ABSTRACT
Recently, emerging evidence has demonstrated that metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), a long non-coding RNAs (lncRNAs), contributes to the initiation and development of tumors, including osteosarcoma (OS). Multiple studies have suggested an oncogenic role of MALAT1 and high-mobility group protein B1 (HMGB1) in OS tumorigenesis and metastasis, but the effects and mechanisms are not unanimous. Here, we showed that MALAT1 and HMGB1 were significantly increased in human OS cell lines and knockdown of MALAT1 reduced HMGB1 expression. By using online tools, we screen out 2 candidate miRNAs, miR-142–3p and miR-129–5p which may be associated with both MALAT1 and HMGB1. Luciferase reporter assay revealed a direct interaction between the 2 miRNAs and MALAT1, respectively, via a putative binding site within MALAT1. Meanwhile, both the 2 miRNAs could bind to HMGB1 3′-untranslated region (3′-UTR) and regulate HMGB1 expression. Moreover, knockdown of MALAT1 decreased HMGB1 expression, inhibited OS cell growth and promoted apoptosis, while miR-142–3p and miR-129–5p inhibitor partly restored the inhibitory effect of MALAT1 knockdown on HMGB1 expression, OS cell growth and the promotion of apoptosis. In OS tissues, the expression of MALAT1 and HMGB1 was upregulated while the expression of miR-142–3p and miR-129–5p was downregulated. Together, our results support a MALAT1/miR-142–3p/miR-129–5p/HMGB1 axis in OS cell proliferation and tumor progression. MALAT1 promoted OS cell growth through inhibition of miR-142–3p or miR-129–5p and by targeting HMGB1.
Introduction
Osteosarcoma (OS) is a highly malignant bone tumor predominantly seen in childhood and adolescence with an annual incidence rate of about 5.6 per million in the USA.Citation1 In young patients, it arises most often in the metaphyses of long bones such as the distal femur, and the major cause of death in osteosarcoma is metastasis to the lungs. To enhance osteosarcoma therapy, new therapeutic targets must be identified, and therapeutic strategies based on the most effective combinatorial approaches must be developed.
It has been demonstrated that more than 90% of the human DNA sequence is actively transcribed but only 2% of it encodes proteins. The majority of transcripts are referred to as non-coding RNAs (ncRNAs).Citation2,3 Small non-coding RNAs, especially, microRNAs, have been studied extensively and their roles in gene regulation and cellular function have been elucidated in numerous cancers.Citation3 For example, our previous research has demonstrated the suppressive function of miR-142–3p on osteosarcoma.Citation4 Recently, long non-coding RNAs (lncRNAs) have been reported to play important roles during development and disease, including cancer.Citation5,6 LncRNAs can be oncogenic or function as a tumor suppressor.Citation7,8 HOTAIR, a cell cycle-associated lncRNA, has been shown to play an oncogenic role in multiple cancers, including breast, gastric, colorectal, and cervical cancers,Citation9 while TUSC7 (tumor suppressor candidate 7), another lncRNA, has been shown to function as a tumor suppressor in hepatocellular carcinoma and gastric cancer.Citation10,11 MALAT1, also a lncRNA, has been reported to be upregulated in several cancers, including lung, breast, pancreas, liver, colon, gastric, uterus, cervix and prostate cancers.Citation12,13 MALAT1 may serve as an independent prognostic biomarker for survival of these cancer cells. MALAT1 expression was recently associated with OS cell fate, as MALAT1 knockdown delayed tumor growth in an OS xenograft model, suggesting its oncogenic role and the potential as a therapeutic target for OS treatment.Citation14 MALAT1 has also been shown to promote OS cell growth and metastasis, possibly via activation of the PI3K/AKT signaling pathway.Citation15 While these findings demonstrated a clear correlation between MALAT1 and OS, the specific effect of MALAT1 on OS tumorigenesis and the mechanisms involved remain to be determined. High mobility group box 1 (HMGB1), as chromatin-binding nuclear protein is highly expressed in many types of cancer cells and linked to cancer development by interfering with several signaling pathway.Citation16
In this study, we report detection of a negative correlation between miR-142–3p/miR-129–5p andMALAT1 or HMGB1 expression in human OS cell lines and tissues. We also showed that miR-142–3p/miR-129–5p dependent HMGB1 activation was required for MALAT1 induced OS cell growth. In addition, direct interaction between miR-142–3p/miR-129–5p and the 3′-untranslated regions (UTRs) of MALAT1 andHMGB1 was observed, suggesting a novel mechanism of MALAT1, miR-142–3p/miR-129–5p, and HMGB1 in regulation of OS cell growth.
