ABSTRACT
Chibby1 (Cby1) was originally isolated as a binding partner for β-catenin, a dual function protein in cell-cell adhesion and in canonical Wnt signaling. The canonical Wnt/β-catenin pathway is dysregulated in various diseases including cancer, most notably of the gastrointestinal origin. To investigate the role of Cby1 in colorectal tumorigenesis, we generated stable Cby1-knockdown (KD) SW480 colon cancer cells. Unexpectedly, we found that Cby1 KD induces mesenchymal-to-epithelial transition (MET)-like changes in SW480 as well as in HEK293 cells. Cby1-KD cells displayed a cuboidal epithelial morphology with tight cell-cell contacts. In Cby1-KD cells, the plasma membrane localization of E-cadherin and β-catenin was dramatically increased with formation of cortical actin rings, while the levels of the mesenchymal marker vimentin were decreased. Consistent with these changes, in wound healing assays, Cby1-KD cells exhibited epithelial cell-like properties as they migrated collectively as epithelial sheets. Furthermore, the anchorage-independent growth of Cby1-KD cells was reduced as determined by soft agar assays. These findings suggest that chronic Cby1 KD in colon cancer cells may counteract tumor progression by promoting the MET process.
Introduction
The epithelial-to-mesenchymal transition (EMT) is a complex biological process in which non-motile epithelial cells acquire a mesenchymal phenotype with migratory and invasive properties.Citation1-4 EMT plays critical roles in multiple biological and pathological processes including embryogenesis, organ development, wound healing, and fibrosis. Importantly, it plays a key role in promoting cancer cell invasion and metastasis. During EMT, epithelial cells lose cell-cell adhesion structures and apical-basal polarity and rearrange their actin cytoskeletons. Tight and adherens junction proteins such as E-cadherin and β-catenin are downregulated, whereas mesenchymal cell-specific markers including vimentin are upregulated. Consequently, epithelial cells lose their characteristics such as cell-cell adhesion and apical-basal polarity and obtain mesenchymal properties including cell motility and invasiveness. This allows them to invade surrounding normal tissues and metastasize. EMT is a highly dynamic and reversible process, and mesenchymal cells undergo mesenchymal-to-epithelial transition (MET). Although relatively little is known about MET, it has been shown to play important roles in development and cancer progression in which it facilitates the formation of secondary tumors in distant organs.Citation1,4
In cell-cell adhesion junctions, E-cadherin interacts with β-catenin, which in turn binds to α-catenin, linking to the actin cytoskeleton.Citation5,6 β-Catenin also plays a pivotal role as a transcriptional co-activator in the canonical Wnt signaling pathway.Citation7-9 In the absence of canonical Wnt signaling, β-catenin is phosphorylated by casein kinase 1 (CK1) and glycogen synthase kinase 3 (GSK3) in the so-called “destruction complex,” containing the tumor suppressors Axin and adenomatous polyposis coli (APC). This results in degradation of β-catenin via the ubiquitin-proteasome pathway. Upon canonical Wnt signaling, the destruction complex becomes inactivated. Consequently, β-catenin accumulates in the cytoplasm and translocates into the nucleus where it binds to the T-cell factor/lymphoid enhancer factor (Tcf/Lef) family of transcription factors to stimulate downstream target genes.Citation7-9 Constitutive activation of Wnt/β-catenin signaling has been linked to a variety of human malignancies including melanoma, breast, colon, and hepatocellular carcinomas.Citation7-10 Notably, more than 70% of colon cancers exhibit increased Wnt/β-catenin activity. The direct target genes of β-catenin include oncogenes such as c-myc and cyclin D1.Citation7,10 The β-catenin-Tcf/Lef complex directly regulates gene expression associated with EMT, particularly snail, which encodes a transcriptional repressor for E-cadherin, thereby driving EMT.Citation11,12
Chibby1 (Cby1) was originally isolated as a β-catenin binding partner in a Ras recruitment screen in yeast.Citation13 It is a 15-kDa coiled-coil protein that is conserved throughout evolution. We previously reported that Cby1 represses canonical Wnt/β-catenin signaling via 2 distinct molecular mechanisms.Citation14,15 Cby1 competes with Tcf/Lef factors for binding to β-catenin. In the nucleus, Cby1 associates with 14–3–3 adaptor proteins to export β-catenin out of the nucleus. 14–3–3 proteins specifically recognize serine 20 within the 14–3–3 binding motif of Cby1. A recent structural analysis of 14–3–3ζ and Cby1 supports the essential role of Cby1 phosphorylation at serine 20 in 14–3–3 binding.Citation16 We showed that Cby1 contains functional nuclear localization signal (NLS) and nuclear export signal (NES) motifs and constitutively shuttles between the nucleus and cytoplasm.Citation17 Direct interactions of 14–3–3 proteins with Cby1 facilitate Cby1 binding to CRM1 export receptor, while suppressing Cby1 binding to the nuclear import receptor importin-1α. This results in re-distribution of Cby1 and β-catenin into the cytoplasm, leading to repression of β-catenin target genes. Cby1 also plays a crucial role during ciliogenesis, likely independent of β-catenin signaling.Citation18-22
Using human colon adenocarcinoma SW480 cells in which APC is mutated, we reported that stable expression of wild-type Cby1, but not a 14–3–3-binding-defective mutant (Cby1S20A), results in cytoplasmic translocation of β-catenin, concomitant with a reduction in β-catenin signaling and cell growth.Citation23 These results suggest a role for Cby1 as a tumor suppressor in colon cancer. Consistent with this notion, Cby1 is expressed at low levels in colon cancer cell lines mainly due to promoter methylation.Citation24 However, no major decrease in Cby1 expression levels was detected in colon cancer samples.Citation24,25 It has been reported that Cby1 expression is significantly downregulated in chronic myeloid leukemiaCitation26 and pediatric ependymomas.Citation27
To gain insights into the function of Cby1 in colorectal tumorigenesis, we created Cby1-knockdown (KD) SW480 clones stably expressing Cby1 small-hairpin (sh) RNAs. Unexpectedly, we found that Cby1 KD induces MET-like changes in SW480 as well as in HEK293 cells, accompanied by increased plasma membrane localization of E-cadherin and β-catenin, formation of cortical actin rings, and decreased levels of vimentin. Consistent with these results, in wound healing assays, Cby1-KD cells migrated as coherent epithelial sheets. Additionally, the anchorage-independent growth of Cby1-KD cells was reduced in soft agar assays. These findings suggest that chronic Cby1 KD may interfere with tumor progression by inducing MET and underscore the complexity of β-catenin regulation by Cby1.
