Cells spend a substantial amount of their resources on protein production during cell growth. Ribosome synthesis is commonly believed to dictate the rate of cell growth, particularly in conditions under which nitrogen levels become rate-limiting for anabolism.Citation1 Transcription of genes that encode ribosome proteins (RPGs), but also tRNA genes, is stimulated strongly by nutrients, including nitrogen. The master nutrient response regulator TORC1 activates several transcription factors to promote transcription of RPGs and tRNA genes. However, the molecular mechanisms by which the cell fine-tunes transcriptional activation of these genes to fuel anabolism are not well understood.
In 2 recent publications, we described how the ubiquitin family member Sumo cooperates with TORC1 to induce full activation of RPGs and tRNA genes in the budding yeast Saccharomyces cerevisiae.Citation2,3 Starting with a ChIP-seq experiment to map the localization of Sumo along the genome, we discovered that Sumo was highly enriched both at promoter regions of RPGs and at tRNA genes. We first determined the physiologic relevance of the sumoylation pathway for expression of RPGs and tRNA genes. We found that inhibiting the Sumo pathway by expressing a partial loss-of-function UBC9 mutant (i.e. ubc9–1), which encodes the sole E2 enzyme in yeast, resulted in strongly reduced transcription of RPGs and tRNAs, indicating that the dominant function of Sumo at these genes is to activate transcription. This was surprising, because Sumo is traditionally viewed as inhibitory to the process of transcription.Citation4 Intrigued by these findings, we aimed to gain insight into the molecular mechanisms by which Sumo drives transcription of RPGs and tRNA genes ().
Figure 1. Sumo bolsters transcription of pro-growth genes. Sumoylation of Rap1 promotes recruitment of the basal transcription factor TFIID, which in turn recruits RNAPII to augment transcription of ribosomal protein genes (RPGs). In parallel, sumoylation of RPc82 facilitates its efficient incorporation into the RNAPIII holoenzyme to promote transcription of tRNA genes.
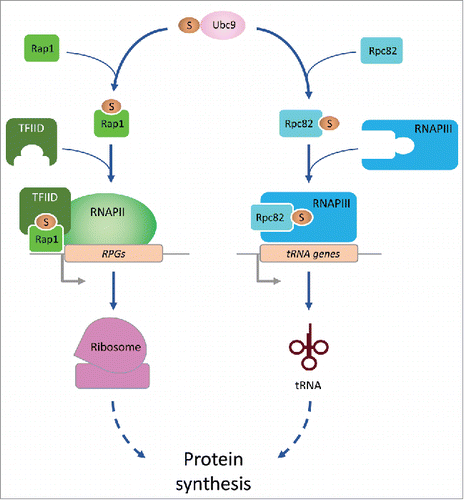
First, we determined the Sumo consensus site at RPG promoters, and discovered it was identical to the consensus site of the transcription factor Rap1, which is well known to be required for transcription of nearly all RPGs. Rap1 was previously shown to promote transcription of RPGs by recruiting TFIID, which in turn helps recruit RNA polymerase II (RNAPII). Interestingly, a non-sumoylatable form of Rap1 interacted much more weakly with TFIID than wild-type Rap1. As a consequence, the levels of both TFIID and RNAPII at RPGs were lower in mutants expressing non-sumoylatable Rap1, and this resulted in a significant reduction in RPG mRNA levels.