Results
The expression of MALAT1 and HMGB1 in human OS cell lines
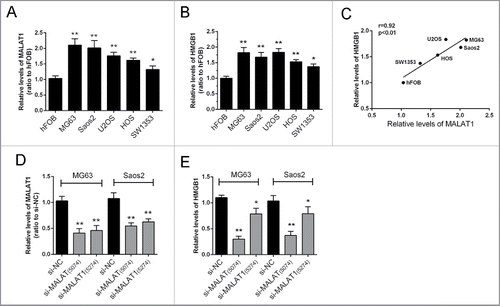
The expression levels of MALAT1and HMGB1 in 5 human OS cell lines, Saos2, MG63, SW1353, U2OS, HOS, and a normal cell line, hFOB were examined using real-time qPCR assay. The results showed that MALAT1 and HMGB1 expression were significantly higher in tumor cells compared with normal cells (). A positive correlation between the relative expression of MALAT1 and HMGB1 in the above 6 cell lines was observed (). To investigate the regulation of HMGB1 by MALAT1, 2 MALAT1 siRNAs were transfected into MG63 and Saos2 cells. The inhibitory efficiency was verified using real-time PCR (); results showed that the expression of MALAT1 could be repressed significantly by either si-MALAT1 (interference site on 5074) or si-MALAT (interference site on 5274). Then the expression of HMGB1 was determined in response to the 2 MALAT1 siRNAs. Results showed that HMGB1 was more significantly repressed by si-MALAT1 compared with si-NC (control) ().
Figure 1. The expression of MALAT1 and HMGB1 in human OS cell lines. (A, B) The expression levels of MALAT1and HMGB1 in 5 OS cell lines, Saos2, MG63, U2OS, SW1353, HOS and normal cells, hFOB were examined by real-time qPCR assay. (C) A positive correlation between MALAT1 and HMGB1 was observed. (D) Two MALAT1 siRNAs, si-MALAT containing the interference site on 5274 and si-MALAT1 containing the interference site on 5074 were generated and transfected into OS cells to achieve MALAT1 knockdown. The inhibitory efficiency was verified using real-time PCR. (E) The expression levels of HMGB1 were monitored in response to 2 MALAT1 siRNAs using real-time PCR. Due to the higher inhibitory efficiency of Si-MALAT1 (5074), it was used for the further study. The data were presented as mean ± SD of 3 independent experiments.
miR-142–3p/miR-129–5p correlated with MALAT1 and HMGB1
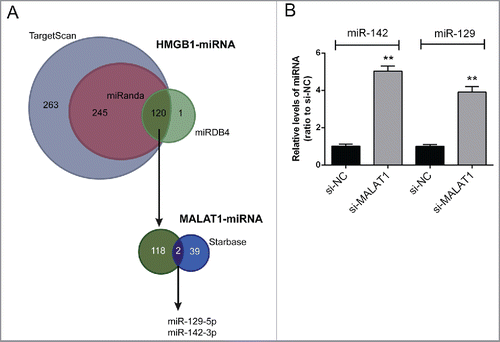
To validate the mechanism by which MALAT1 regulate HMGB1 expression and OS cell proliferation and apoptosis, several online tools were used for scanning out the miRNAs that could be correlated with both MALAT1 and HMGB1. By using TargetScan, miRanda and miRDB4, 120 candidate miRNAs that may bind to HMGB1 3′UTR were scanned out; among which 2 miRNAs were finally chosen for further experiments by using starBase, miR-142–3p and miR-129–5p (). After transfecting si-MALAT1 into Saos2 cells, the expression levels of miR-142–3p and miR-129–5p were significantly upregulated ().
Figure 2. miR-142–3p/miR-129–5p correlated with MALAT1 and HMGB1. (A) By using TargetScan, miRanda and miRDB4 online prediction tools, candidate miRNAs that may bind to HMGB1 3′UTR were scanned out; among which 2 miRNAs were finally chosen for further experiments by using starBase. (B) The expression levels of miR-142–3p and miR-129–5p in Saos2 cells were determined in response to MALAT1 knockdown using real-time PCR. The data were presented as mean ± SD of 3 independent experiments.
miR-142–3p/miR-129–5p binds with MALAT1
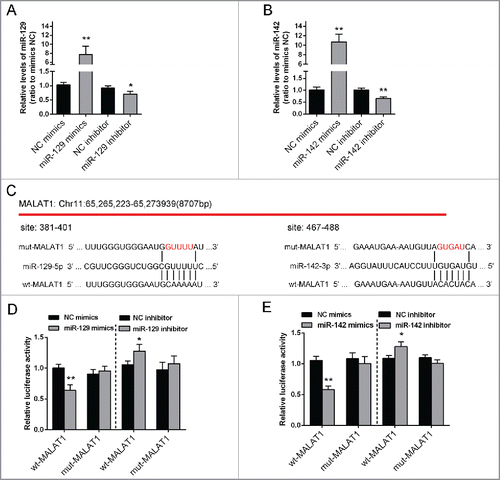
To confirm the correlation between miR-142–3p/miR-129–5p and MALAT1, first, miR-129–5p mimics/miR-129–5p inhibitor or miR-142–3p mimics/miR-142–3p inhibitor was transfected into Saos2 cells, respectively, to verify the transfection efficiency of miR-142–3p/miR-129–5p overexpression or inhibition. (). We successfully generated 2 constructs: a wt-MALAT1 and a mut-MALAT1-containing luciferase reporter vectors. Then mut-MALAT1 contains a 5 bp mutation in the putative miR-142–3p and miR-129–5p binding site within MALAT1 (). These wt-MALAT1/mut-MALAT1 vectors, miR-142–3p mimics or miR-142–3p inhibitor, miR-129–5p mimics or miR-129–5p inhibitor were co-transfected into Saos2 cells respectively. The results showed that compared with the control group, luciferase activity of the wt-MALAT1 vector was significantly reduced in either miR-142–3pmimics or miR-129–5p mimics transfected cells, while promoted in either miR-142–3p inhibitor or miR-129–5p inhibitor transfected cells (). This repression of luciferase activity by miR-142–3p or miR-129–5p was not shown in cells transfected with mut-MALAT1 (). These results suggested a direct interaction betweenmiR-142–3p or miR-129–5p and MALAT1 via the putative miR-142–3p or miR-129–5p binding site within MALAT1.
