ABSTRACT
Androgenetic alopecia is the most common form of hair loss. Minoxidil has been approved for the treatment of hair loss, however its mechanism of action is still not fully clarified. In this study, we aimed to elucidate the effects of 5% minoxidil topical foam on gene expression and activation of signaling pathways in vertex and frontal scalp of men with androgenetic alopecia. We identified regional variations in gene expression and perturbed signaling pathways using in silico Pathway Activation Network Decomposition Analysis (iPANDA) before and after treatment with minoxidil. Vertex and frontal scalp of patients showed a generally similar response to minoxidil. Both scalp regions showed upregulation of genes that encode keratin associated proteins and downregulation of ILK, Akt, and MAPK signaling pathways after minoxidil treatment. Our results provide new insights into the mechanism of action of minoxidil topical foam in men with androgenetic alopecia.
Introduction
Androgenetic alopecia (AGA) is the most common form of hair loss. Men with AGA suffer from hair loss in defined regions of the scalp, namely the frontal hairline as well as the top and vertex scalp. The reproducible pattern of hair loss in AGA can be associated with various factors, such as the differences in the levels of hormone receptorsCitation1 and embryologic scalp patterning.Citation2 The hallmarks of AGA are alterations in hair cycle development,Citation3-5 follicular miniaturization,Citation6-9 and inflammation.Citation10-12 Topical minoxidil and oral finasteride have been approved by the Food and Drug Administration (USA) for the treatment of AGA. It is known that minoxidil stimulates hair growth; however, its mechanism of action is still not fully understood and needs further clarification. Minoxidil is an adenosine triphosphate (ATP) sensitive potassium (K) channel agonist as well as a vasodilator. Studies have also shown the vasodilation effect of topical minoxidil on skin which could justify its activity on the hair follicle.Citation13-15 Moreover, it was demonstrated that only one of 2 forms of K(ATP) channels found in human hair follicles is sensitive to minoxidil.Citation16 The drug's effect seems to be connected to its presence and stops when the treatment is discontinued.
Recent advances in molecular biology and genetic mapping have re-ignited interest in determining the mechanism of hair loss and minoxidil's restorative effects on hair growth. Providing increased insight into minoxidil's mechanism of action may yield novel therapeutic targets and potential points of intervention for the treatment of AGA. Differential signaling pathways perturbation profiles are known to be robust biomarkers of many pathologic and age-related conditions, including cancer.Citation17-23 It has been shown that for different types of cancer, such as bladder cancer, basal cell carcinoma, glioblastoma, hepatocellular carcinoma, etc., pathway activation values showed better area-under-the-curve scores compared with the individual genes that make up the pathways.Citation17 Analyzing potential changes in signaling pathways activation state during the development and progression of hair growth disorders could be a promising strategy for the development of drugs that could target defined signaling pathways related to hair loss, including age-related changes such as senescent alopeciaCitation24 and graying.
We examined the effects of 5% minoxidil topical foam (MTF 5%) on gene expression and activation of signaling pathways in vertex and frontal scalp of men with AGA. We identified regional variations in gene expression and activation of signaling pathways before and after treatment with MTF 5%. Our results provide new insights into the potential mechanism of action of MTF 5% in men with AGA.
Results
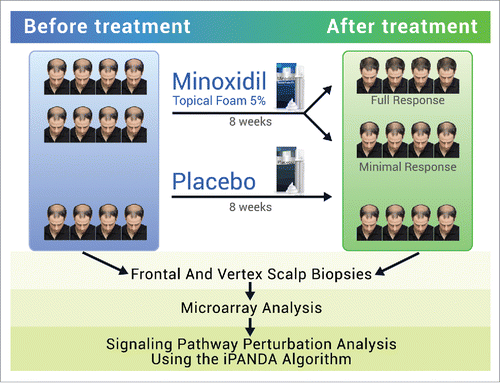
The following results were obtained from an analysis of gene expression data from a minoxidil study conducted by Mirmirani et al.Citation25 The original analysis performed by Mirmirani et al.Citation25 included data from 13 patients; 9 used MTF 5% and 4 used placebo. After 8 weeks of treatment, 9 patients who used MTF 5% were categorized as either responders or non-responders. We reevaluated the original stereotactic photographs of patients and identified 3 groups of patients who used MTF 5%: responders, patients with minimal response, and non-responders. There was only 1 non-responder and his samples were excluded from the analysis. There were 4 clinical responders and 4 patients with minimal response after 8 weeks of treatment with MTF 5%. Thus, the results were based on data from 12 patients; 8 patients used MTF 5% and 4 used placebo. There were a total of 48 samples (4 samples ― frontal and vertex scalp, before and after treatment ― from each of 12 patients). However, 5 out of 48 samples were identified as outliers, thus the final results were based on 29 active and 14 placebo samples. The workflow of the study is shown in .