Results
Cby1 KD alters the morphology of SW480 and HEK293 cells
To investigate the biological functions of Cby1 during colorectal tumorigenesis, we generated stable cell clones expressing Cby1 shRNA in SW480 human colon adenocarcinoma cells. SW480 cells harbor a truncated APC protein, leading to accumulation of β-catenin in the nucleus and constitutive β-catenin signaling.Citation28,29 These cells express Cby1 protein in its wild-type form at low levels.Citation24,30 We transfected these cells with an expression plasmid for Cby1 shRNA or control scrambled shRNA and established 3 independent clones for each Cby1 and scrambled shRNA after selection with puromycin. Western blotting revealed that Cby1 protein was substantially depleted by Cby1 shRNA with a 45% reduction on average (). Similar to wild-type SW480 cells, control scrambled shRNA-expressing cells showed a normal elongated or spindle-shaped morphology (). Cby1-KD SW480 cells appeared to grow as densely packed clones with tight cell-cell contacts, reminiscent of an epithelial phenotype. Similar results were obtained using a pool of stable Cby1 shRNA-expressing SW480 cells (Figure S1).
Figure 1. Cby1 KD alters morphology of SW480 and HEK293 cells. A., C. SW480 (A) or HEK293 (C) cell clones stably expressing control scrambled shRNA or Cby1 shRNA were seeded at the equal density (100 cells/well in 6-well plates), and 7 d later, phase-contrast micrographs were taken. B., D. (Left) Cell lysates were subjected to western blotting with Cby1 and GAPDH (loading control) antibodies to examine the efficiency of Cby1 KD. (Right) The band intensity of Cby1 was quantified and normalized to that of GAPDH. The results are expressed as mean ± SEM from at least 3 independent experiments. The control scrambled shRNA is set as 100. * P < 0.05.
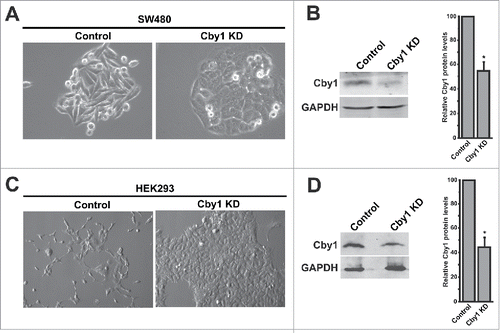
To examine whether the effects of Cby1 KD is a cell type-dependent phenomenon, we next depleted Cby1 in human embryonic kidney (HEK) 293 cells, which express relatively high levels of endogenous Cby1. Stable expression of Cby1 shRNA in HEK293 cells reduced Cby1 protein levels by 56% (). The abnormal growth pattern of the Cby1-KD cells was immediately recognizable compared with the control scrambled shRNA cells (). When seeded at low density, like wild-type HEK293 cells, control scrambled shRNA cells grew as mostly single cells in an elongated spindle-shaped pattern and spread across the entire surface of plates. In marked contrast, Cby1 shRNA cells showed a cuboidal morphology and grew as distinct clones with apparent cell-cell contacts, resembling epithelial cells. The morphological change of HEK293 cells was even more dramatic than that of SW480 cells. In monolayer cultures, no significant differences in growth rate were noted between control and Cby1-KD cells. All 3 independent clones displayed a similar morphology. Similar morphological changes were observed using a second Cby1 shRNA (data not shown).
Cby1 KD cells exhibit elevated levels of β-catenin and E-cadherin at the plasma membrane
The establishment of intercellular contacts in epithelial cells is primarily mediated by E-cadherin.Citation31,32 The cytoplasmic domain of E-cadherin binds β-catenin at adherens junctions, which in turn interacts with α-catenin to link the complex to the actin cytoskeleton. Since Cby1 was initially isolated as a β-catenin-binding protein,Citation13 we analyzed the expression of β-catenin and E-cadherin in Cby1-KD cells. Strikingly, immunofluorescence (IF) staining revealed a significant increase in β-catenin levels at adherens junctions in Cby1-KD cells in comparison with control scrambled shRNA cells (). E-cadherin levels were even more dramatically elevated at cell-cell contacts in Cby1-KD cells (). This dramatic increase in E-cadherin levels was evident throughout the height of cells as revealed by confocal microscopy along the z-axis (Figure S2).