Figure 3. miR-142–3p/miR-129–5p correlated with MALAT1 through binding MALAT1. (A) miR-129–5p mimics or miR-129–5p inhibitor was transfected into Saos2 cells to achieve miR-129–5p overexpression or inhibition. The transfection efficiency was verified using real-time PCR. (B) miR-142–3p mimics or miR-142–3p inhibitor was transfected into OS cells to achieve miR-142–3p overexpression or inhibition. The transfection efficiency was verified using real-time PCR. (C) Two MALAT1 containing luciferase reporter constructs: wt-MALAT1 and a corresponding mut-MALAT1 which contains a 5 bp mutation in putative miR-142–3p and miR-129–5p binding site within MALAT1. These luciferase reporter constructs were co-transfected into Saos2 cells with miR-142–3p mimics/miR-129–5p mimics or miR-142–3p inhibitor/miR-129–5p inhibitor respectively. (D) and (E) The luciferase activity of the wt-MALAT1 reporter and mut-MALAT1 was monitored in different groups, compared with NC mimics or NC inhibitor group. The data were presented as mean ± SD of 3 independent experiments.
miR-142–3p/miR-129–5p regulated HMGB1 expression by binding its 3′ UTR
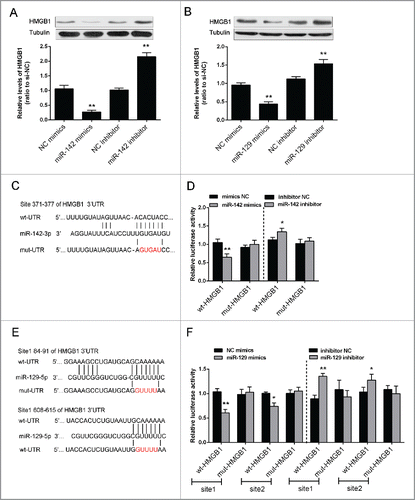
The protein expression of HMGB1 was monitored in response to overexpression or inhibition of the 2 miRNAs using Western blot assays. Results showed that the protein expression of HMGB1 could be upregulated by the inhibition of either miR-142–3p or miR-129–5p, while downregulated by the overexpression of either miR-142–3p or miR-129–5p in Saos2 cells (). To validate the mechanism by which miR-142–3p and miR-129–5p regulated HMGB1, we generated 2 HMGB1 containing luciferase reporter constructs: wt-HMGB1 and a corresponding mut-HMGB1 which contains a 5 bp mutation in putative miR-142–3p binding site or 2 miR-129–5p binding sites within its 3′UTR (). These luciferase reporters were co-transfected intoSaos2 cells with miR-142–3p mimics or miR-142–3p inhibitor, miR-129–5p mimics or miR-129–5p inhibitor respectively. The luciferase activity of the wt-HMGB1 reporterwassignificantly reduced by either miR-142–3p mimics or miR-129–5p mimics transfection, compared with the control groups (). Meanwhile, no significant reduction of the reporter activity was shown in cells co-transfected with miR-142–3p or miR-129–5p and the mut-HMGB1 reporter, suggesting that miR-142–3p and miR-129–5p directly regulates HMGB1 through interaction with its 3′UTR ().
Figure 4. miR-142–3p/miR-129–5p regulated HMGB1 expression by binding its 3′ UTR. (A) and (B) The protein expression of HMGB1 was monitored in response to miR-129–5p or miR-142–3p overexpression or inhibition in Saos2 cells using Western blot assays. The data were presented as mean ± SD of 3 independent experiments. (C) and (E) Two HMGB1 containing luciferase reporter constructs: wt-HMGB1 and a corresponding mut-HMGB1 which contains a 5 bp mutation in one putative miR-142–3p and 2 miR-129–5p binding sites within its 3′-UTR. These luciferase reporter constructs were co-transfected into Saos2 cells with miR-142–3p mimics/miR-129–5p mimics or miR-142–3p inhibitor/miR-129–5p inhibitor respectively. (D) and (F) The luciferase activity of the wt-HMGB1 reporter and mut-HMGB1 was monitored in different groups, compared with NC mimics or NC inhibitor group. The data were presented as mean ± SD of 3 independent experiments.