Next, we focused on the role of Sumo in transcription of tRNA genes. In contrast to RPGs, which are transcribed by RNAPII, tRNA genes are transcribed by RNA polymerase III (RNAPIII). In addition, tRNA genes do not have binding sites for Rap1, and therefore the mechanism by which Sumo promotes tRNA transcription must be fundamentally different from its effect on RPG transcription. To identify potential targets of Sumo at tRNA genes, we mapped the Sumo proteome by purifying Sumo and analyzing its conjugates by mass spectrometry, and found that some of the most prominent Sumo targets were components of RNAPIII, including Rpc82, Ret1 and Rpc53 (also known as C82, C128 and C53, respectively). These findings confirm earlier observations that RNAPIII components are major Sumo targets.Citation5
To understand the effect of Sumo on the function of these proteins we constructed lysine-to-arginine substitution mutants that failed to be sumoylated efficiently. Expression of these mutant alleles appeared to have different effects on tRNA expression levels; non-sumoylatable Rpc82 correlated with a reduction in tRNA levels, non-sumoylatable Rpc53 did not appear to affect tRNA levels, and expression of nonsumoylatable Ret1 resulted in an apparent increase in tRNA levels. We decided to focus on Rpc82, because the effect of non-sumoylatable Rpc82 most closely resembled the phenotype of the ubc9–1 mutant (in which tRNA transcription is diminished).
Interestingly, we found that sumoylation of Rpc82 appeared to be important for its interaction with other RNAPIII subunits and for the efficient incorporation of Rpc82 into the RNAPIII holoenzyme. Structural modeling based on previously published cryo-electron microscopy structures of RNAPIIICitation6 suggested that sumoylation of Rpc82 could stabilize its interaction with other RNAPIII subunits to promote RNAPIII assembly and activity, although clearly more detailed studies are required to determine the exact effect of Rpc82 sumoylation on organization of the RNAPIII holoenzyme. Furthermore, our observation that sumoylation of other components of RNAPIII may have an effect opposite to that of Rpc82 sumoylation (i.e., inhibit rather than promote RNAPIII activity) indicates that regulation of RNAPIII by Sumo is likely to be highly complex. Differential sumoylation of RNAPIII subunits may occur in response to different environmental cues, and the balance between these different sumoylation events likely determines the ultimate effect on RNAPIII activity.
In summary, we have discovered that Sumo promotes transcription of at least 2 classes of pro-growth genes, i.e., RPGs and tRNA genes, and we have unraveled 2 molcular mechanisms by which Sumo activates their transcription. Our findings strongly suggest that Sumo promotes anabolism by augmenting ribosome production, mRNA translation and protein synthesis, although this remains to be formally tested in follow-up experiments.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- Kafri M, Metzl-Raz E, Jona G, Barkai N. The cost of protein production. Cell Rep 2016; 14(1):22-31; PMID:26725116; http://dx.doi.org/10.1016/j.celrep.2015.12.015
- Chymkowitch P, Nguéa PA, Aanes H, Robertson J, Klungland A, Enserink JM. TORC1-dependent sumoylation of Rpc82 promotes RNA polymerase III assembly and activity. Proc Natl Acad Sci U S A 2017; 114(5):1039-1044; PMID:28096404; http://dx.doi.org/10.1073/pnas.1615093114
- Chymkowitch P, Nguéa AP, Aanes H, Koehler CJ, Thiede B, Lorenz S, Meza-Zepeda LA, Klungland A, Enserink JM. Sumoylation of Rap1 mediates the recruitment of TFIID to promote transcription of ribosomal protein genes. Genome Res 2015; 25(6):897-906; PMID:25800674; http://dx.doi.org/10.1101/gr.185793.114
- Chymkowitch P, Nguea PA, Enserink JM. SUMO-regulated transcription: challenging the dogma. Bioessays 2015; 37(10):1095-105; PMID:26354225; http://dx.doi.org/10.1002/bies.201500065
- Panse VG, Hardeland U, Werner T, Kuster B, Hurt E. A proteome-wide approach identifies sumoylated substrate proteins in yeast. J Biol Chem 2004; 279(40):41346-51; PMID:15292183; http://dx.doi.org/10.1074/jbc.M407950200
- Hoffmann NA, Jakobi AJ, Moreno-Morcillo M, Glatt S, Kosinski J, Hagen WJ, Sachse C, Müller CW. Molecular structures of unbound and transcribing RNA polymerase III. Nature 2015; 528(7581):231-6; PMID:26605533; http://dx.doi.org/10.1038/nature16143