Figure 1. Workflow of the original study. Gene expression data were obtained from patients enrolled in a placebo controlled double-blinded study of MTF 5%. Healthy men aged 18–49 with Hamilton-Norwood type IV-V thinning were instructed to apply the treatment (active drug or placebo) topically to the affected area. Scalp biopsies from the frontal and vertex scalp were done at the leading edge of alopecia and global hair photographs were taken before and after 8 weeks of treatment. The effect of minoxidil on gene expression profile and signaling pathways activation state in the frontal and vertex scalp was identified via microarray analysis and in silico Pathway Activation Network Decomposition Analysis (iPANDA).
Gene level analysis
Comparison of frontal and vertex scalp before treatment
summarizes differentially expressed transcripts in frontal scalp compared with vertex. According to our analysis, 14 transcripts were upregulated and 1 gene was downregulated in frontal vs. vertex scalp before MTF 5% treatment. Upregulated transcripts included coding (FOS, FOSB, JUNB, DUSP1, CYR61, NR4A2, ATF3, EGR1, CD69, ZFP36, MMP1, RGS1) as well as non-coding RNAs (SNORD116–26, MIR21). The pseudogene MSL3P1 was downregulated.
Table 1. Differentially expressed genes in frontal vs. vertex scalp before treatment. There were 14 upregulated transcripts and 1 downregulated gene in frontal vs. vertex scalp before treatment. This table reflects baseline difference in gene expression profiles between these 2 regions of the scalp.
Vertex scalp after the use of MTF 5%
We identified 29 dysregulated transcripts in the vertex scalp of patients who were categorized as “responders” to the MTF treatment (). Keratin associated proteins (KRTAP7–1, KRTAP19–3, KRTAP8–1, KRTAP19–5 and KRTAP13–2) were significantly upregulated, while various coding genes (PLCXD1, RNY4P8, DMC1, IFNA10, LOC100506422) and non-coding RNAs (SND1-IT1, LINC01152, FAM99A, MIR99A, INE1 and LINC00028) were downregulated following treatment. These changes in gene expression were unique for the group of responders and were not observed in the samples from either patients with minimal response or patients in the placebo group.
Table 2. Differentially expressed genes in vertex scalp of responders after vs. before MTF 5% treatment. Our analysis revealed 29 dysregulated transcripts in the vertex scalp of patients who were categorized as “responders” to the MTF treatment. Keratin associated proteins (KRTAPs) were among the upregulated genes.
Frontal scalp after the use of MTF 5%
Our analysis revealed 39 dysregulated transcripts in frontal scalp of responders after MTF 5% treatment (). As noted in vertex scalp, upregulation of keratin associated proteins (KRTAP7–1, KRTAP19–3, KRTAP19–5, KRTAP19–1, KRTAP13–2 and KRTAP20–2) was similarly observed in frontal scalp. Late cornified envelope protein 3D (LCE3D), potassium voltage-gated channel modifier protein (KCNS1) as well as other coding genes (SPP1, IGLON5, PLA2G10, ADAMDEC1) and non-coding RNAs (VTRNA1–3) were downregulated in the frontal scalp after treatment. It is interesting to note that downregulation of several other late cornified envelope genes (LCE1D, LCE1F, LCE2C) was also observed in the placebo group.
Table 3. Differentially expressed genes in frontal scalp of responders after vs. before MTF 5% treatment. We identified 39 dysregulated transcripts in frontal scalp of patients in the responders group after MTF 5% treatment. As noted in vertex scalp, upregulation of keratin associated proteins was similarly observed in frontal scalp.