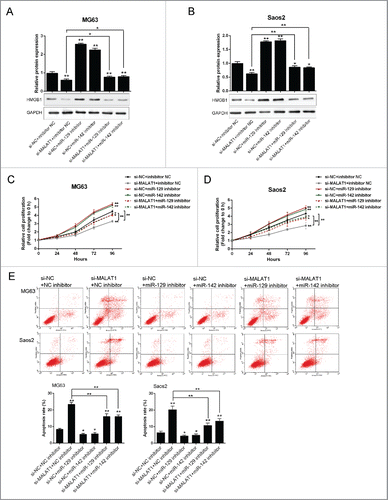
The role of MALAT1 and miR-142–3p/129–5p in OS proliferation and apoptosis
To investigate the exact role of MALAT1 and miR-142–3p/129–5p in OS cell proliferation and apoptosis, further experiments were arranged. As exhibited by Western blot assays, the protein expression of HMGB1 was suppressed after si-MALAT1 transfection while increased by miR-142–3p or miR-129–5p inhibitor transfection; miR-142–3p or miR-129–5p inhibitor could partly restore the inhibition effect of MALAT1 knockdown on HMGB1 expression (). Similar results were observed in MG63 and Saos2 cells. Next, the proliferation of MG63 and Saos2 cells was determined using MTT assays. Results showed that MALAT1 knockdown by si-MALAT1 significantly inhibited the proliferation of both MG63 and Saos2 cells; miR-142–3p or miR-129–5p inhibitor promoted the proliferation of both MG63 and Saos2 cells, and could partly restore the inhibitory effect of MALAT1 knockdown on OS cells (). As exhibited by flow cytometer assay, MALAT1 knockdown by si-MALAT1 significantly promoted the apoptosis of both MG63 and Saos2 cells; miR-142–3p or miR-129–5p inhibitor inhibited the apoptosis of both MG63 and Saos2 cells, and could partly restore MALAT1 knockdown induced cell apoptosis of OS cells ().
Figure 5. The role of MALAT1 and miR-142–3p/129–5p in OS proliferation and apoptosis (A) and (B) The protein expression of HMGB1 in MG63 and Saos2 was monitored in response to co-transfection with si-NC/si-MALAT1 and miR-142–3p/129–5p using Western blot assays. (C) and (D) The proliferation of MG63 and Saos2 cells was monitored in response to co-transfection with si-NC/si-MALAT1 and miR-142–3p/129–5p using MTT assays. (E) The apoptosis of MG63 and Saos2 cells was monitored in response to co-transfection with si-NC/si-MALAT1 and miR-142–3p/129–5p using Flow cytometer assays. The data were presented as mean ± SD of 3 independent experiments.
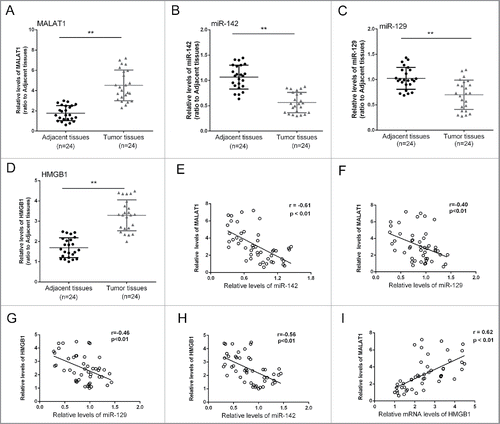
The expression differences of MALAT1/miR-142–3p/miR-129–5p/HMGB1 in tissues and the correlations
To further explore the correlation of MALAT1, miR-142, miR-129 and HMGB1, we obtained 24 pairs of tumor and adjacent normal tissues, determined the expression levels of MALAT1, miR-142, miR-129 and HMGB1, and generated the correlation analysis. Results showed that the expression levels of MALAT1 and HMGB1 were significantly increased in tumor tissues compared with the adjacent normal tissues, while miR-142and miR-129 expression was reduced (). Moreover, an inverse correlation between the expression level of miR-142 and MALAT1, miR-142 and HMGB1, miR-129 and MALAT1, miR-129 and HMGB1, and a positive correlation between the expression level of MALAT1 and HMGB1 was observed ().
Figure 6. The expression differences of MALAT1/miR-142–3p/miR-129–5p/HMGB1 in tissues and the correlations. (A, B, C and D) 24 pairs of tumor and adjacent normal tissues were obtained, the expression levels of MALAT1, miR-142–3p, miR-129–5p and HMGB1 were determined. (D, E, F, G and I) An inverse correlation between the expression level of miR-142–3p and MALAT1, miR-142–3p and HMGB1, miR-129–5p and MALAT1, miR-129–5p and HMGB1, and a positive correlation between the expression level of MALAT1 and HMGB1 was observed. The data were presented as mean ± SD of 3 independent experiments.
Discussion
LncRNA MALAT1 has been shown to be upregulated in many cancers including OS.Citation12,13,17 HMGB1, as chromatin-binding nuclear protein, is also highly expressed in many types of cancer cells and promotes tumor development.Citation16,18,19 In our study, we have shown that MALAT1 and HMGB1 expression were notably higher in all 5 OS cell lines compared with normal cell line. We have also shown that knocking-down ofMALAT1by siRNA resulted in significantly decreased HMGB1 expression in OS cells. Besides, the relative expression of MALAT1 and HMGB1 in 6 cell lines showed to be positively correlated. Multiple molecular mechanisms have been proposed by which MALAT1 promotes cancer cell proliferation in a variety of malignancies. MALAT1 was shown to promote tumor-driven angiogenesis by up-regulating pro-angiogenic gene expression in neuroblastoma.Citation20 MALAT1 may promote CRC development through directly targeting AKAP-9.Citation21 Interestingly, it has been shown that MALAT1 promoted proliferation and metastasis of KIRC, possibly through inhibition of miR-200s, as forced overexpression of miR-200s partially attenuated the promotive effect of MALAT1 on growth and metastasis of KIRC.Citation22 Moreover, a negative correlation between miR-101 or miR-217 and MALAT1 was previously observed in esophageal squamous cell carcinoma, while knockdown ofMALAT1 inhibited cell growth, migration, and invasion.Citation23 These results provide a direct evidence of lncRNA-microRNA interaction in regulation of cancer growth and metastasis. Besides, our previous study has confirmed that HMGB1 was a critical factor in the development of osteosarcoma chemoresistance.Citation24 Several studies have revealed that HMGB1 could be regulated by miRNAs in cancers.Citation25-28 These inspired us to predict that MALAT1 might regulate HMGB1 expression through miRNAs to modulate OS proliferation and apoptosis.
