ABSTRACT
Recent evidence indicates that the accumulation of endogenous DNA damage can induce senescence and limit the function of adult stem cells. It remains elusive whether deficiency in DNA damage repair is associated with the functional alteration of mammary stem cells. In this article, we reported that senescence was induced in mammary epithelial cells during aging along with increased expression of p16Ink4a (p16), an inhibitor of CDK4 and CKD6. Loss of p16 abrogated the age-induced senescence in mammary epithelial cells and significantly increased mammary stem cell function. We showed that loss of Brca1, a tumor suppressor that functions in DNA damage repair, in the mammary epithelium induced senescence with induction of p16 and a decline of stem cell function, which was rescued by p16 loss. These data not only answer the question as to whether deficiency in DNA damage repair is associated with the functional decline of mammary stem cells, but also identify the role of p16 in suppressing Brca1-deficient mammary stem cell function.
Introduction
Recent findings suggest that accumulation of adult stem cells (ASCs) undergoing senescence contribute to the reduced regenerative capacity and aging of tissues.Citation1-3 ASCs are largely maintained in a quiescent state but can be stimulated to re-enter the cell cycle even after prolonged periods of dormancy. A decline in ASC function with age has been detected in many mammalian tissues including mammary glands.Citation4-7 One of the potential mechanisms underlying the age-dependent decline in stem cell function is DNA damage.Citation8,9 It has been shown that endogenous DNA damage resulting from the effects of age or genetic mutations can induce senescence limiting the function of ASCs.Citation10-13 It remains largely elusive whether deficiency in DNA damage repair is associated with the functional decline of mammary stem cells.
Brca1 is a tumor suppressor that plays an important role in DNA damage repair. A deficiency in Brca1 function causes chromosomal abnormalities resulting in the activation of DNA-damage checkpoint pathways. BRCA1 deficiency leads to premature senescence and aging in multiple mouse tissues and/or cells with induction of p53 and p21.Citation14,15 Loss of p53 or its downstream target, p21, partially rescues the premature senescence of Brca1-deficient cells in adult mouse tissues,Citation14,15 suggesting that other pathways may also be involved in mediating premature senescence induced by Brca1 loss. Indeed, it was recently reported that p16Ink4a is induced by Brca1 deficiency in fibroblasts and mammary glands, and that haploid loss of BRCA1 in human mammary epithelial cells (MECs) activates the Rb, not the p53, pathway leading to senescence.Citation16-18 We have recently discovered that heterozygous germline deletion or specific depletion of Brca1 in mouse epithelium induce senescence in MECs.Citation19,20 Whether Brca1 loss affects mammary stem cell function is poorly understood.
The INK4 family of cell cycle inhibitors comprising of p16INK4A, p15INK4B, p18INK4C and p19INK4D (p16, p15, p18 and p19) inhibit CDK4 and CDK6, leading to the functional inactivation of RB.Citation21 Functional inactivation of this INK4-CDK4/6-Rb pathway is a common event in the development of most types of cancers,Citation22 and also causes the loss of senescent states of somatic and adult stem cells.Citation23 Unlike other INK4 members, p16 is not expressed early, but is markedly increased during aging and senescence.Citation24 Experiments in mice with a germline deletion of p16 reveal that p16 controls stem/progenitor cell aging in compartments of the brain, pancreatic islets and blood by inducing senescence.Citation4-6 Early experiments have shown in vitro that human MECs eventually reach a plateau of proliferation and enter a cell cycle arrest with increased expression of p16.Citation25 Furthermore, it has been demonstrated that spontaneous methylation of p16 in breast primary epithelial cells can overcome the senescence barrier.Citation26 It is largely unknown whether p16 controls the function of mammary stem cells and whether p16 plays a role in regulating the function of Brca1-deficient mammary stem cells.
In this report, we generated p16 and Brca1 single and compound mutant mice to determine their function in controlling mammary stem cell function.
