Next generation sequencing methods have revealed that 70 ∼90% of the human genome is transcribed, with the majority of RNA species consisting of long non-coding RNAs (lncRNAs). While early breakthroughs demonstrated roles for nuclear lncRNAs in regulating transcription and genomic organization, it has proven more challenging to assign roles and functions to cytosolic lncRNAs. Since the vast majority of lncRNAs have been identified by high-throughput computational analysis of RNAseq data, rather than by experimental validation, the question remains as to whether these RNAs are truly “non-coding”. Although one of the criteria for classification of an RNA as long non-coding is that it should have minimal open reading frames (ORFs), and no ORF should encode more than 100 amino acids, short predicted ORFs can be found in many lncRNA molecules. Thus, it is possible that some of these putative lncRNAs may encode for hidden polypeptides.
Indeed, there is increasing evidence that suggests that small ORFs within annotated lncRNAs may actually encode for hidden peptides. For example, in 2007 the Kageyama group identified the Drosophila encoded polished rice (pri) lncRNA as encoding 11 or 32 amino acid peptides,Citation1 while in 2014 the Schier group identified a hidden polypeptide encoded by the Toddler lncRNA in zebrafish,Citation2 and in 2015 and 2016 the Olson group identified two lncRNA encoded polypeptides regulating the calcium pump SERCA in mammals.Citation3-4 The identification of these lncRNA encoded polypeptides indicates that there are additional polypeptides still to be discovered encoded by annotated lncRNAs, although approaches and tools available for their systematic discovery are currently lacking.
In order to uncover such hidden polypeptides, we developed a mass spectrometry based pipeline to identify lncRNA translated ORFs (Matsumoto and Pandolfi, manuscript in preparation). This led us to uncover a novel polypeptide, which we termed Small regulatory Polypeptide of Amino Acid Response (SPAR, also known as SPAAR). This polypeptide is encoded by the lncRNA LINC00961, and regulates amino acid dependent mTORC1 signaling.Citation5 LINC00961 is a conserved lncRNA, encoding a polypeptide of 90 amino acids, and we validated endogenous expression by generating anti-SPAR antibodies. Interestingly, SPAR harbors a transmembrane domain at its N terminus and localizes at the late endosome and lysosomal membranes. Using mass spectrometry to identify interaction partners for SPAR, we found it to interact with subunits of the v-ATPase complex.
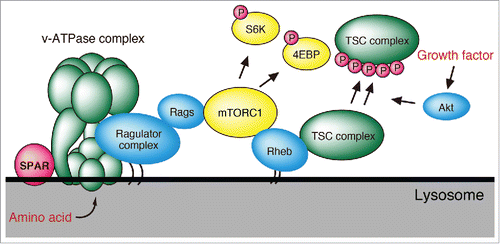
mTORC1, known as mammalian target of rapamycin complex 1, functions as a nutrient sensor that controls protein synthesis and cell growth. The v-ATPase complex is known to be important for mTORC1 activation in response to amino acid stimulation via interaction with the Ragulator complex at the lysosomal surface.Citation6 Amino acid stimulation promotes recruitment of mTORC1 to the lysosomal surface through activation of the v-ATPase and Ragulator/Rag-GTPase complexes ().Citation7 Since SPAR binds to the v-ATPase complex, we examined its effects on mTORC1 activation by amino acid stimulation. SPAR over-expression greatly inhibited mTORC1 activation while knockdown of SPAR promoted mTORC1 activity in response to amino acid stimulation. On the other hand, growth factor signaling results in the activation of the Rheb GTPase, which directly activates mTORC1 at the lysosomal membrane. However, SPAR overexpression failed to inhibit mTORC1 activation mediated by insulin signaling. Similarly, mTORC2 phosphorylation of AKT and EGF mediated phosphorylation of ERK1/2 were not impacted by SPAR overexpression, demonstrating its amino acid specific regulation of mTORC1.
Figure 1. Model for mTORC1 activation and signaling. Amino acids stimulate the v-ATPase/Ragulator super-complex, to activate Rags and recruit mTORC1 to the lysosome. On the other hand, growth factor stimulation activates mTORC1 through AKT signaling. SPAR functions to negatively regulate mTORC1 recruitment, through interaction with v-ATPase, in response to amino acid stimulation, while growth factor activation of mTORC1 remains unaffected by SPAR.