So we used online tools to scan out 2 candidate miRNAs, miR-142–3p and miR-129–5p, which may have binding sites with MALAT1 and the 3′UTR of HMGB1. MiR-142–3p has been regarded as a tumor suppressive microRNA, and has been associated with multiple cancers, including breast cancer, non-small-cell lung cancer, and hepatic cancer,Citation29-31 as well as miR-129–5p.Citation32,33 Luo et al. reported that miR-129–5p attenuates irradiation-induced autophagy and decreases radio-resistance of breast cancer cells by targeting HMGB1.Citation28 MiR-142–3p inhibits chondrocyte apoptosis and inflammation in osteoarthritis by targeting HMGB1.Citation34 In our study, by using a luciferase reporter assay, we detected a direct interaction between miR-142–3p or miR-129–5p and MALAT1, respectively. MiR-142–3p and miR-129–5p may correlate with MALAT1 via the putative binding site within MALAT1. Meanwhile, a significantly increased HMGB1 expression was observed in OS cells in response to either miR-142 or miR-129 inhibition, and a reduced HMGB1 expression in OS cells in response to either miR-142 or miR-129 overexpression. Similarly, by using a luciferase reporter assay, we detected direct interaction between miR-142–3p or miR-129–5p and HMGB1, respectively.
We further investigated whether MALAT1 could regulate OS cell proliferation and apoptosis through modulating miR-142–3p and miR-129–5p expression. Knockdown of MALAT1 decreased HMGB1 expression, inhibited OS cell growth and promoted apoptosis, while miR-142–3p and miR-129–5p inhibitor partly restored the inhibitory effect of MALAT1 knockdown on HMGB1 expression, OS cell growth and the promotion of apoptosis. The effect of MALAT1 on OS cells could be partly restored by inhibitor of miR-142–3p and miR-129–5p, suggesting that MALAT1 functions as an oncogene in OS, and it might exert its functions through promoting HMGB1 expression via inhibiting miR-142–3p and miR-129–5p. The similar functions of MALAT1 in tumor growth and cell apoptosis also have been reported in cervical cancer and renal cell carcinoma.Citation35,36
In 24 pairs of tumor and adjacent normal tissues, the expression of MALAT1, HMGB1, miR-142–3p and miR-129–5p was monitored. A significantly increased expression of MALAT1 and HMGB1 was observed in OS tissues, compared with adjacent normal tissues; a repressed expression of miR-142–3p and miR-129–5p was observed in OS tissues, compared with adjacent normal tissues, consistent with our in vitro experiment results. Both miR-142–3p and miR-129–5p was inversely correlated with MALAT1 and HMGB1, while MALAT1 positively correlated with HMGB1, indicating that MALAT1 could affect OS cell proliferation and apoptosis through miR-142–3p or miR-129–5p independent HMGB1 regulation.
In conclusion, we detected varied expression of MALAT1, miR-142–3p or miR-129–5p and HMGB1 in OS cell lines and tissues. All 4 genes have been associated with OS tumor progression. We have shown that HMGB1 distinctly correlated with MALAT1 and miR-142–3p or miR-129–5p expression in OS. More importantly, direct interaction between miR-142–3p or miR-129–5p and MALAT1 or HMGB1 was demonstrated. Our results support a MALAT1/miR-142–3p/miR-129–5p/HMGB1 axis in OS cell proliferation and tumor progression. MALAT1 may promote OS cell growth through inhibition of miR-142–3p or miR-129–5p and by targeting HMGB1, although other mechanisms could not be excludedCitation37
Materials and methods
Cell lines
Human OS cell lines, Saos2, MG63, U2OS, SW1353, HOS and normal cells, hFOB, were purchased from American Type Culture Collection.
Tissue specimens
24 paired OS specimens and corresponding adjacent non-tumor tissues were collected from tumor surgical resection in The Second Xiangya Hospital of Central South University (Changsha, China). All the human tissues were obtained with informed consent and this study was approved by the Clinical Research Ethics Committee of The Second Xiangya Hospital of Central South University.
Plasmid construction
Full length human HMGB1 cDNA generated by reverse transcription-polymerase chain reaction (using olig 5′- CCC AAG CTT ATG GGC AAA GGA GAT CCT-3′ and 5′- CGC GGA TCC TTA ATC ATC ATC ATC ATC-3′) with HindIII and BamH I sites at the 5′ and 3′ ends was cloned into the pCMV expression vector.