Results
Increased expression of p16 is associated with age-induced senescence in mammary glands
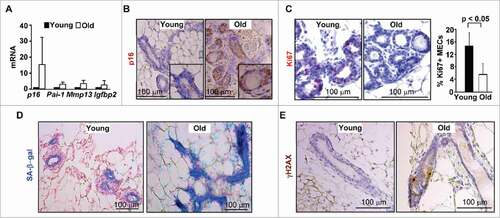
We determined the expression of p16 in both young (2–4-month-old) and old (12–24-month-old) virgin mammary glands in wild-type (WT) mice. We found that mRNA levels of p16 along with senescence markers, Pai-1, Mmp13, and Igfbp2 were increased in old mammary glands relative to young (), suggesting that expression of p16 increases with age-induced senescence. Notably, immunohistochemical staining of virgin mammary glands revealed that p16 was predominantly expressed in the MECs of the old mice, whereas, it was hardly detected in those of the young (). Consistently, mammary epithelia from aged mice exhibited significantly fewer Ki67-positive cells and a stronger positive staining of senescence-associated β-galactosidase (SA-β-gal) than young epithelia (). To explore the age-related increase of DNA damage in mammary epithelial cells, we determined the expression of γH2AX, a marker for DNA double-strand breaks. We found that on average, a positive MEC was detected in every 10–15 old glands while γH2AX-positive cells were rarely seen in young glands (), confirming enhanced DNA damage in old MECs.
Figure 1. p16 expression is increased in MECs during age-induced senescence. (A) mRNA expression of mammary tissue was analyzed by q-RT-PCR in WT young (2–4-month-old) and old (12–24-month-old) virgin mice. Data are expressed as the mean ± SD from triplicates of each 2 separate mice. (B-E) Representative immunohistochemical staining of p16 (B), Ki67 (C) and γH2AX (E) in 3 mice per group, and SA-β-gal assay in 2 animals per group (D), in mammary glands. The percentages of Ki67-positive cells in (C) were calculated from cells situated in clear duct/gland structure, and the results represent the mean ± SD of 3 animals per group. Note the significant decrease of Ki67-positive cells and strong p16 and SA-β-gal staining in MECs from old mammary glands.
We examined mammosphere forming potential for primary MECs and found a decreased number of mammospheres formed by MECs from old mice than those from young mice (145.0 ± 73.9 vs 200.0 ± 40.6, p >0.05, Suppl. Fig. 1A), though it was not statistically significant. Consistently, we detected that the percentage of the mammary fat pad filled by the outgrowths reconstituted with old MECs was marginally less compared with that from young MECs (% of fat pad filled: 30.3% ± 19.6% vs 40.3% ± 19.7%, p >0.05, Suppl. Fig. 1B), when primary MECs were transplanted into cleared mammary fat pads (MFPs) of NSG recipient mice.
Taken together, this data suggests that expression of p16 is increased during aging, along with the senescence of MECs.
Loss of p16 rescues age-induced senescence of MECs and enhances mammary stem cell function
Given that age-induced senescence of MECs is associated with the increased expression of p16, we determined the role of p16 loss in controlling MEC senescence during aging. We found that p16 loss drastically reduced the expression of senescence markers () and enhanced MEC proliferation in old mice (Ki67-positive cells, 14.65% in p16−/− vs. 5.7% in WT, , C). These data indicate that loss of p16 rescues the age-induced senescence of MECs.
Figure 2. Loss of p16 rescues the age-induced senescence of MECs, and increases mammary stem cell function. (A) mRNA expression was analyzed by q-RT-PCR from WT virgin mammary tissue of old mice. q-RT-PCR data are expressed as the mean ± SD from duplicates of each of 3 separate mice in each age group. (B, C) Representative immunohistochemical staining of Ki67 in age-matched mammary glands. Results for the quantification of Ki67-positive MECs. Results represent the mean ± SD of 3 animals per group. (D, E) Mammary cells freshly isolated from both WT and p16−/− old mice were analyzed by colony formation and mammosphere assays. The number of colonies and spheres larger than 20µm was quantified. The bar graphs represent the mean ± SD from duplicates of each 3 separate mice. (F) Mammary cells isolated from old mice of the indicated genotypes were transplanted into the cleared MFPs of 3-week-old NSG mice. The mammary glands were dissected after 8 weeks and analyzed for the percentage of mammary fat pad filled. The bar graph represents the mean ± SD of N = 8 for WT and N = 4 for p16−/− mice examined for each group respectively.