Comparison of frontal and vertex scalp after the use of MTF 5%
We compared differential gene expression of frontal and vertex scalp of the responders after MTF 5% treatment and observed 40 differentially expressed transcripts (). Genes including collagen type VI α 6 chain (COL6A6) and non-coding RNAs (LOC100131796, MIR331, SND1-IT1) were upregulated in frontal scalp compared with vertex. Downregulated genes included keratins (KRT31, KRT33B, KRT39, KRT75, KRT82, KRT83, KRT85), keratin associated proteins (KRTAP1–1, KRTAP1–3, KRTAP3–1), potassium channel protein (KCNK10), matrix metallopeptidase 7 (MMP7), and a pore-forming subunit of a voltage-gated ion channel (FAM26D).
Table 4. Differentially expressed genes in frontal vs. vertex scalp of responders after MTF 5% treatment. There were 40 differentially expressed transcripts in frontal scalp compared with vertex after MTF 5% treatment. Regional variations in gene expression changed after the treatment with MTF 5%.
Signaling pathways analysis
We performed signaling pathways activation analysis using information regarding pathways from various databases. The results are summarized in .
Table 5. iPANDA signaling pathways activation analysis. Information about signaling pathways was obtained from various pathway databases.
Comparison of frontal and vertex scalp before treatment
We observed variations in signaling pathways activation between frontal and vertex scalp before treatment. Interleukin 2 (IL-2), integrin-linked kinase (ILK), mitogen activated protein kinase (MAPK), transforming growth factor β (TGF-β), Janus kinase/signal transducers and activators of transcription (JAK/STAT), phosphatase and tensin homolog (PTEN), and Akt (v-Akt Murine Thymoma Viral Oncogene)/PKB (protein kinase B) pathways were upregulated, while Presenilin action in Notch and Wnt signaling and IL-6 pathways were downregulated in frontal vs. vertex scalp before treatment.
Vertex scalp after the use of MTF 5%
Vertex scalp of responders exhibited upregulation of PTEN and Cellular apoptosis pathways and downregulation of ILK, Akt, mechanistic target of rapamycin (mTOR), JAK/STAT, MAPK, and Ras pathways after treatment with MTF 5%.
Frontal scalp after the use of MTF 5%
The following pathways were upregulated in frontal scalp after the use of MTF 5%: Protein digestion and absorption (KEGG), Ras, mTOR, and Wingless-related integration site (Wnt) pathways. Downregulated pathways included Akt, PTEN, MAPK, and ILK pathways.
Comparison of frontal and vertex scalp after the use of MTF 5%
Regional variations between frontal and vertex scalp were also observed after the treatment with MTF 5% on a pathway level. Ras, IL-2, mTOR, Protein digestion and absorption (KEGG) pathways were upregulated, while Akt, ILK, MAPK, PTEN pathways were downregulated in frontal vs. vertex scalp of the responders.
Discussion
Our analysis revealed that both genes and non-coding RNAs were differentially expressed between vertex and frontal scalp before and after treatment with MTF 5%.
We observed variations in gene expression in the 2 scalp regions before treatment. Several genes, including genes induced by oxidative stress and growth factors (DUSP1, CYR61),Citation26-28 and non-coding RNAs were upregulated in frontal compared with vertex scalp, while the expression of pseudogene MSL3P1 that may be involved in chromatin remodeling and regulation of transcription was decreased. These results suggest that gene expression in hair follicles in vertex versus frontal scalp of patients with AGA may not be completely identical and exhibit different molecular signatures.
In general, vertex and frontal scalp showed similar molecular response to MTF 5% treatment. The strong upregulation of keratin-associated genes in both the vertex and frontal regions was a distinctive feature of responding patients, while patients who showed minimal response to MTF 5% treatment did not exhibit a similar level of upregulation of keratin-associated genes. The expression of several non-coding RNAs was significantly decreased in both scalp regions after treatment. It should be noted that keratinization-related genes were downregulated in the frontal scalp of both responders and placebo control patients. Thus, it is not clear whether the decreased expression of late cornified envelope protein 3D (LCE3D) was caused by the MTF 5% treatment or other as yet unidentified factors.