Figure 2. Cby1 KD leads to profound plasma membrane localization of β-catenin and E-cadherin. A.-B. Localization of β-catenin (A) or E-cadherin (B) in control or Cby1-KD SW480 or HEK293 cells as indicated. Nuclei were visualized with DAPI. Scale bars, 20 μm. C. (Left) Cell lysates from control or Cby1-KD HEK293 cells were subjected to western blotting with E-cadherin and GAPDH antibodies. (Right) The band intensity of E-cadherin was quantified and normalized to that of GAPDH. The results are expressed as mean ± SEM from at least 3 independent experiments. The control scrambled shRNA is set as 1. * P < 0.05. D. E-cadherin mRNA levels were measured in control or Cby1-KD HEK293 cells using RT-qPCR and normalized against GAPDH mRNA levels. The data are mean ± SEM from triplicate measurements, and the control scrambled shRNA is set as 1. * P < 0.05. E. TopFlash assays. Control or Cby1-KD SW480 cells were transfected with TopFlash or mutant FopFlash luciferase reporter. Luciferase activities were measured 48 h post-transfection, and normalized to Renilla luciferase used as an internal control. The normalized FopFlash baseline values were subtracted from the normalized TopFlash values. All transfections were carried out in triplicates, and the results are expressed as mean ± SEM from at least 3 independent experiments. The control scrambled shRNA is set as 100. ** P < 0.001.
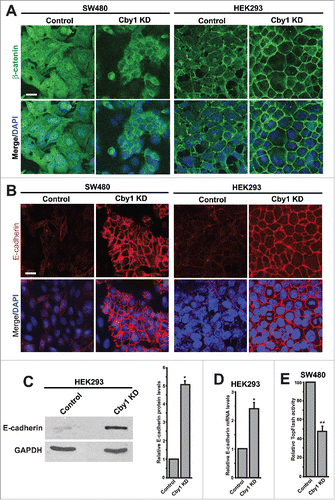
Consistent with our IF staining data, western blotting demonstrated a 5-fold increase in E-cadherin protein levels in Cby1-KD HEK293 cells (). To examine if E-cadherin expression is upregulated at the mRNA level, we performed real-time RT-qPCR. E-cadherin mRNA expression was moderately increased by 2.4-fold in Cby1-KD HEK293 cells compared with scrambled shRNA cells (). Similar trends of increased E-cadherin protein and mRNA levels were observed in SW480 cells (data not shown). These data suggest that Cby1 KD results in accumulation of β-catenin and E-cadherin at adherens junctions, leading to an epithelial-like morphology in SW480 and HEK293 cells.
SW480 cells exhibit elevated levels of endogenous β-catenin signaling due to truncated APC. To examine the effects of Cby1 KD on β-catenin signaling activity, we performed β-catenin-dependent luciferase reporter (TopFlash) assays.Citation14,30 As shown in , there was a mild but statistically significant reduction in TopFlash activity in Cby1-KD SW480 cells. This suggests that chronic depletion of Cby1 sequesters β-catenin at the plasma membrane, resulting in diminished β-catenin signaling in SW480 cells.
Cby1 KD induces mesenchymal-to-epithelial transition-like changes
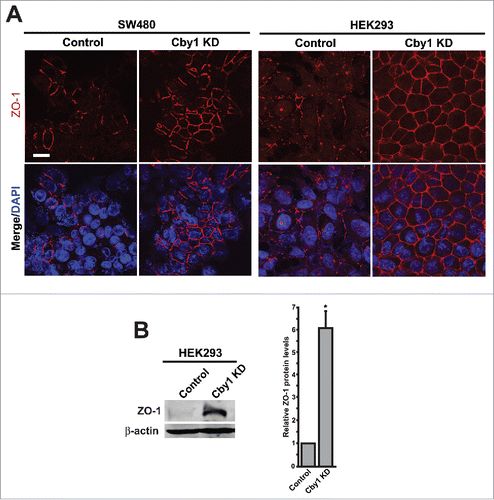
Given the significantly elevated levels of E-cadherin in Cby1-KD cells, we next examined whether epithelial and mesenchymal markers are altered. Assembly of E-cadherin-mediated adherens junctions is thought to initialize the establishment of cell-cell contacts, triggering the subsequent formation of other cell-cell junctions.Citation31,32 We therefore investigated if tight junctions are also affected in Cby1-KD cells by IF staining for ZO-1, a widely used tight junction marker. As shown in , ZO-1 staining at cell-cell junctions was significantly increased and more discrete in Cby1-KD SW480 and HEK293 cells. In agreement with this, ZO-1 protein levels were elevated by 6-fold in HEK293 cells ().
Figure 3. Cby1 KD results in a dramatic increase in ZO-1 protein levels at the plasma membrane. A. IF staining for the epithelial marker ZO-1 in control or Cby1-KD SW480 or HEK293 cells as indicated. Nuclei were visualized with DAPI. Scale bar, 20 μm. B. (Left) Cell lysates from control or Cby1-KD HEK293 cells were subjected to western blotting with ZO-1 and β-actin (loading control) antibodies. (Right) The band intensity of ZO-1 was quantified and normalized to that of β-actin. The results are expressed as mean ± SEM from triplicate samples in a single representative experiment. The control scrambled shRNA is set as 1. * P < 0.05.