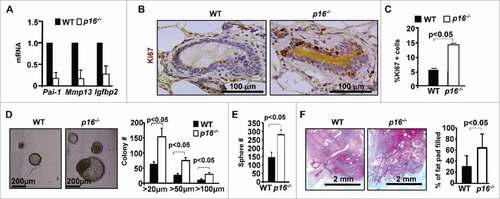
We then determined the role of p16 loss in the regulation of stem cell function. We found that MECs from p16−/− mice produced significantly more and larger colonies than WT cells (155.8 ± 30.6 vs 63.3 ± 8.9, for colonies greater than 20 µm, 76.8 ± 6.0 vs 27.8 ± 6.0, for colonies greater than 50 µm, 30.6 ± 4.7 vs 9.9 ± 2.4, for colonies greater than 100 µm, )
Correspondingly, MECs from p16−/− mice formed more and larger mammospheres than those from WT cells at similar ages (sphere number: 280.4 ± 30.9 vs 145.5 ± 28.3, sphere size: 136.0 µm ± 37.7 vs 71.7µm ± 15.6, p<0.05, ). We then performed transplantation assays for MECs isolated from old WT and p16−/− mice and found that the percentage of mammary fat pad filled by the outgrowths derived from p16−/− MECs was greater than that from WT cells (), indicating increased reconstitution potential in old p16−/− MECs.
Taken together, these results suggest that p16 loss mitigates MEC senescence and enhances mammary stem cell function during aging.
Deletion of Brca1 in mammary epithelium results in senescence with an increase of p16 expression and decline of stem cell function
We previously demonstrated that heterozygous germline deletion or specific depletion of Brca1 in mice induces senescence in MECs.Citation19,20 To directly determine the role of loss of function of Brca1 in controlling MECs, we used Brca1f/−;MMTV-Cre (Brca1f/−;MC) mice, in which MMTV-cre is active in virgin epithelia and mammary glands from these mice express <5% of Brca1 protein and mRNA relative to the levels in Brca1f/+;MC mice, as we described previously.Citation19,27 We compared p16 expression in Brca1f/−;MC mice with age-matched WT animals and found a substantial increase of p16 in Brca1f/−;MC MECs compared with WT (). Consistently, mRNA levels of p16 and senescence markers were increased in Brca1f/−;MC mammaries relative to WT counterparts (). These data suggest that specific depletion of Brca1 in the mammary epithelium induces senescence with increased p16 expression.
Figure 3. Brca1 depletion in the mammary epithelium leads to senescence, and a functional decline of mammary stem cells. (A) Representative immunohistochemical staining of p16 in mammary glands of WT and Brca1f/−;MC mice in 3 mice per group. (B) mRNA expression of mammary tissue was analyzed by q-RT-PCR in WT and Brca1f/−;MC mice. Data represent the mean ± SD from duplicates of 2 mice per genotype. (C, D) Mammary cells from age-matched WT and Brca1f/−;MC mice were isolated and analyzed by colony formation and mammosphere assays. Colonies and spheres larger than 20 µm were quantified. Bar graph, mean ± SD from duplicates of each 2 separate mice per genotype. (E) Representative mammary outgrowths from transplanted MECs of age-matched WT and Brca1f/−;MC mice. The mammary glands were dissected after 8 weeks and analyzed for the percentage of mammary fat pad filled. The bar graph represents the mean ± SD of N = 8 for WT and N = 5 for WT and Brca1f/−;MC mice respectively.