Cell transfection
Both the miRNA mimics and siRNAs were synthesized by Genepharma Company (Shanghai, China). Cells were seeded in 6-well plates at a concentration of 2 × 105 cells/well. When cells reached 40–60% confluence, 2 µg overexpression plasmid, 150nMmiR-142–3p mimics and miR-129–5p mimics or negative control (mimics NC) was transfected by using the Lipofectamine™ 2000 transfection reagent (Invitrogen, USA) and followed the protocol recommended by the manufacturer. After 48 h transfection, the cells were collected and used for further analysis as described below.
MTT assay
Cell proliferation assay using the MTT kit (Promega Corporation, Madison, WI, USA) was performed according to the manufacturer's instructions. Briefly, cells were seeded into 96-well plates at a density of 5000 cells per well and grown for 24 hours. The cells were then transfected with 100 nM miR-142–3p mimics/miR-142–3p NC or si-NC/si-MALAT1 or si-NC/si-HMGB1. After 24h's transfection, 20 μl of 5 mg/mL MTT was added and further incubated for 4 h in a humidified incubator. 200 μL of DMSO was added to dissolve the formazan after supernatant removed. The optical density (OD) was measured at 490 nm.
Flow cytometer assay
For apoptosis analysis, quantification of apoptotic cells was performed with Annexin V-FITC apoptosis detection kit (Keygen, China). Briefly, the cell samples were harvested with 0.25% trypsin without EDTA after 48 hours of infection and then washed twice with ice-cold PBS and re-suspended in 500 μl binding buffer. Then cells were incubated with 5 μl Annexin V-FITC specific antibodies and 5 μl propidium iodide (PI) then incubated for 15–20 minutes in dark and detected by BD Accuri C6 flow cytometer (BD, USA) with the excitation wavelength of Ex = 488 nm and emission wavelength of Em = 530 nm. Each experiment was repeated 3 times in triplicate.
Western blot
RIPA buffer (Sigma-Aldrich, USA) was used to lyse cell lysates with Complete Protease Inhibitor Cocktail (Roche, USA). Cell lysates were transferred to 1.5 mL tube and kept at −20°C before use. SDS-PAGE was conducted to separate the cellular proteins. Proteins were separated by 5% stacking gel and 10% running gel. The molecular weight of candidate proteins was referred to the information of the Pre-stained See Blue rainbow marker (Invitrogen, USA) loaded in parallel. The following antibodies were used: MALAT1 (Santa Cruz, USA), HMGB1 (Abcam, MA, USA), and β-actin (Sigma, USA). The blots were detected on Kodak film developer (Fujifilm, Japan).
RNA extraction and real-time PCR
Total RNAs were extracted using the TRIZOL reagent (Invitrogen, USA) and followed the manufacturer's instructions. High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, USA) was used to reversely transcribe RNA samples. The quantitative RT-PCR was performed using the Fast Start Universal SYBR Green Master (Roche, USA). The primers were showed in . The relative folds changes of candidate genes were analyzed using the 2−ΔΔCt method.
Table 1. The primers used for real-time PCR.
Luciferase reporter assay
The wild type or mutant type of MALAT1 containing the putative target site for miR-142–3p was chemically synthesized and inserted downstream of the luciferase gene in the internal control pRSV-β-Galactosidase vector. Saos2 cells cultured in 24 well plates were co-transfected with luciferase reporter plasmids and miRNA mimics as well as the internal control pRSV-β-Galactosidase vector. After transfection for 48 h, Saos2 cells were lysed using the lysis buffer (25 mM Tris-phosphate, 1% Triton X-100, 1 mM DTT, 2 mM EDTA, 10% Glycerol, pH = 27.8). Cells were then collected and centrifuged at 14,000 rpm for 3 minutes, and the supernatant transferred to a new 1.5ml tube. Luciferase reporter activity was monitored by mixing 50 μl supernatant with 50 μl luciferase assay buffer using the Gloxmax 20/20 Luminometer (Promega). O-nitrophenyl- β-galactoside (ONPG) colorimetric assays were performed to measure the β-Galactosidase activity from the pRSV-β-Galactosidase vector, which was used fornormalization of the luminescence levels. 50 μl supernatant from aforementioned cell extract was mixed with 100 μl of ONPG solution (0.666 mg/ml ONPG, 40 mM NaH2PO4, 60 mM Na2HPO4, 10 mM KCl, 1 mM MgSO4, 2% β-mercaptoethanol) and β-Galactosidase activity was measured using the ELISA plate reader (Bio-Rad, USA) at the wavelength of 490nm.
Statistical analysis
Experimental results are presented as mean ± SD. Comparisons between 2 groups were conducted using 2-tailed Student's T-test and differences were considered to be statistically significant when P value is less than 0.05.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by The National Science Foundation of China (number 81302338, 81272947and 81402247).