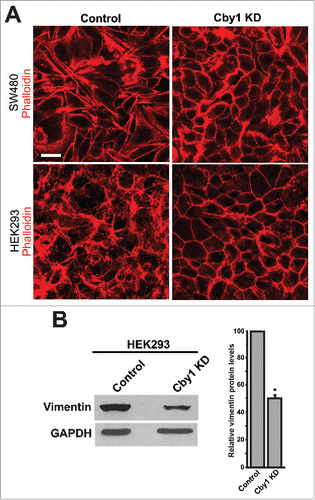
In addition to changes in cell-cell junctions, the epithelialization is associated with changes in actin cytoskeletal arrangements from stress fibers toward a cortical actin ring.Citation33,34 Consistent with this notion, Cby1-KD SW480 and HEK293 cells showed formation of cortical actin rings (). We next examined the expression level of vimentin, a marker for mesenchymal cells. Western blotting demonstrated that vimentin was reduced by 50% compared with control scrambled shRNA-expressing HEK293 cells (). These results support the notion that Cby1 KD leads to MET-like changes.
Figure 4. Cby1-KD cells exhibit epithelial characteristics. A. F-actin was visualized by phalloidin in control or Cby1-KD SW480 or HEK293 cells as indicated. Scale bar, 20 μm. B. (Left) Cell lysates from control or Cby1-KD HEK293 cells were subjected to western blotting with the mesenchymal marker vimentin and GAPDH antibodies. The band intensity of vimentin was quantified and normalized to that of GAPDH. The results are expressed as mean ± SEM from at least 3 independent experiments. The control scrambled shRNA is set as 100. * P < 0.05.
Cby1 KD influences the migration patterns of SW480 cells in wound healing assays
The MET-like changes in Cby1-KD cells prompted us to investigate the effects of Cby1-KD on cell migration. To this end, Cby1-KD and control scrambled shRNA SW480 cells were seeded at equal density (15×105 cells/well in 6-well plates) and grown to confluence. A wound was introduced with a plastic pipette tip. The ability of cells to migrate into the wound area was evaluated at 24, 48, and 72 h. No clear differences in the time course of wound closure were detected between Cby1-KD and control scrambled shRNA cells (). Wound healing assays were also performed in serum-free culture conditions to detect possible differences in cell migration. While overall cell migration was slower in serum-free media, no obvious difference was noticeable between Cby1-KD and control SW480 cells (data not shown).
Figure 5. Cby1 KD causes changes in cell morphology during cell migration. A. Cell migration of control or Cby1-KD SW480 cells was analyzed using wound healing assays. SW480 cells were grown to confluence, and a scratch was applied with a plastic pipette tip. Phase-contrast images were taken at 0 and 72 h after wounding. B. SW480 cells were seeded onto coverslips, fixed at 48 h after wounding, and stained with phalloidin and DAPI. Scale bar, 20 μm. C. Quantification of cell morphology at the wound edge. SW480 cells were fixed and stained with phalloidin and DAPI at 48 h after wounding. The numbers of cuboid- and spindle-shaped cells as well as single cells near the wound edge were counted. The results are expressed as mean ± SEM, and the numbers of control cells are set as 100. * P < 0.05; ** P < 0.001.
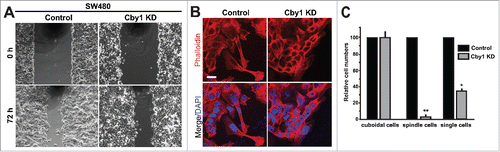
During migration, as revealed by phalloidin staining, control SW480 cells displayed a typical elongated spindle-shaped morphology with lamellipodia at the leading edge (). Intriguingly, however, most of Cby1-KD cells appeared cuboidal and migrated into the wound area as a sheet rather than single cells. Cby1-KD SW480 cells maintained their epithelial actin cytoskeletal rings even 72 h after application of the wound. Quantification of cells at the wound edge revealed a dramatic reduction in the numbers of spindle-shaped cells as well as single cells upon Cby1 KD (). These observations are consistent with the idea that Cby1 KD results in an epithelial phenotype and suggest that the tighter cell-cell adhesion of Cby1-KD cells interferes with single cell, but not sheet, movement into the wound area.
Cby1 KD inhibits anchorage-independent growth
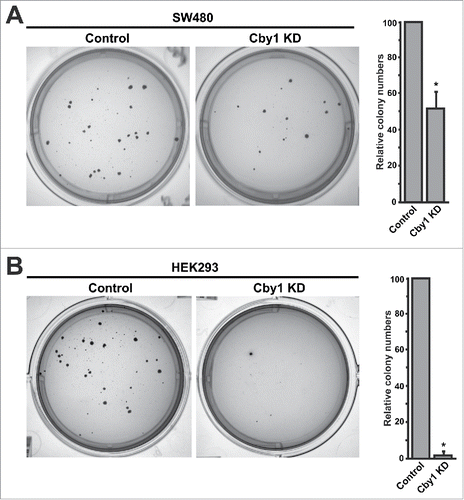
We next performed soft agar colony formation assays to assess the effects of Cby1 KD on anchorage-independent growth. As predicted from their epithelial characteristics, Cby1-KD SW480 cells showed a 48% reduction in colony formation in comparison with control shRNA cells (). Typically, the size of individual Cby1-KD colonies was smaller. Consistent with the more dramatic phenotype of Cby1-KD HEK293 cells, they exhibited a marked decrease in colony formation by 97% (). Taken together, these results suggest that Cby1 KD inhibits anchorage-independent growth by promoting MET-like changes in SW480 and HEK293 cells.
Figure 6. Cby1 KD suppresses anchorage-independent growth. A.-B. (Left) Representative images of colonies of control and Cby1-KD SW480 cells (A) or Cby1-KD HEK293 cells (B) in soft agar assays. (Right) The quantification of colony numbers. The results are expressed as mean ± SEM, and the number of control cells was set as 100. * P < 0.05.