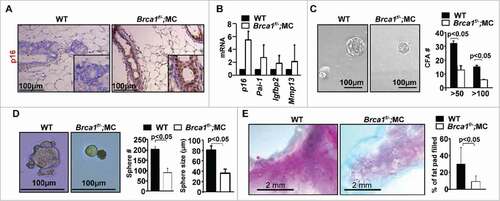
We performed colony formation and mammosphere assays for primary MECs and found that the number of Brca1f/−;MC colonies was significantly reduced when compared with colonies from WT MECs (13.3 ± 3.3 vs 32.7 ± 1.7, for colonies greater than 50 µm, 6.0 ± 0.5 vs 15.8 ± 0.8, for colonies greater than 100 µm, ). Correspondingly, Brca1f/−;MC mammospheres were reduced relative to WT counterparts (sphere number: 91.7 ± 21.2 vs 205.9 ± 12.5, sphere size: 36.2 ± 8.3 µm vs 80.2 ± 8.6 µm, ). We then transplanted primary MECs into the MFPs of NSG mice and found a significant reduction in the percentage of mammary fat pad filled by the outgrowths derived from Brca1f/−;MC MECs when compared with that from WT cells (). Together, these data suggest that Brca1 loss in mammary epithelium results in premature senescence of MECs with a functional decline of mammary stem cells, and an increase of p16 expression.
Loss of p16 rescues MEC senescence and decline of stem cell function induced by Brca1 deficiency
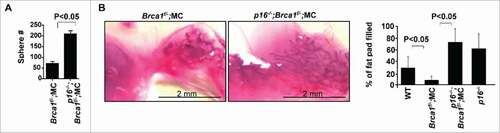
Considering that loss of p16 stimulates proliferation and mitigates senescence in MECs during aging, and that Brca1 loss induces senescence with an increase of p16 expression, we were inspired to determine the role of p16 in controlling Brca1 deficiency-induced senescence. To this end, we generated p16−/−;Brca1f/−;MC mice. We performed mammosphere and transplantation assays and found a significant increase in mammosphere-forming potential from p16−/−;Brca1f/−;MC MECs (212.0 ± 18.0) when compared with Brca1f/−;MC MECs (72.0 ± 15.5, ). We also performed transplantation assays and found a significant increase in the percentage of mammary fat pad filled by the outgrowths derived from p16−/−;Brca1f/−;MC MECs relative to that from Brca1f/−;MC counterparts (). These data confirm that p16 loss rescues the decline of mammary stem cell function caused by Brca1 insufficiency
Figure 4. Loss of p16 rescues the decline in mammary stem cell function caused by Brca1 deficiency. (A) Mammary cells freshly isolated from WT, Brca1f/−;MC, p16−/−;Brca1f/−;MC and p16−/− mice were analyzed by mammosphere assay. After 2 weeks the number of spheres larger than 20µm were quantified. Bar graph, mean ± SD from duplicates of each 3 separate mice. (B) Mammary cells isolated from mice of the indicated genotypes were transplanted into the cleared MFPs of 3-week-old NSG mice. The mammary glands were dissected after 8 weeks and analyzed for the percentage of mammary fat pad filled. The bar graph represents the mean ± SD of N = 8 for WT, N = 5 for Brca1f/−;MC, N = 4 for p16−/−;Brca1f/−;MC and N = 4 for p16−/− mice examined for each group respectively.
Discussion
In this article, we reported that during aging mammary epithelial cells undergo an increase of DNA damage and senescence along with increased expression of p16. Furthermore, loss of p16 attenuated mammary cell senescence and enhanced mammary stem cell function. Disruption of Brca1 in mammary epithelium resulted in senescence with an increase of p16 expression and the decline of stem cell function, which was also rescued by p16 loss. These data not only answer the question as to whether deficiency in DNA damage repair is associated with the functional decline of mammary stem cells, but also identify the role of p16 in suppressing Brca1-deficient function of mammary stem cells.
It has only been demonstrated in a few systems including the haematopoietic and skeletal muscle systems in which stem cell functional decline can be resulted from endogenous DNA damage that is accumulated with age and genetic deficiency in DNA damage repair.Citation10-13 The inability to purify homogeneous resident stem cell populations due to the lack of suitable mouse models, makes it difficult to determine if the function of stem cells in solid organs is also limited by deficiency in DNA damage repair.Citation1,2 Our finding that the reconstitution potential of MECs from Brca1-deficient mice was significantly reduced relative to WT counterparts, suggests that deficiency in DNA damage repair is also associated with functional decline of mammary stem cells, one type of solid organ stem cells.