We also observed regional variations in gene expression after treatment with MTF 5%. Collagen type VI α 6 chain (COL6A6), fatty acid and lipid metabolism genes (ACOT4, CIDEA) and several non-coding RNAs were upregulated in frontal scalp compared with vertex. The downregulated genes included keratin-encoding and keratin-associated genes as well as genes that play a role in cation channel activity and extracellular matrix binding. It is tempting to speculate that these differences in gene expression reflect the underlying mechanism of the response of hair follicles from 2 scalp regions to MTF 5% treatment.
Baseline regional variations between frontal and vertex scalp were also visible upon pathway level examination. Thus, IL-2, which is being studied in the context of alopecia areata,Citation29 and ILK, a key mediator in integrin signal transduction, were upregulated pathways identified in frontal compared with vertex scalp. The following pathways were also upregulated in frontal vs. vertex scalp before treatment: MAPK (facilitates the survival of dermal papilla cellsCitation30), TGF-β (induces catagenCitation31), JAK/STAT (prevents anagen reentryCitation32), PTEN, Akt (promotes dermal papilla cells survival and anagen initiationCitation33,34). The downregulated pathways included IL-6, which may provoke inflammation, and Presenilin action in Notch and Wnt signaling (NCI) pathways. It has been shown that activation of Notch signaling is necessary for keratinocyte differentiation,Citation35,36 and Wnt signaling drives hair follicle morphogenesis, hair shaft differentiation, hair cycling induction and maintenance.Citation37-42
ILK, Akt, and MAPK pathways became downregulated in both vertex and frontal scalp of responders after treatment with MTF 5%. Previous studies have shown that Akt and MAPK pathways promote dermal papilla cell survival,Citation30,33,34 so the effect of minoxidil on these pathway requires further clarification.
JAK/STAT signaling and Ras pathways were downregulated in vertex scalp of patients who responded to MTF 5% treatment. JAK/STAT inhibition should promote hair growth, while Ras pathway regulates cellular proliferation, differentiation, and senescence by stimulating various parallel effector pathways.Citation43
Protein digestion and absorption pathway, which includes several collagen-encoding genes, was significantly upregulated in the frontal scalp of responders. MTF 5% treatment also caused upregulation of mTOR pathway in the frontal scalp region. It is known that mTOR pathway promotes hair follicle stem cells proliferation and activation, and is activated at the moment of telogen-to-anagen transition.Citation44,45
The results of our analysis suggest that vertex and frontal scalp of AGA patients showed generally similar response to minoxidil treatment implying that minoxidil can be recommended for both vertex and frontal scalp. However, there were some differences in response to the treatment at the level of gene expression and signaling pathways response. Both scalp regions showed upregulation of genes that encode keratin-associated proteins and downregulation of ILK, Akt, and MAPK signaling pathways after MTF 5% treatment, suggesting that control of inflammation in conjunction with keratin stimulation may contribute to the improvement of hair growth disorders. To our knowledge this is the first time that regional variations in the activation of signaling pathways before and after treatment with MTF 5% has received significant attention. These results were based on available data from a study involving a minoxidil foam product,Citation25 and as such may not be representative of all products containing minoxidil.
Methods
We analyzed the microarray gene expression data from a minoxidil study conducted by the Skin Study Center at Case Western Reserve University, Cleveland, OH. Gene expression data were obtained from patients enrolled in a placebo controlled double-blinded study of a formulated foam product containing 5% minoxidil (Men's Rogaine Hair Loss & Hair Thinning Treatment Minoxidil Foam, Johnson & Johnson Consumer Products, Skillman, NJ). Healthy men aged 18–49 with Hamilton-Norwood type IV-V thinning were instructed to apply the treatment (active drug or placebo) topically to the affected area. Scalp biopsies from the frontal and vertex scalp were done from the leading edge of alopecia and global hair photographs were taken before and after 8 weeks of treatment. Based on a blinded review of stereotactic photographs patients were categorized as having either full or minimal clinical response. А responder was defined as a subject who showed significant hair growth based on stereotactic photographs after 8 weeks treatment with 5% MTF. Microarray analysis was done using the Affymetrix GeneChip HG U133 Plus 2.0. The original results were published by Mirmirani et al.Citation25
For raw gene expression data preprocessing we used the Frozen RMA (fRMA) method implemented in frma R package.Citation46-48 We only considered transcripts with a Benjamini q-value < 0.05 and log2(fold change) > 0.4, which is equivalent to a more than 30% change in expression level, to be significant. If a value of expression lied outside the mean level of variation observed in the study, the sample was labeled as an outlier and was excluded from the analysis. Pathway activation analysis was performed using in silico Pathway Activation Network Decomposition Analysis (iPANDA).Citation49 The top signaling pathways with cutoff equal to 30 arb. units on pathway score scale was chosen based on the distribution of scores in the pathway collection. Such threshold corresponds to approximately 15% perturbed pathways for each experimental group. SABiosciences, KEGG, Reactome and NCI pathway collections containing 2071 pathways and 9502 unique genes were used for the analysis (http://www.sabiosciences.com/pathwaycentral.php.).Citation50-52
Disclosure of potential conflicts of interest
The authors declare no conflict of interest. The authors are employed by the companies that may benefit from new insights into the androgenic alopecia and the mechanism of action of minoxidil in AGA.