Discussion
In this report, we demonstrated that Cby1 KD in SW480 and HEK293 cells results in MET-like changes such as increased levels of β-catenin and E-cadherin () and ZO-1 () at the plasma membrane, and formation of cortical actin rings and reduced vimentin levels (). Both SW480 and HEK293 Cby1-KD cells maintained coherent epithelial properties during wound closure in wound healing assays (). Moreover, Cby1-KD cells were unable to efficiently form colonies in an anchorage-independent manner (). Thus, our findings suggest that Cby1 KD induces epithelial features.
How does Cby1 affect cell-cell contacts and cell morphology? Cby1 is known to physically interact with β-catenin.Citation13,14 We did not detect a physical interaction of Cby1 with E-cadherin, nor a competition of Cby1 with E-cadherin for β-catenin binding (Figure S3). Therefore, it is possible that the morphological changes induced by Cby1 KD are exerted, at least in part, through its ability to interact with β-catenin. One plausible scenario would be that Cby1, likely together with 14–3–3 proteins, retains a pool of β-catenin in the cytoplasm. In the absence of Cby1, β-catenin is targeted to the plasma membrane or translocates into the nucleus, depending on the cellular context. In agreement with this, the most common mode of 14–3–3 functions is to sequester their interacting partners into the cytoplasm.Citation35,36
Cby1 localizes to the base of cilia and plays a crucial role in ciliogenesis.Citation18-22 More recently, we demonstrated that, during differentiation of airway ciliated cells, Cby1 localizes to nascent centrioles and interacts with Rabin8, a guanine nucleotide exchange factor (GEF) for the small GTPase Rab8 to promote Rab8 recruitment for efficient assembly of membranous structures, called ciliary vesicles, at the distal end of centrioles.Citation18 Intriguingly, Rab8 has been shown to regulate cell shape, and expression of dominant-negative Rab8 mutants or depletion of Rab8 by small interfering (si) RNAs promotes an epithelial cell-like morphology including tight cell-cell contacts and actin stress fiber formation.Citation37 Therefore, it is possible that Cby1 KD suppresses Rab8 activity, leading to MET-like changes, independent of or in parallel to its association with β-catenin. Clearly, further study will be required to understand the mechanistic details of how loss of Cby1 leads to epithelial phenotypes.
Cby1 was originally isolated as a β-catenin binding protein. We demonstrated that, in the nucleus, Cby1 represses β-catenin-mediated transcriptional activation downstream of canonical Wnt signaling. Acute Cby1 KD using morpholino oligonucleotides in HEK293T cells stimulated TopFlash activity.Citation13 It was therefore surprising to find that stable Cby1-KD SW480 cells exhibited diminished TopFlash activity (). One possibility is that the acute KD and chronic KD of Cby1 elicit differential effects on Wnt signaling, which suggests a complex nature of β-catenin regulation by Cby1. Suppression of β-catenin signaling in SW480 cells by Cby1 KD is most likely attributable to the robust recruitment of E-cadherin and β-catenin to the plasma membrane (). In support of this notion, it was reported that E-cadherin expression alone is sufficient to recruit β-catenin to the plasma membrane and reduce nuclear β-catenin signaling in SW480 cells.Citation38 We speculate that, in cancer cells, elevated expression of Cby1 inhibits their growth as a Wnt/β-catenin antagonist, whereas reduction or loss of Cby1 suppresses metastasis.
In summary, our study revealed that Cby1 KD induces MET-like changes in both SW480 and HEK293 cells with elevated levels of E-cadherin and β-catenin at the plasma membrane. Loss of E-cadherin function through genetic or epigenetic alterations is considered to be a major driver for tumor progression.Citation39,40 Conversely, overexpression of E-cadherin in cancer cells impedes their growth and metastasis.Citation38,39 Therefore, it is conceivable that silencing of Cby1 levels may serve as a potential tool for cancer therapy.
Materials and methods
Cell culture, transfection, and plasmids
HEK293, SW480, and HEK293T cells were obtained from the American Tissue Culture Collection (ATCC) and maintained in DMEM with 10% FBS (Denville Scientific) with 100 U/ml penicillin-streptomycin (Life Technologies) at 37°C with 5% CO2. To generate stable Cby1-knockdown lines, cells were transfected with a SureSilencing pGeneClip shRNA plasmid with a puromycin resistance gene for a scrambled shRNA (negative control) or a Cby1 shRNA (GGCTGAAAGTGGACATCTTAT) (SABiosciences). For transfection, Expressfect (Denville Scientific) was used according to the manufacturer's instructions. Transfected cells were selected with 2.5 μg/ml puromycin (Gemini Bioproducts). Puromycin-resistant single or pooled colonies were isolated, tested for Cby1-knockdown by western blotting, and maintained with 1.25 μg/ml puromycin. At least 3 independent stable cell lines for the control or Cby1 shRNA were characterized to ensure consistent and reproducible results.
pCS2+Cby1 and β-cateninS33Y-Flag were described previously.Citation14 The HA-E-cadherin expression construct was generated by PCR amplification of a full-length human E-cadherin cDNA, followed by subcloning into pCS2+HA.
TopFlash assays
SW480 cells were seeded onto 12-well plates and, 24 h later, cells were transfected with 25 ng of TopFlash or mutant FopFlash luciferase plasmid in triplicate. Luciferase activities were measured 48 h post-transfection using the Dual-Luciferase Reporter Assay System (Promega) with a luminometer (Berthold Technologies) with MikroWin 2000 software (Mikrotek Laborsysteme) as described previously.Citation14,30 A Renilla luciferase expression plasmid (pRL-TK) (25 ng/well) was cotransfected to normalize transfection efficiency. The normalized FopFlash baseline values were subtracted from the corresponding normalized TopFlash values.