BRCA1 deficiency causes chromosomal abnormalities, leading to the activation of DNA-damage checkpoint pathways and premature senescence and aging.Citation14-17 The finding that loss of p16 rescues the functional decline of mammary stem cells and MEC senescence with Brca1 deficiency suggests that p16 blocks these cells from entering an active cell cycle. These results indicate that, in addition to the p53-p21 pathway that is activated by BRCA1 loss,Citation14-16 p16 is also a critical downstream target of BRCA1 in controlling mammary cell proliferation and senescence. In line with these data, it was recently reported in vitro that BRCA1 knockdown enhances the association of BRG1, a chromatin-remodeling factor that interacts with BRCA1, with the p16 promoter, leading to activation of its transcription and senescence.Citation16 However, whether BRCA1 directly interacts with p16 promoter remains to be determined. More recently, it was found that human mammary epithelial cells from BRCA1-mutation carriers exhibit senescence, which is triggered by pRb pathway activation.Citation17 Since deregulation of p53 alone induces DNA damage, p16;Brca1 compound mutant mice and cells offer a unique opportunity to investigate the role of Brca1 in DNA damage repair under a genetically p53 intact background.
Of the 4 INK4 genes, p16 is frequently deleted and inactivated in breast cancers,Citation21,22,28-30 and it is not expressed early, but is markedly increased during aging and senescence in most organs or cells.Citation24 It has been shown that loss of p16 in mice rescues the functional decline of adult stem/progenitor cells in old mice in compartments of the brain, pancreatic islets and blood by suppressing age-associated cell senescence.Citation4-6 Consistently, we and others have shown that p16 along with senescence markers are induced in mouse and human during aging [ref. Citation31 and this study]. Though we detect a marginal reduced reconstitution potential in aged MECs, no statistical significance was achieved. This may be mainly resulted from micro-environmental factors in the mammary stroma that are specific to aging and affect mammary stem cell function in a cell non-autonomous manner.Citation32 Importantly, we demonstrated that loss of p16 does stimulate reconstitution potential of MECs in aged mice, confirming the role of p16 in increasing mammary stem cell function in aging mammary glands.
Materials and methods
Mice
Brca1f/−;MMTV-Cre mutant mice in a Balb/c-B6 mixed background were generated as described previously.Citation27 Tg(MMTV-Cre)4Mam mice in B6 a background were obtained from the NCI Mouse Repository and JAX laboratory respectively. p16 mutant mice in a FVB background were gifted by our collaborators, Dr. Norman Sharpless.Citation33 All mice used in the study were virgin female. All animal procedures were approved by the Institutional Animal Care and Use Committee at the University of North Carolina and University of Miami.
Histopathology, immunohistochemical stainning, and qRT-PCR
Histopathology and immunohistochemical stainning were performed as described previously.Citation20,27 Primary antibodies used are as follows: p16 (Santa Cruz Biotechnology), Ki67 (Novocastra Laboratories, Newcastle upon Tyne, UK), γH2AX (Cell Signaling). The IACUC (Institutional Animal Care and Use Committee) at the University of Miami approved all procedures. QRT-PCR was performed as described previously.Citation20,27
Mammary cell preparation, mammosphere assay, colony-formation assay, mammary transplantation assay
Mammary glands were isolated from mice at the indicated ages and genotypes, the tissue was processed, the mammosphere and colony-formation assays were performed as we described previously.Citation20,27,34 Briefly, for mammosphere assays, freshly isolated primary mammary cells (20,000 cells per well) were cultured in triplicate onto 24-well, ultra-low attachment plates. Mammosphere number was counted under microscope after 2 weeks. For colony-formation assays, freshly isolated cells (20,000 cells per well) were cultured in triplicate onto 24-well, Matrigel-coated plates. Colony number was counted under microscope after 2 weeks.
