PMID:17156911; http://dx.doi.org/10.1016/j.pneurobio.2006.10.002
- Wilms H, Claasen J, Rohl C, Sievers J, Deuschl G, Lucius R. Involvement of benzodiazepine receptors in neuroinflammatory and neurodegenerative diseases: evidence from activated microglial cells in vitro. Neurobiol Dis 2003; 14:417-24; PMID:14678758; http://dx.doi.org/10.1016/j.nbd.2003.07.002
- Tu LN, Zhao AH, Stocco DM, Selvaraj V. PK11195 effect on steroidogenesis is not mediated through the translocator protein (TSPO). Endocrinology 2015; 156:1033-39; PMID:25535830; http://dx.doi.org/10.1210/en.2014-1707
- Costa B, Salvetti A, Rossi L, Spinetti F, Lena A, Chelli B, Rechichi M, Da Pozzo E, Gremigni V, Martini C. Peripheral Benzodiazepine Receptor: Characterization in Human T-Lymphoma Jurkat Cells. Mol Pharmacol 2006; 69:37-44; PMID:16189298
- Choi J, Ifuku M, Noda M, Guilarte TR. Translocator protein (18 kDa)/peripheral benzodiazepine receptor specific ligands induce microglia functions consistent with an activated state. Glia 2011; 59:219-30; PMID:21125642; http://dx.doi.org/10.1002/glia.21091
- Eckert GP, Schiborr C, Hagl S, Abdel-Kader R, Muller WE, Rimbach G, Frank J. Curcumin prevents mitochondrial dysfunction in the brain of the senescence-accelerated mouse-prone 8. Neurochem Int 2013; 62:595-602; PMID:23422877; http://dx.doi.org/10.1016/j.neuint.2013.02.014
- Krestinina OV, Grachev DE, Odinokova IV, Reiser G, Evtodienko YV, Azarashvili TS. Effect of peripheral benzodiazepine receptor (PBR/TSPO) ligands on opening of Ca2+-induced pore and phosphorylation of 3.5-kDa polypeptide in rat brain mitochondria. Biochemistry (Mosc) 2009; 74:421-9; PMID:19463096; http://dx.doi.org/10.1134/S0006297909040105
- Rosenberg N, Rosenberg O, Weizman A, Veenman L, Gavish M. In vitro catabolic effect of protoporphyrin IX in human osteoblast-like cells: possible role of the 18 kDa mitochondrial translocator protein. J Bioenerg Biomembr 2013; 45:333-41; PMID:23475134; http://dx.doi.org/10.1007/s10863-013-9501-4
- Seneviratne MS, Faccenda D, De Biase V, Campanella M. PK11195 inhibits mitophagy targeting the F1Fo-ATPsynthase in Bcl-2 knock-down cells. Curr Mol Med 2012; 12:476-82; PMID:22348615
- Veenman L, Gavish M, Kugler W. Apoptosis induction by erucylphosphohomocholine via the 18 kDa mitochondrial translocator protein: implications for cancer treatment. Anticancer Agents Med Chem 2014; 14:559-77; PMID:24628235; http://dx.doi.org/10.2174/1871520614666140309230338
- Ko YH, Delannoy M, Hullihen J, Chiu W, Pedersen PL. Mitochondrial ATP synthasome. Cristae-enriched membranes and a multiwell detergent screening assay yield dispersed single complexes containing the ATP synthase and carriers for Pi and ADP/ATP. J Biol Chem 2003; 278:12305-9; PMID:12560333; http://dx.doi.org/10.1074/jbc.C200703200
- McEnery MW, Snowman AM, Trifiletti RR, Snyder SH. Isolation of the mitochondrial benzodiazepine receptor: association with the voltage-dependent anion channel and the adenine nucleotide carrier. Proc Natl Acad Sci U S A 1992; 89:3170-4; PMID:1373486; http://dx.doi.org/10.1073/pnas.89.8.3170
- Ruderman NB, Xu XJ, Nelson L, Cacicedo JM, Saha AK, Lan F, Ido Y. AMPK and SIRT1: a long-standing partnership? Am J Physiol Endocrinol Metab 2010; 298:E751-60; PMID:20103737; http://dx.doi.org/10.1152/ajpendo.00745.2009