Acknowledgments
We would like to thank Nvidia for providing the high-performance computing equipment used in this study.
Funding
The described analysis was funded by Johnson & Johnson Consumer Companies, Inc. The original clinical study was supported by an independent investigator grant from Johnson & Johnson Consumer Companies, Inc., and in part by National Institutes of Health grant NIH R01 AR056245.
References
- Sawaya ME, Price VH. Different levels of 5alpha-reductase type I and II, aromatase, and androgen receptor in hair follicles of women and men with androgenetic alopecia. J Invest Dermatol 1997; 109:296-300; PMID:9284093; https://doi.org/10.1111/1523-1747.ep12335779
- Dawber RP. The embryology and development of human scalp hair. Clin Dermatol 1988; 6:1-6; PMID:3063365; https://doi.org/10.1016/0738-081X(88)90059-4
- Courtois M, Loussouarn G, Hourseau C, Grollier JF. Hair cycle and alopecia. Skin Pharmacol 1994; 7:84-9; PMID:8003330; https://doi.org/10.1159/000211279
- Guarrera M, Rebora A. Anagen hairs may fail to replace telogen hairs in early androgenic female alopecia. Dermatology 1996; 192:28-31; PMID:8832948; https://doi.org/10.1159/000246309
- Ellis JA, Sinclair R, Harrap SB. Androgenetic alopecia: Pathogenesis and potential for therapy. Expert Rev Mol Med 2002; 4:1-11; PMID:14585162; https://doi.org/10.1017/S1462399402005112
- Oliver RF, Jahoda CAB. The dermal papilla and maintenance of hair growth. In: Rogers GE, Reis PJ, Ward KA, Marshall RC, editors. The Biology of Wool and Hair. Netherlands: Springer; 1988; page 51-67
- Obana NJ, Uno H. Dermal papilla cells in macaque alopecia trigger a testosterone-dependent inhibition of follicular cell proliferation. Hair research in the next millennium Amsterdam: Elsevier; 1996; 307-10
- Randall VA. The use of dermal papilla cells in studies of normal and abnormal hair follicle biology. Dermatol Clin 1996; 14:585-94; PMID:9238318; https://doi.org/10.1016/S0733-8635(05)70386-7
- Sinclair R, Torkamani N, Jones L. Androgenetic alopecia: New insights into the pathogenesis and mechanism of hair loss. F1000Res 2015; 4:585; PMID:26339482; https://doi.org/10.12688/f1000research.6401.1
- Jaworsky C, Kligman AM, Murphy GF. Characterization of inflammatory infiltrates in male pattern alopecia: Implications for pathogenesis. Br J Dermatol 1992; 127:239-46; PMID:1390168; https://doi.org/10.1111/j.1365-2133.1992.tb00121.x
- Sueki H, Stoudemayer T, Kligman AM, Murphy GF. Quantitative and ultrastructural analysis of inflammatory infiltrates in male pattern alopecia. Acta Derm Venereol 1999; 79:347-50; PMID:10494708; https://doi.org/10.1080/000155599750010238
- Vogt A, Pfannes EKB, Fimmel S, Hadam S, Andruck A, Kottner J, Blume-Peytavi U. Infundibular protein and RNA-microarray analyses from affected and clinically non-affected scalp in male androgenetic alopecia patients. Exp Dermatol [Internet] 2017; 26(6):518-521; PMID:28266729; https://doi.org/10.1111/exd.13326
- Wester RC, Maibach HI, Guy RH, Novak E. Minoxidil stimulates cutaneous blood flow in human balding scalps: Pharmacodynamics measured by laser Doppler velocimetry and photopulse plethysmography. J Invest Dermatol 1984; 82:515-7; PMID:6239893; https://doi.org/10.1111/1523-1747.ep12261084