For mammary transplantation assays, 2×106 mammary cells isolated from mice of indicated genotypes were injected into the MFPs precleared 3-week old NSG mice. After 8 weeks, the resultant outgrowths were dissected and stained with carmine alum for wholemount analysis. The percentage of total ductal outgrowth into the MFP was quantified as described.Citation35
Statistical analysis
All data are presented as the mean ± SD for at least 3 repeated individual experiments for each group unless otherwise specified. Quantitative results were analyzed by 2-tailed Student's t-test. P<0.05 was considered statistically significant.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
KCCY_A_1295185_Supplemental.pdf
Download PDF (155.8 KB)Acknowledgments
We thank Drs. Norman Sharpless, Beverly Koller, Chuxia Deng, and Lothar Hennighausen for p16, Brca1 mutant and MMTV-cre mice, and the DVR core facility for animal husbandry.
Funding
This study was supported by DOD Idea Award (W81XWH-10–1–0302), DOD Idea Expansion Award (W81XWH-13–1–0282), Sylvester BFBCI Developmental Grant, IRG-98–277–13 from the American Cancer Society, and funds from the University of Miami Department of Surgery and Sylvester Cancer Center to Xin-Hai Pei
References
- Mandal PK, Blanpain C, Rossi DJ. DNA damage response in adult stem cells: Pathways and consequences. Nat Rev Mol Cell Biol 2011; 12:198-202; PMID:21304553; http://dx.doi.org/10.1038/nrm3060
- Sharpless NE, DePinho RA. How stem cells age and why this makes us grow old. Nat Rev Mol Cell Biol 2007; 8:703-13; PMID:17717515; http://dx.doi.org/10.1038/nrm2241
- Lombard DB, Chua KF, Mostoslavsky R, Franco S, Gostissa M, Alt FW. DNA repair, genome stability, and aging. Cell 2005; 120:497-512; PMID:15734682; http://dx.doi.org/10.1016/j.cell.2005.01.028
- Molofsky AV, Slutsky SG, Joseph NM, He S, Pardal R, Krishnamurthy J, Sharpless NE, Morrison SJ. Increasing p16INK4a expression decreases forebrain progenitors and neurogenesis during ageing. Nature 2006; 443:448-52; PMID:16957738; http://dx.doi.org/10.1038/nature05091
- Krishnamurthy J, Ramsey MR, Ligon KL, Torrice C, Koh A, Bonner-Weir S, Sharpless NE. p16INK4a induces an age-dependent decline in islet regenerative potential. Nature 2006; 443:453-7; PMID:16957737; http://dx.doi.org/10.1038/nature05092
- Janzen V, Forkert R, Fleming HE, Saito Y, Waring MT, Dombkowski DM, Cheng T, DePinho RA, Sharpless NE, Scadden DT. Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature 2006; 443:421-6; PMID:16957735; http://dx.doi.org/10.1038/nature05159
- Jackson HW, Waterhouse P, Sinha A, Kislinger T, Berman HK, Khokha R. Expansion of stem cells counteracts age-related mammary regression in compound Timp1/Timp3 null mice. Nat Cell Biol 2015; 17:217-27; PMID:25706237; http://dx.doi.org/10.1038/ncb3118
- Sperka T, Wang J, Rudolph KL. DNA damage checkpoints in stem cells, ageing and cancer. Nat Rev Mol Cell Biol 2012; 13:579-90; PMID:22914294; http://dx.doi.org/10.1038/nrm3420
- Behrens A, van Deursen JM, Rudolph KL, Schumacher B. Impact of genomic damage and ageing on stem cell function. Nat Cell Biol 2014; 16:201-7; PMID:24576896; http://dx.doi.org/10.1038/ncb2928
