ABSTRACT
Melanoma is cancer of melanin-containing melanocyte cells. This neoplasm is one of the most deadly forms of skin cancer, and currently available therapeutic options are insufficient in significantly improve outcomes for many patients. Therefore, novel targets are required to effectively manage this neoplasm. Several sirtuins have previously been found to be upregulated in melanoma, so in this study, the expression profile of SIRT2 was determined. Employing a tissue microarray containing benign nevi, primary melanomas, and lymph node metastases, we have found that the tissue from lymph node metastases appears to have a significant upregulation of SIRT2 relative to primary tumors across the nuclear, cytoplasmic, and whole cell data. Additionally, SIRT2 staining was found to be higher in the nucleus of metastatic melanomas compared to cytoplasmic staining. As SIRT2 is considered to be a predominantly cytoplasmic protein, this is a novel and very interesting finding. This, combined with previous studies that show other sirtuins are increased in melanoma and involved in cellular proliferation and survival, leads to the suggestion that exploring pan-sirtuin inhibitors may be the best target for the next iteration of melanoma chemotherapeutics.
Introduction
In the United States, it is estimated that over 87,000 new cases of melanoma will be diagnosed in 2017 alone, with almost 10,000 people dying from this deadly neoplasm this year.Citation1 Because the risk of melanoma increases with age and ultraviolet radiation exposure, it is expected that cases and deaths will increase in coming years due to the aging population in the United States and frequent tanning. In its early stages, melanoma is generally able to be surgically removed without further difficulties for the patient. However, if the cancer is not diagnosed in time, or if the operation is difficult due to tumor location or patient health, it is much more likely that there will be significant complications or even death. Currently, non-surgical treatments include radiation therapy or pharmaceutical compounds designed to block targeted signaling pathways, including the BRAF-MEK-ERK pathway inhibitor vemurafenib. These are successful in some patients, but there are significant adverse events associated with them.Citation2,3 Additionally, many tumors develop resistance to currently available therapies, which is a common problem with the use of vemurafenib.Citation4,5 Thus, novel cancer therapeutics are needed to work alone or in combination with existing agents to combat this lethal malignancy.
The sirtuins are a class of enzymes that are involved in a myriad of cellular processes. Traditionally classified as histone deacetylases (HDAC), these enzymes share a common requirement for nicotinamide adenine dinucleotide (NAD) as a co-substrate for their deacetylase activity. Because of this, sirtuins occupy a class of their own in the HDAC hierarchy; class III. The first sirtuin, SIR2, was originally characterized in yeast in 1984 as a transcriptional silencer gene.Citation6 Since then, numerous other sirtuins have been discovered and characterized, including seven genes in humans.Citation7 In addition to their traditional HDAC activity, it has also been found that many sirtuins target non-histone proteins throughout the cell, and are capable of catalyzing several additional post-translational modification reactions.Citation8,9 Given the diversity of the biological processes they modulate, it comes as no surprise that this class of enzymes has been found to be involved in many different diseases in humans, including cancer (reviewed in 10).
In our previous research, we evaluated one of the sirtuin family members, SIRT1, as a new potential target in melanoma therapy.Citation11-13 We observed that several chemical inhibitors of SIRT1 are capable of inhibiting melanoma cell growth and clonogenic survival in vitro. However, the inhibitors that were used in these studies, while specific for sirtuins, had the unintended effect of targeting both SIRT1 and SIRT2. As the sirtuins share a conserved catalytic domain, it is difficult to develop more specific chemical inhibitors, although many efforts have been made by other groups to accomplish this task. In one such effort, Zhang et al developed a novel small molecule SIRT2 inhibitor, AC-93253, which is more potent at inhibiting SIRT2 than SIRT1 or 3 by 7.5- and 4-fold.Citation14 Importantly, AC-93253 demonstrated selective cytotoxic effects in the tested metastatic melanoma cells, suggesting that SIRT2 may play a role in melanoma survival. Further, it has also been shown that SIRT2 may play a role in BRAF and MEK inhibitor resistance in BRAF mutant melanoma.Citation15 In addition, a recent study has shown that SIRT2 levels were higher in uveal melanoma compared to normal melanocytes.Citation16 Based on the known functions of SIRT1 in cancer, it is presumed that a specific inhibitor of SIRT1 would be the preferable treatment, but since the status and role of SIRT2 in cutaneous melanoma are currently unknown, further investigation is required. Therefore, as an adjunct to our published studies regarding the use of SIRT1/SIRT2 combination therapies in melanoma, here, we evaluate the endogenous expression of SIRT2 protein in a small array of melanoma tissue samples.