We next generated Spar-deficient mice to determine its functional relevance in vivo. In order to maintain expression of the host lncRNA while ablating polypeptide expression, we introduced a ΔATG mutation in the murine Spar ORF by CRISPR/Cas9. Spar-deficient mice are viable with no gross abnormalities observed up to 8 weeks of age, indicating that Spar is dispensable for normal development. Given that Spar expression is enriched in skeletal muscle, we looked closer at its role in this tissue. mTORC1 is highly activated during muscle regeneration, and regeneration can be blocked by rapamycin. Notably, Spar expression is substantially downregulated upon muscle injury, and its expression gradually restored upon regeneration. Thus, we hypothesized that Spar down regulation is required to ensure maximal mTORC1 activation for regeneration. Indeed, Spar-deficient mice experiencing muscle injury demonstrated an increased mTORC1 activation, which in turn promotes increased stem cell proliferation, differentiation, and myofiber maturation.
Thus, these data demonstrate the importance of lncRNA-encoded peptides, and reveals a manner by which such peptides can fine-tune the activity of larger ubiquitous macromolecule complexes. The fact that lncRNAs are frequently enriched in specific tissue types, highlights a manner by which tissues and organs can adapt to enable the most appropriate utilization of such protein complexes for their needs. Both our data and that of othersCitation1-4 emphasize the need to re-examine carefully those RNAs categorized as lncRNAs, as it is likely a number of them encode functional polypeptides. Additional, high-throughput approaches will be required towards their identification. To this end, significant challenges exist that still need to be addressed. These include technical challenges associated with mass spectrometry and computational analysis of data collected, as analytic peptides generated by tryptic digestion of lncRNA encoded polypeptides are frequently low in number and abundance as compared with analytic peptides generated from coding mRNAs, while bioinformatics ORF predictions should be run also taking into account alternative starting codon. Additionally, biological challenges exist due to the fact that lncRNAs and their encoded polypeptides are often enriched in specific cell and tissue types, making it difficult to obtain a comprehensive overview of lncRNA encoded polypeptides using single cell or tissue sources. Thus, further optimization of protocols to detect hidden polypeptides will be essential to expanding this field of study. Finally, the tissue specific enrichment of lncRNAs and their encoded polypeptides may offer unique opportunities for clinical application and therapeutic intervention.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- Kondo T, Hashimoto Y, Kato K, Inagaki S, Hayashi S, Kageyama Y. Small peptide regulators of actin-based cell morphogenesis encoded by a polycistronic mRNA. Nat Cell Biol 2007; 9:660-5; PMID:17486114; http://dx.doi.org/10.1038/ncb1595
- Pauli A, Norris ML, Valen E, Chew GL, Gagnon JA, Zimmerman S, Mitchell A, Ma J, Dubrulle J, Reyon D, Tsai SQ, Joung JK, Saghatelian A, Schier AF. Toddler: An embryonic signal that promotes cell movement via Apelin receptors. Science 2014; 343:1248636; PMID:24407481; http://dx.doi.org/10.1126/science.1248636
- Anderson DM, Anderson KM, Chang CL, Makarewich CA, Nelson BR, McAnally JR, Kasaragod P, Shelton JM, Liou J, Bassel-Duby R, Olson EN. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 2015; 160:595-606; PMID:25640239; http://dx.doi.org/10.1016/j.cell.2015.01.009
- Nelson BR, Makarewich CA, Anderson DM, Winders BR, Troupes CD, Wu F, Reese AL, McAnally JR, Chen X, Kavalali ET, Cannon SC, Houser SR, Bassel-Duby R, Olson EN. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science 2016; 351:271-275; PMID:26816378; http://dx.doi.org/10.1126/science.aad4076
- Matsumoto A, Pasut A, Matsumoto M, Yamashita R, Fung J, Monteleone E, Saghatelian A, Nakayama KI, Clohessy JG, Pandolfi PP. mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature 2017; 541:228-232; PMID:28024296; http://dx.doi.org/10.1038/nature21034
- Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science 2011; 334:678-683; PMID:22053050; http://dx.doi.org/10.1126/science.1207056
- Bar-Peled L, Sabatini DM. Regulation of mTORC1 by amino acids. Trends Cell Biol 2014; 24:400-406; PMID:24698685; http://dx.doi.org/10.1016/j.tcb.2014.03.003