- Rossi DJ, Bryder D, Seita J, Nussenzweig A, Hoeijmakers J, Weissman IL. Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature 2007; 447:725-9; PMID:17554309; http://dx.doi.org/10.1038/nature05862
- Beerman I, Seita J, Inlay MA, Weissman IL, Rossi DJ. Quiescent hematopoietic stem cells accumulate DNA damage during aging that is repaired upon entry into cell cycle. Cell Stem Cell 2014; 15:37-50; PMID:24813857; http://dx.doi.org/10.1016/j.stem.2014.04.016
- Sinha M, Jang YC, Oh J, Khong D, Wu EY, Manohar R, Miller C, Regalado SG, Loffredo FS, Pancoast JR, et al. Restoring systemic GDF11 levels reverses age-related dysfunction in mouse skeletal muscle. Science 2014; 344:649-52; PMID:24797481; http://dx.doi.org/10.1126/science.1251152
- Blau HM, Cosgrove BD, Ho AT. The central role of muscle stem cells in regenerative failure with aging. Nat Med 2015; 21:854-62; PMID:26248268; http://dx.doi.org/10.1038/nm.3918
- Cao L, Li W, Kim S, Brodie SG, Deng CX. Senescence, aging, and malignant transformation mediated by p53 in mice lacking the Brca1 full-length isoform. Genes Dev 2003; 17:201-13; PMID:12533509; http://dx.doi.org/10.1101/gad.1050003
- Cao L, Kim S, Xiao C, Wang RH, Coumoul X, Wang X, Li WM, Xu XL, De Soto JA, Takai H, et al. ATM-Chk2-p53 activation prevents tumorigenesis at an expense of organ homeostasis upon Brca1 deficiency. EMBO J 2006; 25:2167-77; PMID:16675955; http://dx.doi.org/10.1038/sj.emboj.7601115
- Tu Z, Zhuang X, Yao YG, Zhang R. BRG1 is required for formation of senescence-associated heterochromatin foci induced by oncogenic RAS or BRCA1 loss. Mol Cell Biol 2013; 33:1819-29; PMID:23438604; http://dx.doi.org/10.1128/MCB.01744-12
- Sedic M, Skibinski A, Brown N, Gallardo M, Mulligan P, Martinez P, Keller PJ, Glover E, Richardson AL, Cowan J, et al. Haploinsufficiency for BRCA1 leads to cell-type-specific genomic instability and premature senescence. Nat Commun 2015; 6:7505; PMID:26106036; http://dx.doi.org/10.1038/ncomms8505
- Gorrini C, Baniasadi PS, Harris IS, Silvester J, Inoue S, Snow B, Joshi PA, Wakeham A, Molyneux SD, Martin B, et al. BRCA1 interacts with Nrf2 to regulate antioxidant signaling and cell survival. J Exp Med 2013; 210:1529-44; PMID:23857982; http://dx.doi.org/10.1084/jem.20121337
- Scott A, Bai F, Chan HL, Liu S, Ma J, Slingerland JM, Robbins DJ, Capobianco AJ, Pei XH. p16INK4a suppresses BRCA1-deficient mammary tumorigenesis. Oncotarget 2016; 7:84496-507; PMID:27811360; http://dx.doi.org/10.18632/oncotarget.13015
- Bai F, Smith MD, Chan HL, Pei XH. Germline mutation of Brca1 alters the fate of mammary luminal cells and causes luminal-to-basal mammary tumor transformation. Oncogene 2013; 32:2715-25; PMID:22777348; http://dx.doi.org/10.1038/onc.2012.293
- Pei XH, Xiong Y. Biochemical and cellular mechanisms of mammalian CDK inhibitors: A few unresolved issues. Oncogene 2005; 24:2787-95; PMID:15838515; http://dx.doi.org/10.1038/sj.onc.1208611
- Sherr CJ, McCormick F. The RB and p53 pathways in cancer. Cancer Cell 2002; 2:103-12; PMID:12204530; http://dx.doi.org/10.1016/S1535-6108(02)00102-2
- Orford KW, Scadden DT. Deconstructing stem cell self-renewal: Genetic insights into cell-cycle regulation. Nat Rev Genet 2008; 9:115-28; PMID:18202695; http://dx.doi.org/10.1038/nrg2269