Results & discussion
A tissue microarray (TMA) containing 18 benign nevi controls, 62 primary melanoma cores, and 20 melanoma lymph node metastases was obtained from US Biomax (Fig. S1A). Tissue cores within the array varied by patient age, gender, tumor location, and stage. The TMA was stained with SIRT2 antibody with hematoxylin counterstain, and scanned using the Vectra Imaging System. Scanned tissue cores were then blindly analyzed by an experienced pathologist using inForm software, with data recorded as an average SIRT2 staining intensity within the nucleus, the cytoplasm, or across the entire cell. shows the entire stained TMA, while provides representative stained sections of the TMA at 20x magnification. Detailed sample information and full results can be found in Supplementary Table 1.
Figure 1. SIRT2 staining is increased in human metastatic melanoma samples. (A) Whole image of the SIRT2/AEC stained slide with hematoxylin counterstaining. ME1004d indicates the catalog number from US Biomax, Inc. that was used for the study. (B) Representative 20x images of individual cores with low, medium, and high SIRT2 expression are shown. Scale bar = 200 μm. (C) The mean whole cell SIRT2 protein levels of each core as determined by Vectra analysis are plotted. Error bars represent mean ± SEM.
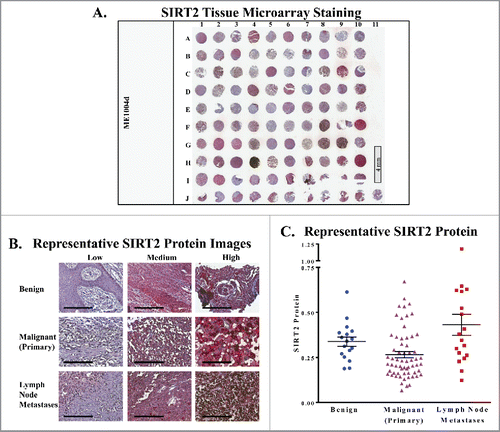
Our results show no significant difference in SIRT2 expression between benign and malignant melanoma samples (). While the mean of the malignant group is slightly lower than that of the benign samples, we are limited by a low number of benign samples available for analysis, and a larger sample size would be necessary to draw any conclusions with statistical significance. However, we did find that the group of tissue from lymph node metastases showed a significant upregulation of SIRT2 relative to primary tumors across the nuclear, cytoplasmic, and whole cell data (). This is an interesting finding, as the therapeutic need for new metastatic treatments is very high. As mentioned earlier, primary melanoma tumors can generally be successfully treated through surgical excision. However, 82% of patients with distant metastases are deceased within 5 years.Citation1 A difference in SIRT2 expression between primary melanomas and metastases could indicate that this protein plays a role in the changes required for tumor cell migration, i.e. processes such as the epithelial mesenchymal transition (EMT), cytoskeletal reorganization, or the development of filopodia. Thus, it shows potential as a therapeutic target, and further exploration into these possibilities is warranted. If SIRT2 is indeed shown to play a role in melanoma metastasis, it could indicate that the development of specific SIRT1 chemical inhibitors is unnecessary, as the currently available SIRT1/SIRT2 combination inhibitors would be not only acceptable, but preferable alternatives.
Figure 2. (A) Mean SIRT2 protein levels of whole cells, cytoplasmic, and nuclear cellular compartments are compared. (B) The ratio of the nuclear and cytoplasmic staining of the cells in each core was determined and the points are plotted by cancer status. Error bars represent mean ± SEM. * P ≤ 0.05; ** P ≤ 0.01 *** P ≤ 0.001.