Co-immunoprecipitation and western blot analysis
For co-immunoprecipitation assays, transfected HEK293T cells were harvested in ice-cold lysis buffer (20 mM Tris-HCl [pH 8.0], 135 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 1% Triton X-100, and 10% glycerol) with protease inhibitor cocktail (Sigma) and incubated for 20 min on ice with intermittent agitation. Cell lysates were cleared by centrifugation at 12,000 rpm for 30 min at 4°C. The supernatants were then incubated with 1 μg of primary antibody as indicated for 1 h at 4°C, followed by the addition of protein A/G beads (Sigma) and rotation for 1 h. The beads were collected and washed twice with 1 ml of ice-cold lysis buffer before SDS-PAGE. For western blotting, cells were lysed in RIPA or lysis buffer (20 mM Tris-HCl, pH 8.0, 135 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 1% Triton X-100, 10% glycerol) with complete protease inhibitor cocktail (Roche) and rotated at 4°C for 15 min. Cell lysates were then cleared by centrifugation at 12,000 rpm for 10 min at 4°C. Protein concentrations were measured using the DC protein assay (Bio-Rad). Western blotting was performed as described previously.Citation14,17 The primary antibodies used were as follows: rabbit anti-Cby113; mouse anti-GAPDH (Meridian Life Sciences); mouse anti-E-cadherin (BD Transduction Laboratories); mouse anti-ZO-1 (BD Transduction Laboratories); mouse anti-vimentin (Neomarkers); mouse anti-Flag M2 (Sigma); mouse anti-HA (Proteintech). All HRP-conjugated secondary antibodies were purchased from Jackson ImmunoResearch Laboratories. Relative band intensities on western blots were quantified using ImageJ (NIH).
RT-qPCR assays
For quantitative analysis of E-cadherin mRNA levels, total RNA was isolated using the RNeasy Mini kit (Qiagen), and cDNA was synthesized using the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems), following the manufacturer's instructions. qPCR analysis was performed using the Fast SYBR Green Master Mix (Applied Biosystem) on the StepOne Plus Real-Time PCR System (Applied Biosystems). Primer sequences used were from the public RTPrimerDB database (http://www.rtprimerdb.org/index.php)41: E-cadherin ID#1018 and GAPDH ID#813. All samples were analyzed in triplicate, and the relative gene expression was calculated according to the comparative threshold cycle (ΔΔCt) method.
Immunofluorescence and phase-contrast microscopy
For immunofluorescence staining, cells were seeded on glass coverslips and cultured for 48 to 72 h. Cells were then washed with PBS and fixed with fresh 4% paraformaldehyde in PBS for 20 min at room temperature and permeabilized with 0.5% Triton X-100 in PBS for 5 min at room temperature or fixed with ice-cold methanol/acetone (1:1, v/v) for 5 min at room temperature. Cells were blocked with 1% BSA (Sigma) and probed with primary antibodies as indicated. The primary antibodies used were as follows: mouse anti-β-catenin and mouse anti-E-cadherin (BD Transduction Laboratories); mouse anti-ZO-1 (BD Transduction Laboratories). Alexa Fluor 568-conjugated phalloidin was obtained from Molecular Probes. Fluorophore-conjugated secondary antibodies were purchased from Jackson ImmunoResearch Laboratories. Nuclei were stained with DAPI (Sigma), and coverslips were mounted onto glass slides with Fluoromount-G (Southern Biotech). Immunostained cells were analyzed using a Zeiss LSM 510 META confocal laser scanning microscope with LSM Image Browser software (Carl Zeiss). All phase-contrast images were obtained using a Leica DMIL inverted microscope with Leica DFC300 FX camera and Application Suite software.
Wound healing assays
Cells were grown to confluence in 6-well plates in triplicate. The cells were washed carefully with PBS, and a wound was applied with a plastic pipette tip. The cells were washed with PBS, imaged (t = 0 h), and then incubated in media to evaluate cell migration. Phase-contrast images were captured every 24 h to document the closure of the wounded area.
Soft agar assays
SW480 cells were suspended in growth medium containing 0.3% agarose at 1×104 cells/well and layered over 0.6% agarose in growth media in 6-well plates in triplicate. For HEK293 cells, 104 and 5×104 cells/well were used. The agar was left at room temperature for 45 min to solidify before incubation at 37°C with 5% CO2, and growth medium was added 4 h later. Cells were fed 3 times weekly. Four weeks after plating, colonies were stained with 0.005% crystal violet (Sigma) in PBS for 1 h at room temperature, thoroughly washed, and destained with PBS for several hours. Colonies were photographed using a FluorChem 8900 Imaging System with αEase FC software. Colonies were counted under a dissecting microscope. For wells containing more than 1,000 colonies, 5 fields marked on graph paper were counted, and total colony numbers were calculated.
Statistical analysis
Statistical significance was determined by the Student's t-test, and a value of P < 0.05 was considered statistically significant.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
KCCY_A_1281478_Supplementary_material.zip
Download Zip (84 MB)Acknowledgments
We would like to thank Drs. Howard Crawford and Barry Gumbiner for reagents and Ben Cyge for productive discussions.
Funding
This work was supported by a Carol M. Baldwin Breast Cancer Research Award to FQL and an NIH/NHLBI grant (R01HL107493) to KIT.