- Zindy F, Quelle DE, Roussel MF, Sherr CJ. Expression of the p16INK4a tumor suppressor versus other INK4 family members during mouse development and aging. Oncogene 1997; 15:203-11; PMID:9244355; http://dx.doi.org/10.1038/sj.onc.1201178
- Brenner AJ, Stampfer MR, Aldaz CM. Increased p16 expression with first senescence arrest in human mammary epithelial cells and extended growth capacity with p16 inactivation. Oncogene 1998; 17:199-205; PMID:9674704; http://dx.doi.org/10.1038/sj.onc.1201919
- Feijoo P, Terradas M, Soler D, Dominguez D, Tusell L, Genesca A. Breast primary epithelial cells that escape p16-dependent stasis enter a telomere-driven crisis state. Breast Cancer Res 2016; 18:7; PMID:26758019; http://dx.doi.org/10.1186/s13058-015-0667-z
- Bai F, Chan HL, Scott A, Smith MD, Fan C, Herschkowitz JI, Perou CM, Livingstone AS, Robbins DJ, Capobianco AJ, et al. BRCA1 suppresses epithelial-to-mesenchymal transition and stem cell dedifferentiation during mammary and tumor development. Cancer Res 2014; 74:6161-72; PMID:25239453; http://dx.doi.org/10.1158/0008-5472.CAN-14-1119
- Kamb A, Gruis NA, Weaver-Feldhaus J, Liu Q, Harshman K, Tavtigian SV, Stockert E, Day RS, Johnson BE, 3rd, Skolnick MH. A cell cycle regulator potentially involved in genesis of many tumor types. Science 1994; 264:436-40; PMID:8153634; http://dx.doi.org/10.1126/science.8153634
- Herman JG, Merlo A, Mao L, Lapidus RG, Issa J-PJ, Davidson NE, Sidransky D, Baylin SB. Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res 1995; 55:4525-30; PMID:7553621
- Koboldt DC, Fulton RS, McLellan MD, Schmidt H, Kalicki-Veizer J, McMichael JF, Fulton LL, Dooling DJ, Ding L, Mardis ER, et al. Comprehensive molecular portraits of human breast tumours. Nature 2012; 490:61-70; PMID:23000897; http://dx.doi.org/10.1038/nature11412
- Pirone JR, D'Arcy M, Stewart DA, Hines WC, Johnson M, Gould MN, Yaswen P, Jerry DJ, Smith Schneider S, Troester MA. Age-associated gene expression in normal breast tissue mirrors qualitative age-at-incidence patterns for breast cancer. Cancer Epidemiol Biomarkers Prev 2012; 21:1735-44; PMID:22859400; http://dx.doi.org/10.1158/1055-9965.EPI-12-0451
- Young LJ, Medina D, DeOme KB, Daniel CW. The influence of host and tissue age on life span and growth rate of serially transplanted mouse mammary gland. Exp Gerontol 1971; 6:49-56; PMID:5572739; http://dx.doi.org/10.1016/0531-5565(71)90048-9
- Sharpless NE, Bardeesy N, Lee KH, Carrasco D, Castrillon DH, Aguirre AJ, Wu EA, Horner JW, DePinho RA. Loss of p16Ink4a with retention of p19Arf predisposes mice to tumorigenesis. Nature 2001; 413:86-91; PMID:11544531; http://dx.doi.org/10.1038/35092592
- Pei XH, Bai F, Smith MD, Usary J, Fan C, Pai SY, Ho IC, Perou CM, Xiong Y. CDK inhibitor p18(INK4c) is a downstream target of GATA3 and restrains mammary luminal progenitor cell proliferation and tumorigenesis. Cancer Cell 2009; 15:389-401; PMID:19411068; http://dx.doi.org/10.1016/j.ccr.2009.03.004
- Huo Y, Macara IG. The Par3-like polarity protein Par3L is essential for mammary stem cell maintenance. Nat Cell Biol 2014; 16:529-37; PMID:24859006; http://dx.doi.org/10.1038/ncb2969