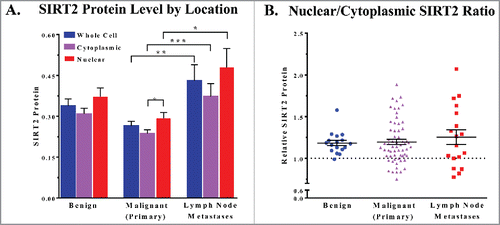
Finally, our results also show both nuclear and cytoplasmic SIRT2 expression in all samples. Furthermore, most samples show an average nuclear staining level that is higher than the cytoplasmic staining level (). As SIRT2 is considered to be a predominantly cytoplasmic protein, this is an interesting finding. Whether this distribution across the cell is tissue specific or nuclear SIRT2 expression is more common than previously thought is unclear. However, considering the size difference between nuclear and cytoplasmic cellular compartments, we speculate that the differences in mean staining likely still indicate a higher overall amount of SIRT2 within the cell resides in the cytoplasm for most of our samples. Further study into the cellular distribution of SIRT2 is needed to evaluate these inconsistencies.
Conclusion
Because melanoma has a high potential to become deadly if not diagnosed early, new treatments are needed for individuals who cannot undergo surgery or whose disease has already become metastatic. Our study suggests that SIRT2 levels may be increased in metastatic human melanoma tissues, compared to primary tumors. Although this data is limited, taken in conjunction with previous research, it strongly indicates that SIRT2 could be involved in metastatic progression. This information, combined with previous research indicating that other sirtuins are involved in cellular proliferation and survival, suggests that using a combination approach targeting multiple sirtuins may be the best course for their successful use as chemotherapeutics in melanoma.
Materials and methods
Immunohistochemical staining
Human melanoma tissue microarray purchased from US Biomax (Cat #ME1004d) was deparaffinized with a standard xylene and ethanol series followed with H2O2 incubation. Antigen unmasking was performed using Antigen Unmasking Solution (Cat #H-3300; Vector Laboratories) by simmering in a microwave for 30 minutes. Slides were blocked in 2.5% normal horse blocking serum for 30 minutes at room temperature followed by incubation with SIRT2 antibody (Cat #HPA011165; Sigma-Aldrich; 1:25) for 1 h at room temperature. After washing in PBS, slides were then incubated for 30 minutes with ImmPRESS HRP Anti-Rabbit IgG (Cat #MP-7401, Vector Laboratories) and developed with AEC Peroxidase (HRP) Substrate Kit (Cat #SK-4200, Vector Laboratories), counterstained with hematoxylin and mounted using VectaMount AQ (Cat #H-5501, Vector Laboratories). Single stained control slides of AEC and hematoxylin were made as controls for developing spectral libraries for Vectra analysis (below).
Image acquisition
The Vectra® automated quantitative tissue imaging system (PerkinElmer) was used for quantitative staining analysis. This system includes an automated slide scanner and state-of-the-art software (Nuance® and inForm®; PerkinElmer). Each chromogen has unique spectral characteristics (curve), which is the basis for spectral imaging. To acquire TMA images, the stained TMA slide was loaded onto the Vectra slide scanner. A scanning protocol including the spectral library was created based on the TMA core size and layout. An 8-bit image cube was acquired from each of the TMA tissue cores for analysis. This data was then used to analyze the protein levels in the individual cores using inForm software.
Image analysis
inForm software (PerkinElmer) was used to segment tissues (melanoma vs. others, etc.) and cells (nucleus vs. cytoplasm) using the data acquired as described above. The spectral library was used to unmix the signals on the acquired image cubes stained with AEC and hematoxylin by recognizing their unique spectral curves. Then, the target -SIRT2 signals (AEC staining) were quantitated within the tissue and subcellular compartment(s) of interest. By such, signal noises and cross-talk are eliminated. Continuous signal intensity data (mean optical density per pixel) was generated for each TMA core or cellular compartment.
Statistical analysis
Data were analyzed using GraphPad Prism software (GraphPad Software). Statistical significance was determined by one-way ANOVA with Tukey's post hoc test or by two-tailed Student's t- test. Data is shown as mean ± SEM unless otherwise specified. A p-value < 0.05 was considered to be statistically significant.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
1288323_Supplemental_Material.docx
Download MS Word (71.1 KB)Funding
This work was supported by funding from the NIH under Grants #R01AR059130 and #R01CA176748 to NA, and the Department of Veterans Affairs under the VA Merit Review Award #I01BX001008 to NA. We also acknowledge the core facilities supported by the Skin Diseases Research Center (SDRC) Core Grant P30AR066524 from NIH/NIAMS and UW Carbone Comprehensive Cancer Center Core Grant P30CA014520 from NIH/NCI.