References
- Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res 2009; 152:3-13; PMID:20213383; http://dx.doi.org/10.1007/978-1-4419-0284-9_1
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature 2012; 489:101-8; PMID:22955620; http://dx.doi.org/10.1038/nature11233
- Martens-Uzunova ES, Bottcher R, Croce CM, Jenster G, Visakorpi T, Calin GA. Long noncoding RNA in prostate, bladder, and kidney cancer. Eur Urol 2014; 65:1140-51; PMID:24373479; http://dx.doi.org/10.1016/j.eururo.2013.12.003
- Zheng Z, Ding M, Ni J, Song D, Huang J, Wang J. MiR-142 acts as a tumor suppressor in osteosarcoma cell lines by targeting Rac1. Oncol Rep 2015; 33:1291-9; PMID:25530132; http://dx.doi.org/10.3892/or.2014.3687
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell 2009; 136:629-41; PMID:19239885; http://dx.doi.org/10.1016/j.cell.2009.02.006
- Di Gesualdo F, Capaccioli S, Lulli M. A pathophysiological view of the long non-coding RNA world. Oncotarget 2014; 5:10976-96; PMID:25428918; http://dx.doi.org/10.18632/oncotarget.2770
- Zhou S, Wang J, Zhang Z. An emerging understanding of long noncoding RNAs in kidney cancer. J Cancer Res Clin Oncol 2014; 140:1989-95; PMID:24816785; http://dx.doi.org/10.1007/s00432-014-1699-y
- Malek E, Jagannathan S, Driscoll JJ. Correlation of long non-coding RNA expression with metastasis, drug resistance and clinical outcome in cancer. Oncotarget 2014; 5:8027-38; PMID:25275300; http://dx.doi.org/10.18632/oncotarget.2469
- Bhan A, Mandal SS. LncRNA HOTAIR: A master regulator of chromatin dynamics and cancer. Biochim Biophys Acta 2015; 1856:151-64; PMID:26208723; http://dx.doi.org/10.1016/j.bbcan.2015.07.001
- Wang Y, Liu Z, Yao B, Dou C, Xu M, Xue Y, Ding L, Jia Y, Zhang H, Li Q, et al. Long non-coding RNA TUSC7 acts a molecular sponge for miR-10a and suppresses EMT in hepatocellular carcinoma. Tumour Biol 2016; 37:11429-41; doi: http://dx.doi.org/10.1007/s13277-016-4892-6
- Qi P, Xu MD, Shen XH, Ni SJ, Huang D, Tan C, Weng WW, Sheng WQ, Zhou XY, Du X. Reciprocal repression between TUSC7 and miR-23b in gastric cancer. Int J Cancer 2015; 137:1269-78; PMID:25765901; http://dx.doi.org/10.1002/ijc.29516
- Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: A new frontier in the study of human diseases. Cancer Lett 2013; 339:159-66; PMID:23791884; http://dx.doi.org/10.1016/j.canlet.2013.06.013
- Li T, Mo X, Fu L, Xiao B, Guo J. Molecular mechanisms of long noncoding RNAs on gastric cancer. Oncotarget 2016; 7:8601-12; PMID:26788991; http://dx.doi.org/10.18632/oncotarget.6926
- Cai X, Liu Y, Yang W, Xia Y, Yang C, Yang S, Liu X. Long noncoding RNA MALAT1 as a potential therapeutic target in osteosarcoma. J Orthop Res 2016; 34:932–41; PMID:26575981; http://dx.doi.org/10.1002/jor.23105
- Dong Y, Liang G, Yuan B, Yang C, Gao R, Zhou X. MALAT1 promotes the proliferation and metastasis of osteosarcoma cells by activating the PI3K/Akt pathway. Tumour Biol 2015; 36:1477-86; PMID:25431257; http://dx.doi.org/10.1007/s13277-014-2631-4
- Huang J, Liu K, Yu Y, Xie M, Kang R, Vernon P, Cao L, Tang D, Ni J. Targeting HMGB1-mediated autophagy as a novel therapeutic strategy for osteosarcoma. Autophagy 2012; 8:275-7; PMID:22301993; http://dx.doi.org/10.4161/auto.8.2.18940
- Gutschner T, Hammerle M, Diederichs S. MALAT1 – a paradigm for long noncoding RNA function in cancer. J Mol Med (Berl) 2013; 91:791-801; PMID:23529762; http://dx.doi.org/10.1007/s00109-013-1028-y
- Shrivastava S, Mansure JJ, Almajed W, Cury F, Ferbeyre G, Popovic M, Seuntjens J, Kassouf W. The role of HMGB1 in radioresistance of bladder cancer. Mol Cancer Ther 2016; 15:471-9; PMID:26719575; http://dx.doi.org/10.1158/1535-7163.MCT-15-0581
- Yang GL, Zhang LH, Bo JJ, Huo XJ, Chen HG, Cao M, Liu DM, Huang YR. Increased expression of HMGB1 is associated with poor prognosis in human bladder cancer. J Surg Oncol 2012; 106:57-61; PMID:22237993; http://dx.doi.org/10.1002/jso.23040
- Tee AE, Liu B, Song R, Li J, Pasquier E, Cheung BB, Jiang C, Marshall GM, Haber M, Norris MD, et al. The long noncoding RNA MALAT1 promotes tumor-driven angiogenesis by up-regulating pro-angiogenic gene expression. Oncotarget 2016; 7:8663-75; PMID:26848616; http://dx.doi.org/10.18632/oncotarget.6675