References
- Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol 2014; 15:178-96; PMID:24556840; http://dx.doi.org/10.1038/nrm3758
- Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest 2009; 119:1420-8; PMID:19487818; http://dx.doi.org/10.1172/JCI39104
- Tsai JH, Yang J. Epithelial-mesenchymal plasticity in carcinoma metastasis. Genes Dev 2013; 27:2192-206; PMID:24142872; http://dx.doi.org/10.1101/gad.225334.113
- Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell 2009; 139:871-90; PMID:19945376; http://dx.doi.org/10.1016/j.cell.2009.11.007
- Nelson WJ, Dickinson DJ, Weis WI. Roles of cadherins and catenins in cell-cell adhesion and epithelial cell polarity. Prog Mol Biol Transl Sci 2013; 116:3-23; PMID:23481188
- Shapiro L, Weis WI. Structure and biochemistry of cadherins and catenins. Cold Spring Harb Perspect Biol 2009; 1:a003053; PMID:20066110; http://dx.doi.org/10.1101/cshperspect.a003053
- Takemaru KI, Ohmitsu M, Li FQ. An oncogenic hub: beta-catenin as a molecular target for cancer therapeutics. In Klussmann E, Scott J, editors. Protein-Protein Interactions as New Drug Targets. Berlin, Germany: Springer; 2008:261-84; PMID:18491056; http://dx.doi.org/10.1007/978-3-540-72843-6_11
- Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer 2013; 13:11-26; PMID:23258168; http://dx.doi.org/10.1038/nrc3419
- Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell 2012; 149:1192-205; PMID:22682243; http://dx.doi.org/10.1016/j.cell.2012.05.012
- Klaus A, Birchmeier W. Wnt signalling and its impact on development and cancer. Nat Rev Cancer 2008; 8:387-98; PMID:18432252; http://dx.doi.org/10.1038/nrc2389
- Yook JI, Li XY, Ota I, Fearon ER, Weiss SJ. Wnt-dependent regulation of the E-cadherin repressor snail. J Biol Chem 2005; 280:11740-8; PMID:15647282; http://dx.doi.org/10.1074/jbc.M413878200
- Yook JI, Li XY, Ota I, Hu C, Kim HS, Kim NH, Cha SY, Ryu JK, Choi YJ, Kim J, et al. A Wnt-Axin2-GSK3beta cascade regulates Snail1 activity in breast cancer cells. Nat Cell Biol 2006; 8:1398-406; PMID:17072303; http://dx.doi.org/10.1038/ncb1508
- Takemaru K, Yamaguchi S, Lee YS, Zhang Y, Carthew RW, Moon RT. Chibby, a nuclear beta-catenin-associated antagonist of the Wnt/Wingless pathway. Nature 2003; 422:905-9; PMID:12712206; http://dx.doi.org/10.1038/nature01570
- Li FQ, Mofunanya A, Harris K, Takemaru K. Chibby cooperates with 14-3-3 to regulate beta-catenin subcellular distribution and signaling activity. J Cell Biol 2008; 181:1141-54; PMID:18573912; http://dx.doi.org/10.1083/jcb.200709091
- Takemaru K, Fischer V, Li FQ. Fine-tuning of nuclear-catenin by Chibby and 14-3-3. Cell Cycle 2009; 8:210-3; PMID:19158508; http://dx.doi.org/10.4161/cc.8.2.7394
- Killoran RC, Fan J, Yang D, Shilton BH, Choy WY. Structural analysis of the 14-3-3zeta/Chibby interaction involved in Wnt/beta-Catenin Signaling. PLoS One 2015; 10:e0123934; PMID:25909186; http://dx.doi.org/10.1371/journal.pone.0123934
- Li FQ, Mofunanya A, Fischer V, Hall J, Takemaru K. Nuclear-cytoplasmic shuttling of Chibby controls beta-catenin signaling. Mol Biol Cell 2010; 21:311-22; PMID:19940019; http://dx.doi.org/10.1091/mbc.E09-05-0437
- Burke MC, Li FQ, Cyge B, Arashiro T, Brechbuhl HM, Chen X, Siller SS, Weiss MA, O'Connell CB, Love D, et al. Chibby promotes ciliary vesicle formation and basal body docking during airway cell differentiation. J Cell Biol 2014; 207:123-37; PMID:25313408; http://dx.doi.org/10.1083/jcb.201406140
- Voronina VA, Takemaru K, Treuting P, Love D, Grubb BR, Hajjar AM, Adams A, Li FQ, Moon RT. Inactivation of Chibby affects function of motile airway cilia. J Cell Biol 2009; 185:225-33; PMID:19364920; http://dx.doi.org/10.1083/jcb.200809144
- Lee YL, Sante J, Comerci CJ, Cyge B, Menezes LF, Li FQ, Germino GG, Moerner WE, Takemaru K, Stearns T. Cby1 promotes Ahi1 recruitment to a ring-shaped domain at the centriole-cilium interface and facilitates proper cilium formation and function. Mol Biol Cell 2014; 25:2919-33; PMID:25103236; http://dx.doi.org/10.1091/mbc.E14-02-0735
- Steere N, Chae V, Burke M, Li FQ, Takemaru K, Kuriyama R. A Wnt/beta-catenin pathway antagonist Chibby binds Cenexin at the distal end of mother centrioles and functions in primary cilia formation. PLoS One 2012; 7:e41077; PMID:22911743; http://dx.doi.org/10.1371/journal.pone.0041077
- Siller SS, Burke MC, Li FQ, Takemaru K. Chibby functions to preserve normal ciliary morphology through the regulation of intraflagellar transport in airway ciliated cells. Cell Cycle 2015; 14:3163-72; PMID:26266958; http://dx.doi.org/10.1080/15384101.2015.1080396