References
- Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017; 67(1):7-30; http://dx.doi.org/10.3322/caac.21387
- Mavropoulos JC, Wang TS. Managing the skin toxicities from new melanoma drugs. Curr Treat Options Oncol 2014; 15(2):281-301; PMID: 24867225; http://dx.doi.org/10.1007/s11864-014-0284-6
- Peuvrel L, Quereux G, Saint-Jean M, Brocard A, Nguyen JM, Khammari A, Knol AC, Varey E, Dreno B. Profile of vemurafenib-induced severe skin toxicities. J Eur Acad Dermatol Venereol 2016; 30(2):250-7; PMID: 26524690; http://dx.doi.org/10.1111/jdv.13443
- McArthur GA, Chapman PB, Robert C, Larkin J, Haanen JB, Dummer R, Ribas A, Hogg D, Hamid O, Ascierto PA, et al. Safety and efficacy of vemurafenib in BRAF(V600E) and BRAF(V600K) mutation-positive melanoma (BRIM-3): extended follow-up of a phase 3, randomised, open-label study. Lancet Oncol 2014; 15(3):323-32; PMID: 24508103; http://dx.doi.org/10.1016/S1470-2045(14)70012-9
- Spagnolo F, Ghiorzo P, Queirolo P. Overcoming resistance to BRAF inhibition in BRAF-mutated metastatic melanoma. Oncotarget 2014; 5(21):10206-21; PMID: 25344914; http://dx.doi.org/10.18632/oncotarget.2602
- Shore D, Squire M, Nasmyth KA. Characterization of two genes required for the position-effect control of yeast mating-type genes. EMBO J. 1984; 3(12):2817-23; PMID: 6098447
- Costantini S, Sharma A, Raucci R, Costantini M, Autiero I, Colonna G. Genealogy of an ancient protein family: the Sirtuins, a family of disordered members. BMC Evol Biol 2013; 13:60; PMID: 23497088
- Osborne B, Bentley NL, Montgomery MK, Turner N. The role of mitochondrial sirtuins in health and disease. Free Radic Biol Med 2016; pii: S0891-5849(16):30217-9
- Yuan H, Su L, Chen WY. The emerging and diverse roles of sirtuins in cancer: a clinical perspective. Onco Targets Ther 2013; 6:1399-416
- Bedalov A, Chowdhury S, Simon JA. Biology, Chemistry, and Pharmacology of Sirtuins. Methods Enzymol 2016; 574:183-211; PMID: 27423863
- Singh CK, George J, Nihal M, Sabat G, Kumar R, Ahmad N. Novel downstream molecular targets of SIRT1 in melanoma: a quantitative proteomics approach. Oncotarget 2014; 5(7):1987-99; PMID: 24743044; http://dx.doi.org/10.18632/oncotarget.1898
- Wilking MJ, Singh C, Nihal M, Zhong W, Ahmad N. SIRT1 deacetylase is overexpressed in human melanoma and its small molecule inhibition imparts anti-proliferative response via p53 activation. Arch Biochem Biophys 2014; 563:94-100; PMID: 24751483
- Wilking MJ, Singh CK, Nihal M, Ndiaye MA, Ahmad N. Sirtuin deacetylases: a new target for melanoma management. Cell Cycle 2014; 13(18):2821-6; PMID: 25486469; http://dx.doi.org/10.4161/15384101.2014.949085
- Zhang Y, Au Q, Zhang M, Barber JR, Ng SC, Zhang B. Identification of a small molecule SIRT2 inhibitor with selective tumor cytotoxicity. Biochem Biophys Res Commun 2009; 386(4):729-33; PMID: 19559674; http://dx.doi.org/10.1016/j.bbrc.2009.06.113
- Bajpe PK, Prahallad A, Horlings H, Nagtegaal I, Beijersbergen R, Bernards R. A chromatin modifier genetic screen identifies SIRT2 as a modulator of response to targeted therapies through the regulation of MEK kinase activity. Oncogene 2015; 34(4):531-6; PMID: 24469059; http://dx.doi.org/10.1038/onc.2013.588
- Halfed DG, Zoroquiain P, Wood HA, Blanco P, Al-Saati N, Aldrees S, Bravo-Filho V, Burnier MN. SIRT2 Expression Is Higher in Uveal Melanoma than In Ocular Melanocytes. Ocul Oncol Pathol 2015; 2(2):100-4; PMID: 27171429; http://dx.doi.org/10.1159/000439309