- Casellas P, Galiegue S, Basile AS. Peripheral benzodiazepine receptors and mitochondrial function. Neurochem Int 2002; 40:475-86; PMID:11850104; http://dx.doi.org/10.1016/S0197-0186(01)00118-8
- Corsi L, Geminiani E, Avallone R, Baraldi M. Nuclear location-dependent role of peripheral benzodiazepine receptor (PBR) in hepatic tumoral cell lines proliferation. Life Sci 2005; 76:2523-33; PMID:15769477; http://dx.doi.org/10.1016/j.lfs.2004.08.040
- Mendonca-Torres MC, Roberts SS. The translocator protein (TSPO) ligand PK11195 induces apoptosis and cell cycle arrest and sensitizes to chemotherapy treatment in pre- and post-relapse neuroblastoma cell lines. Cancer Biol Ther 2013; 14:319-26; PMID:23358477; http://dx.doi.org/10.4161/cbt.23613
- Rechichi M, Salvetti A, Chelli B, Costa B, Da Pozzo E, Spinetti F, Lena A, Evangelista M, Rainaldi G, Martini C, et al. TSPO over-expression increases motility, transmigration and proliferation properties of C6 rat glioma cells. Biochim Biophys Acta 2008; 1782:118-25; PMID:18190798; http://dx.doi.org/10.1016/j.bbadis.2007.12.001
- Wu X, Gallo KA. The 18-kDa translocator protein (TSPO) disrupts mammary epithelial morphogenesis and promotes breast cancer cell migration. PLoS One 2013; 8:e71258; PMID:23967175; http://dx.doi.org/10.1371/journal.pone.0071258
- Liu GJ, Simpson AM, Swan MA, Tao C, Tuch BE, Crawford RM, Jovanovic A, Martin DK. ATP-sensitive potassium channels induced in liver cells after transfection with insulin cDNA and the GLUT 2 transporter regulate glucose-stimulated insulin secretion. Faseb J 2003; 17:1682-4; PMID:12958175
- Tamarina NA, Kuznetsov A, Fridlyand LE, Philipson LH. Delayed-rectifier (KV2.1) regulation of pancreatic beta-cell calcium responses to glucose: inhibitor specificity and modeling. Am J Physiol Endocrinol Metab 2005; 289:E578-85; PMID:16014354; http://dx.doi.org/10.1152/ajpendo.00054.2005
- Liu Y, Sato T, O'Rourke B, Marban E. Mitochondrial ATP-dependent potassium channels: novel effectors of cardioprotection? Circulation 1998; 97:2463-9; PMID:9641699; http://dx.doi.org/10.1161/01.CIR.97.24.2463
- O'Rourke B. Evidence for mitochondrial K+ channels and their role in cardioprotection. Circ Res 2004; 94:420-32; PMID:15001541; http://dx.doi.org/10.1161/01.RES.0000117583.66950.43
- Hatty CR, Le Brun AP, Lake V, Clifton LA, Liu GJ, James M, Banati RB. Investigating the interactions of the 18kDa translocator protein and its ligand PK11195 in planar lipid bilayers. Biochim Biophys Acta 2014; 1838:1019-30; PMID:24374318; http://dx.doi.org/10.1016/j.bbamem.2013.12.013
- Cheng KT, Hou WC, Huang YC, Wang LF. Baicalin induces differential expression of cytochrome C Oxidase in human lung H441 Cell. J Agric Food Chem 2003; 51:7276-9; PMID:14640570; http://dx.doi.org/10.1021/jf0301549
- Dveksler GS, Basile AA, Dieffenbach CW. Analysis of gene expression: use of oligonucleotide primers for glyceraldehyde-3-phosphate dehydrogenase. PCR Methods Appl 1992; 1:283-5; PMID:1477664; http://dx.doi.org/10.1101/gr.1.4.283
- Liu GJ, Nagarajah R, Banati RB, Bennett MR. Glutamate induces directed chemotaxis of microglia. Eur J Neurosci 2009; 29:1108-18; PMID:19302147; http://dx.doi.org/10.1111/j.1460-9568.2009.06659.x
- Milakovic T, Johnson GVW. Mitochondrial respiration and ATP production are significantly impaired in striatal cells expressing mutant Huntingtin. J Biol Chem 2005; 280:30773-82; PMID:15983033; http://dx.doi.org/10.1074/jbc.M504749200