- Uno H, Cappas A, Brigham P. Action of topical minoxidil in the bald stump-tailed macaque. J Am Acad Dermatol 1987; 16:657-68; PMID:3558911; https://doi.org/10.1016/S0190-9622(87)70084-X
- Sakita S, Kagoura M, Toyoda M, Morohashi M. The induction by topical minoxidil of increased fenestration in the perifollicular capillary wall. Br J Dermatol 1999; 140:294-6; PMID:10233226; https://doi.org/10.1046/j.1365-2133.1999.02666.x
- Shorter K, Farjo NP, Picksley SM, Randall VA. Human hair follicles contain two forms of ATP-sensitive potassium channels, only one of which is sensitive to minoxidil. FASEB J 2008; 22:1725-36; PMID:18258787; https://doi.org/10.1096/fj.07-099424
- Borisov NM, Terekhanova NV, Aliper AM, Venkova LS, Smirnov PY, Roumiantsev S, Korzinkin MB, Zhavoronkov AA, Buzdin AA. Signaling pathways activation profiles make better markers of cancer than expression of individual genes. Oncotarget 2014; 5:10198-205; PMID:25415353; https://doi.org/10.18632/oncotarget.2548
- Lezhnina K, Kovalchuk O, Zhavoronkov AA, Korzinkin MB, Zabolotneva AA, Shegay PV, Sokov DG, Gaifullin NM, Rusakov IG, Aliper AM, et al. Novel robust biomarkers for human bladder cancer based on activation of intracellular signaling pathways. Oncotarget 2014; 5:9022-32; PMID:25296972; https://doi.org/10.18632/oncotarget.2493
- Makarev E, Cantor C, Zhavoronkov A, Buzdin A, Aliper A, Csoka AB. Pathway activation profiling reveals new insights into age-related macular degeneration and provides avenues for therapeutic interventions. Aging 2014; 6:1064-75; PMID:25543336; https://doi.org/10.18632/aging.100711
- Zhu Q, Izumchenko E, Aliper AM, Makarev E, Paz K, Buzdin AA, Zhavoronkov AA, Sidransky D. Pathway activation strength is a novel independent prognostic biomarker for cetuximab sensitivity in colorectal cancer patients. Hum Genome Var 2015; 2:15009; PMID:27081524; https://doi.org/10.1038/hgv.2015.9
- Aliper AM, Csoka AB, Buzdin A, Jetka T, Roumiantsev S, Moskalev A, Zhavoronkov A. Signaling pathway activation drift during aging: Hutchinson-Gilford Progeria Syndrome fibroblasts are comparable to normal middle-age and old-age cells. Aging 2015; 7:26-37; PMID:25587796; https://doi.org/10.18632/aging.100717
- Shepelin D, Korzinkin M, Vanyushina A, Aliper A, Borisov N, Vasilov R, Zhukov N, Sokov D, Prassolov V, Gaifullin N, et al. Molecular pathway activation features linked with transition from normal skin to primary and metastatic melanomas in human. Oncotarget 2016; 7:656-70; PMID:26624979; https://doi.org/10.18632/oncotarget.6394
- Zhavoronkov A, Kanherkar RR, Izumchenko E, Teka M, Cantor C, Manaye K, Sidransky D, West MD, Makarev E, Csoka AB. Pro-fibrotic pathway activation in trabecular meshwork and lamina cribrosa is the main driving force of glaucoma. Cell Cycle 2016; 15:1643-52; PMID:27229292; https://doi.org/10.1080/15384101.2016.1170261
- Karnik P, Shah S, Dvorkin-Wininger Y, Oshtory S, Mirmirani P. Microarray analysis of androgenetic and senescent alopecia: Comparison of gene expression shows two distinct profiles. J Dermatol Sci 2013; 72:183-6; PMID:23886704; https://doi.org/10.1016/j.jdermsci.2013.06.017