- Fischer V, Brown-Grant D-A, Li F-Q. Chibby suppresses growth of human SW480 colon adenocarcinoma cells through inhibition of beta-catenin signaling. J Mol Signal 2012; 7:6; PMID:22651859; http://dx.doi.org/10.1186/1750-2187-7-6
- Schuierer MM, Graf E, Takemaru K, Dietmaier W, Bosserhoff AK. Reduced expression of beta-catenin inhibitor Chibby in colon carcinoma cell lines. World J Gastroenterol 2006; 12:1529-35; PMID:16570344; http://dx.doi.org/10.3748/wjg.v12.i10.1529
- Gad S, Teboul D, Lievre A, Goasguen N, Berger A, Beaune P, Laurent-Puig P. Is the gene encoding Chibby implicated as a tumour suppressor in colorectal cancer? BMC Cancer 2004; 4:31; PMID:15245581; http://dx.doi.org/10.1186/1471-2407-4-31
- Leo E, Mancini M, Aluigi M, Luatti S, Castagnetti F, Testoni N, Soverini S, Santucci MA, Martinelli G. BCR-ABL1-associated reduction of beta catenin antagonist Chibby1 in chronic myeloid leukemia. PLoS One 2013; 8:e81425; PMID:24339928; http://dx.doi.org/10.1371/journal.pone.0081425
- Karakoula K, Suarez-Merino B, Ward S, Phipps KP, Harkness W, Hayward R, Thompson D, Jacques TS, Harding B, Beck J, et al. Real-time quantitative PCR analysis of pediatric ependymomas identifies novel candidate genes including TPR at 1q25 and CHIBBY at 22q12-q13. Genes Chromosomes Cancer 2008; 47:1005-22; PMID:18663750; http://dx.doi.org/10.1002/gcc.20607
- Henderson BR. Nuclear-cytoplasmic shuttling of APC regulates beta-catenin subcellular localization and turnover. Nat Cell Biol 2000; 2:653-60; PMID:10980707; http://dx.doi.org/10.1038/35023605
- Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, Vogelstein B, Clevers H. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/- colon carcinoma. Science 1997; 275:1784-7; PMID:9065401; http://dx.doi.org/10.1126/science.275.5307.1784
- Fischer V, Brown-Grant DA, Li FQ. Chibby suppresses growth of human SW480 colon adenocarcinoma cells through inhibition of beta-catenin signaling. J Mol Signal 2012; 7:6; PMID:22651859; http://dx.doi.org/10.1186/1750-2187-7-6
- Cavey M, Lecuit T. Molecular bases of cell-cell junctions stability and dynamics. Cold Spring Harb Perspect Biol 2009; 1:a002998; PMID:20066121; http://dx.doi.org/10.1101/cshperspect.a002998
- Hartsock A, Nelson WJ. Adherens and tight junctions: structure, function and connections to the actin cytoskeleton. Biochim Biophys Acta 2008; 1778:660-9; PMID:17854762; http://dx.doi.org/10.1016/j.bbamem.2007.07.012
- Zavadil J, Bitzer M, Liang D, Yang Y-C, Massimi A, Kneitz S, Piek E, Boettinger EP. Genetic programs of epithelial cell plasticity directed by transforming growth factor-beta. Proc Natl Acad Sci U S A 2001; 98:6686-91; PMID:11390996; http://dx.doi.org/10.1073/pnas.111614398
- Das S, Becker B, Hoffmann F, Mertz J. Complete reversal of epithelial to mesenchymal transition requires inhibition of both ZEB expression and the Rho pathway. BMC Cell Biol 2009; 10:94; PMID:20025777; http://dx.doi.org/10.1186/1471-2121-10-94
- Hermeking H. The 14-3-3 cancer connection. Nat Rev Cancer 2003; 3:931-43; PMID:14737123; http://dx.doi.org/10.1038/nrc1230
- Reinhardt HC, Yaffe MB. Phospho-Ser/Thr-binding domains: navigating the cell cycle and DNA damage response. Nat Rev Mol Cell Biol 2013; 14:563-80; PMID:23969844; http://dx.doi.org/10.1038/nrm3640
- Hattula K, Furuhjelm J, Tikkanen J, Tanhuanpaa K, Laakkonen P, Peranen J. Characterization of the Rab8-specific membrane traffic route linked to protrusion formation. J Cell Sci 2006; 119:4866-77; PMID:17105768; http://dx.doi.org/10.1242/jcs.03275
- Gottardi CJ, Wong E, Gumbiner BM. E-cadherin suppresses cellular transformation by inhibiting beta-catenin signaling in an adhesion-independent manner. J Cell Biol 2001; 153:1049-60; PMID:11381089; http://dx.doi.org/10.1083/jcb.153.5.1049
- Jeanes A, Gottardi CJ, Yap AS. Cadherins and cancer: how does cadherin dysfunction promote tumor progression? Oncogene 2008; 27:6920-9; PMID:19029934; http://dx.doi.org/10.1038/onc.2008.343
- Hajra KM, Fearon ER. Cadherin and catenin alterations in human cancer. Genes Chromosomes Cancer 2002; 34:255-68; PMID:12007186; http://dx.doi.org/10.1002/gcc.10083
- Lefever S, Vandesompele J, Speleman F, Pattyn F. RTPrimerDB: the portal for real-time PCR primers and probes. Nucleic Acids Res 2009; 37:D942-5; PMID:18948285; http://dx.doi.org/10.1093/nar/gkn777