- Yang MH, Hu ZY, Xu C, Xie LY, Wang XY, Chen SY, Li ZG. MALAT1 promotes colorectal cancer cell proliferation/migration/invasion via PRKA kinase anchor protein 9. Biochim Biophys Acta 2015; 1852:166-74; PMID:25446987; http://dx.doi.org/10.1016/j.bbadis.2014.11.013
- Xiao H, Tang K, Liu P, Chen K, Hu J, Zeng J, Xiao W, Yu G, Yao W, Zhou H, et al. LncRNA MALAT1 functions as a competing endogenous RNA to regulate ZEB2 expression by sponging miR-200s in clear cell kidney carcinoma. Oncotarget 2015; 6:38005-15; PMID:26461224; http://dx.doi.org/10.18632/oncotarget.5357
- Wang X, Li M, Wang Z, Han S, Tang X, Ge Y, Zhou L, Zhou C, Yuan Q, Yang M. Silencing of long noncoding RNA MALAT1 by miR-101 and miR-217 inhibits proliferation, migration, and invasion of esophageal squamous cell carcinoma cells. J Biol Chem 2015; 290:3925-35; PMID:25538231; http://dx.doi.org/10.1074/jbc.M114.596866
- Huang J, Ni J, Liu K, Yu Y, Xie M, Kang R, Vernon P, Cao L, Tang D. HMGB1 promotes drug resistance in osteosarcoma. Cancer Res 2012; 72:230-8; PMID:22102692; http://dx.doi.org/10.1158/0008-5472.CAN-11-2001
- Zhang C, Ge S, Hu C, Yang N, Zhang J. MiRNA-218, a new regulator of HMGB1, suppresses cell migration and invasion in non-small cell lung cancer. Acta Biochim Biophys Sin (Shanghai) 2013; 45:1055-61; PMID:24247270; http://dx.doi.org/10.1093/abbs/gmt109
- Yang Y, Huang JQ, Zhang X, Shen LF. MiR-129-2 functions as a tumor suppressor in glioma cells by targeting HMGB1 and is down-regulated by DNA methylation. Mol Cell Biochem 2015; 404:229-39; PMID:25772485; http://dx.doi.org/10.1007/s11010-015-2382-6
- Xiao P, Liu WL. MiR-142-3p functions as a potential tumor suppressor directly targeting HMGB1 in non-small-cell lung carcinoma. Int J Clin Exp Pathol 2015; 8:10800-7; PMID:26617792
- Luo J, Chen J, He L. mir-129-5p attenuates irradiation-induced autophagy and decreases radioresistance of breast cancer cells by targeting HMGB1. Med Sci Monit 2015; 21:4122-9; PMID:26720492; http://dx.doi.org/10.12659/MSM.896661
- Schwickert A, Weghake E, Bruggemann K, Engbers A, Brinkmann BF, Kemper B, Seggewiss J, Stock C, Ebnet K, Kiesel L, et al. microRNA miR-142-3p inhibits breast cancer cell invasiveness by synchronous targeting of WASL, integrin alpha V, and additional cytoskeletal elements. PLoS One 2015; 10:e0143993; PMID:26657485; http://dx.doi.org/10.1371/journal.pone.0143993
- Lei Z, Xu G, Wang L, Yang H, Liu X, Zhao J, Zhang HT. MiR-142-3p represses TGF-beta-induced growth inhibition through repression of TGFbetaR1 in non-small cell lung cancer. FASEB J 2014; 28:2696-704; PMID:24558198; http://dx.doi.org/10.1096/fj.13-247288
- Chai S, Tong M, Ng KY, Kwan PS, Chan YP, Fung TM, Lee TK, Wong N, Xie D, Yuan YF, et al. Regulatory role of miR-142-3p on the functional hepatic cancer stem cell marker CD133. Oncotarget 2014; 5:5725-35; PMID:25015418; http://dx.doi.org/10.18632/oncotarget.2167
- Bijnsdorp IV, Hodzic J, Lagerweij T, Westerman B, Krijgsman O, Broeke J, Verweij F, Nilsson RJ, Rozendaal L, van Beusechem VW, et al. miR-129-3p controls centrosome number in metastatic prostate cancer cells by repressing CP110. Oncotarget 2016; 7:16676-87; PMID:26918338; http://dx.doi.org/10.18632/oncotarget.7572
- Zhang Y, Wang Y, Wei Y, Li M, Yu S, Ye M, Zhang H, Chen S, Liu W, Zhang J. MiR-129-3p promotes docetaxel resistance of breast cancer cells via CP110 inhibition. Sci Rep 2015; 5:15424; PMID:26487539; http://dx.doi.org/10.1038/srep15424
- Wang X, Guo Y, Wang C, Yu H, Yu X. MicroRNA-142-3p inhibits chondrocyte apoptosis and inflammation in osteoarthritis by targeting HMGB1. Inflammation 2016; 39:1718–28; PMID:27447821; http://dx.doi.org/10.1007/s10753-016-0406-3
- Yang L, Bai HS, Deng Y, Fan L. High MALAT1 expression predicts a poor prognosis of cervical cancer and promotes cancer cell growth and invasion. Eur Rev Med Pharmacol Sci 2015; 19:3187-93.
- Hirata H, Hinoda Y, Shahryari V, Deng G, Nakajima K, Tabatabai ZL, Ishii N, Dahiya R. Long noncoding RNA MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and interacts with miR-205. Cancer Res 2015; 75:1322-31; PMID:25600645; http://dx.doi.org/10.1158/0008-5472.CAN-14-2931
- Yang G, Zhang P, Lv A, Liu Y, Wang G. MiR-205 functions as a tumor suppressor via targeting TGF-alpha in osteosarcoma. Exp Mol Pathol 2016; 100:160-6; PMID:26708425; http://dx.doi.org/10.1016/j.yexmp.2015.12.010