- Mirmirani P, Consolo M, Oyetakin-White P, Baron E, Leahy P, Karnik P. Similar response patterns to topical minoxidil foam 5% in frontal and vertex scalp of men with androgenetic alopecia: A microarray analysis. Br J Dermatol 2015; 172:1555-61; PMID:25204361; https://doi.org/10.1111/bjd.13399
- Keyse SM, Emslie EA. Oxidative stress and heat shock induce a human gene encoding a protein-tyrosine phosphatase. Nature 1992; 359:644-7; PMID:1406996; https://doi.org/10.1038/359644a0
- Charles CH, Sun H, Lau LF, Tonks NK. The growth factor-inducible immediate-early gene 3CH134 encodes a protein-tyrosine-phosphatase. Proc Natl Acad Sci U S A 1993; 90:5292-6; PMID:8389479; https://doi.org/10.1073/pnas.90.11.5292
- Chin L-H, Hsu S-P, Zhong W-B, Liang YC. Involvement of cysteine-rich protein 61 in the epidermal growth factor-induced migration of human anaplastic thyroid cancer cells. Mol Carcinog 2016; 55:622-32; PMID:25773758; https://doi.org/10.1002/mc.22308
- Gregoriou S, Papafragkaki D, Kontochristopoulos G, Rallis E, Kalogeromitros D, Rigopoulos D. Cytokines and other mediators in alopecia areata. Mediators Inflamm 2010; 2010:928030; PMID:20300578; https://doi.org/10.1155/2010/928030
- Han JH, Kwon OS, Chung JH, Cho KH, Eun HC, Kim KH. Effect of minoxidil on proliferation and apoptosis in dermal papilla cells of human hair follicle. J Dermatol Sci 2004; 34:91-8; PMID:15033191; https://doi.org/10.1016/j.jdermsci.2004.01.002
- Hibino T, Nishiyama T. Role of TGF-beta2 in the human hair cycle. J Dermatol Sci 2004; 35:9-18; PMID:15194142; https://doi.org/10.1016/j.jdermsci.2003.12.003
- Harel S, Higgins CA, Cerise JE, Dai Z, Chen JC, Clynes R, Christiano AM. Pharmacologic inhibition of JAK-STAT signaling promotes hair growth. Sci Adv 2015; 1:e1500973; PMID:26601320; https://doi.org/10.1126/sciadv.1500973
- Murayama K, Kimura T, Tarutani M, Tomooka M, Hayashi R, Okabe M, Nishida K, Itami S, Katayama I, Nakano T. Akt activation induces epidermal hyperplasia and proliferation of epidermal progenitors. Oncogene 2007; 26:4882-8; PMID:17297448; https://doi.org/10.1038/sj.onc.1210274
- Kwon OS, Han JH, Yoo HG, Chung JH, Cho KH, Eun HC, Kim KH. Human hair growth enhancement in vitro by green tea epigallocatechin-3 gallate (EGCG). Phytomedicine 2007; 14:551-5; PMID:17092697; https://doi.org/10.1016/j.phymed.2006.09.009
- Rangarajan A, Talora C, Okuyama R, Nicolas M, Mammucari C, Oh H, Aster JC, Krishna S, Metzger D, Chambon P, et al. Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J 2001; 20:3427-36; PMID:11432830; https://doi.org/10.1093/emboj/20.13.3427
- Lin HY, Kao CH, Lin KMC, Kaartinen V, Yang LT. Notch signaling regulates late-stage epidermal differentiation and maintains postnatal hair cycle homeostasis. PLoS One 2011; 6:e15842; PMID:21267458; https://doi.org/10.1371/journal.pone.0015842
- Andl T, Reddy ST, Gaddapara T, Millar SE. WNT signals are required for the initiation of hair follicle development. Dev Cell 2002; 2:643-53; PMID:12015971; https://doi.org/10.1016/S1534-5807(02)00167-3
- Heilmann S, Kiefer AK, Fricker N, Drichel D, Hillmer AM, Herold C, Tung JY, Eriksson N, Redler S, Betz RC, et al. Androgenetic alopecia: Identification of four genetic risk loci and evidence for the contribution of WNT signaling to its etiology. J Invest Dermatol 2013; 133:1489-96; PMID:23358095; https://doi.org/10.1038/jid.2013.43
- Wodarz A, Nusse R. Mechanisms of Wnt signaling in development. Annu Rev Cell Dev Biol 1998; 14:59-88; PMID:9891778; https://doi.org/10.1146/annurev.cellbio.14.1.59
- Shimizu H, Morgan BA. Wnt signaling through the beta-catenin pathway is sufficient to maintain, but not restore, anagen-phase characteristics of dermal papilla cells. J Invest Dermatol 2004; 122:239-45; PMID:15009701; https://doi.org/10.1046/j.0022-202X.2004.22224.x
- Li Y-H, Zhang K, Ye J-X, Lian X-H, Yang T. Wnt10b promotes growth of hair follicles via a canonical Wnt signalling pathway. Clin Exp Dermatol 2011; 36:534-40; PMID:21392083; https://doi.org/10.1111/j.1365-2230.2011.04019.x
- Kwack MH, Kang BM, Kim MK, Kim JC, Sung YK. Minoxidil activates β-catenin pathway in human dermal papilla cells: A possible explanation for its anagen prolongation effect. J Dermatol Sci 2011; 62:154-9; PMID:21524889; https://doi.org/10.1016/j.jdermsci.2011.01.013
- Rishikaysh P, Dev K, Diaz D, Qureshi WMS, Filip S, Mokry J. Signaling involved in hair follicle morphogenesis and development. Int J Mol Sci 2014; 15:1647-70; PMID:24451143; https://doi.org/10.3390/ijms15011647
- Deng Z, Lei X, Zhang X, Zhang H, Liu S, Chen Q, Hu H, Wang X, Ning L, Cao Y, et al. mTOR signaling promotes stem cell activation via counterbalancing BMP-mediated suppression during hair regeneration. J Mol Cell Biol 2015; 7:62-72; PMID:25609845; https://doi.org/10.1093/jmcb/mjv005
- Castilho RM, Squarize CH, Chodosh LA, Williams BO, Gutkind JS. mTOR mediates Wnt-induced epidermal stem cell exhaustion and aging. Cell Stem Cell 2009; 5:279-89; PMID:19733540; https://doi.org/10.1016/j.stem.2009.06.017
- McCall MN, Bolstad BM, Irizarry RA. Frozen robust multiarray analysis (fRMA). Biostatistics 2010; 11:242-53; PMID:20097884; https://doi.org/10.1093/biostatistics/kxp059
- McCall MN, Uppal K, Jaffee HA, Zilliox MJ, Irizarry RA. The Gene Expression Barcode: Leveraging public data repositories to begin cataloging the human and murine transcriptomes. Nucleic Acids Res 2011; 39:D1011-5; PMID:21177656; https://doi.org/10.1093/nar/gkq1259
- McCall MN, Jaffee HA, Irizarry RA. fRMA ST: Frozen robust multiarray analysis for Affymetrix Exon and Gene ST arrays. Bioinformatics 2012; 28:3153-4; PMID:23044545; https://doi.org/10.1093/bioinformatics/bts588
- Ozerov IV, Lezhnina KV, Izumchenko E, Artemov AV, Medintsev S, Vanhaelen Q, Aliper A, Vijg J, Osipov AN, Labat I, et al. In silico Pathway Activation Network Decomposition Analysis (iPANDA) as a method for biomarker development. Nat Commun 2016; 7:13427; PMID:27848968; https://doi.org/10.1038/ncomms13427
- Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000; 28:27-30; PMID:10592173; https://doi.org/10.1093/nar/28.1.27
- Croft D, Mundo AF, Haw R, Milacic M, Weiser J, Wu G, Caudy M, Garapati P, Gillespie M, Kamdar MR, et al. The Reactome pathway knowledgebase. Nucleic Acids Res 2014; 42:D472-7; PMID:24243840; https://doi.org/10.1093/nar/gkt1102
- Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, Hannay T, Buetow KH. PID: The pathway interaction database. Nucleic Acids Res 2009; 37:D674-9; PMID:18832364; https://doi.org/10.1093/nar/gkn653
