ABSTRACT
Recently, long non-coding RNAs (lncRNAs) have emerged as new gene regulators and prognostic markers in several types of cancer, including renal cell carcinoma (RCC). In this study, we identified an upregulated lncRNA, DLX6-AS1, in RCC tumor tissues compared with normal kidney tissues. Our data suggested that DLX6-AS1 promoted RCC cell growth and tumorigenesis via targeting miR-26a. In addition, we observed that PTEN overexpression restored the renal cancer cell growth and also rescued the RCC tumorigenesis. In summary, we conclude that DLX6-AS1 promotes renal cell carcinoma development via regulation of miR-26a/PTEN axis.
Introduction
Renal cell carcinoma (RCC) is one of the most common urinary tract malignancies in adults. In renal cell carcinoma (RCC), surgical resection, either via nephron-sparing or radical nephrectomy, can be curative in patients with early stage, localized disease. However, no adjuvant treatment has been proven to be beneficial.Citation1,2 A significant proportion of patients eventually develop tumor recurrence, and subsequent therapeutic options in the advanced or metastatic setting are limited. Renal cell carcinoma is the most common subtype of RCC and is responsible for nearly 85% of all RCC cases. In China, an estimated of 66.8 ‰ new cases and 23.4 ‰ deaths from renal cancer occurred in 2015.Citation3 Despite numerous studies that have shown that many oncogenic factors are associated with the progression of RCC, the molecular mechanism of renal cancer pathogenesis and the prognosis still remains unclear.
Long noncoding RNAs (lncRNAs) are a newly discovered class of noncoding RNAs (ncRNA) that are longer than 200 nucleotides and are not translated into proteins.Citation4 Numerous evidence has indicated that lncRNAs play important roles in diverse biologic processes, such as cell growth, cell death, tumor cell metastasis, tumorigenesis and development.Citation5 It has been wildly investigated that lncRNAs modulate many types of tumor progression.Citation6-8 For example, lncRNA LOC100129148 and EWSAT1 have been reported to act as oncogenic role in nasopharyngeal carcinoma,Citation9,10 lncRNA 00858 functions as a competing endogenous RNA for miR-422a to facilitate the cell growth in non-small cell lung cancerCitation11 and Increased expression of long-noncoding RNA ZFAS1 is associated with epithelial-mesenchymal transition of gastric cancer.Citation12 It also has been reported that high DLX6-AS1 expression levels were significantly associated with both histological differentiation and TNM stage of lung cancer. Down-regulation of lncRNA DLX6-AS1 expression decreased the DLX6 mRNA and protein levels.Citation13 However, the role of DLX6-AS1 in renal cell cancer is still unclear.
Recently, accumulating articles revealed that one potential function of lncRNAs was to directly interact with microRNAs (miRNAs) as a sponge and regulate their expression and activity.Citation14,15 MiRNAs, a class of short (18–24 nt), single stranded and noncoding RNA molecules, are involved in regulating gene transcription and expression via directly binding with the target mRNAs.Citation16 miR-26a and miR-26b inhibit esophageal squamous cancer cell proliferation via suppression of MYC signaling.Citation17 It has also been reported miR-26 was significantly reduced in colorectal cancer via targeting Rb1.Citation18
In summary, our study revealed that lncRNA DLX6-AS1 was increased in renal cell cancer tissues compared with paired adjacent normal kidney tissues via suppressing miR-26a expression. Luciferase reporter assays and western blot assays showed that PTEN was the direct target of miR-26a in renal cancer cells. Taken together, we conclude that DLX6-AS1 mediates the progression of renal cell carcinoma via miR-26a/PTEN axis.
Materials and methods
Clinic samples
From 2014 to 2017, 52 RCC patients were recruited in tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology. Inform content was obtained from every patient before the surgery. Tumor tissues and paired normal kidney tissues were collected immediately after resection and was stored in liquid nitrogen before further use.
Cell culture
The human renal cancer cell lines (A498, ACHN, Caki-1, Caki-2, 786-O and G401) and normal kidney HK-2 cells were obtained from ATCC. Cells were cultured in RPMI-1640 medium (Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum and 100 U/ml penicillin/streptomycin (Life Technologies, USA) in a humidified incubator at 37°C with 5% CO2.
MTT assays
Exponentially growing cells were seeded at 10,000 cells (100 μl culture medium) per well in 96-well plates and incubated for 12 h. The cells were then transfected with DLX6-AS1 siRNAs or miR-26a mimics, then 20 μl of MTT (Sigma Chemicals, St. Louis, MO, USA; 5 mg/ml in PBS) was added to each well, and the cells were cultured for an additional 4 h. Subsequently, 200 μl of DMSO was added to each well to dissolve the crystals. The values of the optical density at 490 nm were then measured using a micro-plate reader.
Flow cytometry
RCC cells were transfected with miR-26a mimics or DLX6-AS1 siRNAs or PTEN overexpression plasimids for flow cytometry analysis using an Annexin V Apoptosis Detection Kit (Becton Dickinson, NJ, USA), untreated group, miR-nc or siNC were considered as control. Cells were stained with Annexin V-fluorescein isothiocyanate (FITC), Propidium Iodide (PI) for 25 min, and then analyzed by flow cytometry (BD CantoII). FACS data were analyzed using FlowJo (Tree Star, Inc.). A498 cells were transfected with DLX6-AS1 siRNAs or miR-26a mimics for 48h and then the cells were analyzed by flow cytometry as the following procedures: after trypsinization, 1 × 106 cells were centrifuged for 5 min at 500 g. The cellular pellet was fixed with ice-cold 70% ethanol and incubated for 16 h at −20°C. The cells were centrifuged at 500 g at 4°C for 5 min, washed once with PBS and stained in the dark for 40 min at 37°C with a 500μl propidium iodide (PtdIns) solution (0.05% Triton X-100, 5 U/ml RNaseA and 50μg/ml propidium iodide in PBS). Afterward, cells were centrifuged for 5 min at 500 g, resuspended in 1 ml PBS, filtered using a cell strainer snap-cap tube (BD Falcon™) and immediately acquired in a Flow Cytometer (Beckman Coulter, Inc.). These data were analyzed by Modifit software.
Quantitative PCR
Total RNAs were extracted from cancer cells by using RNAiso Plus reagent (Takara Biotechnology Co., Ltd, DALIAN). To detect miR-26a expression, total RNAs were reversed using MMLV reverse transcriptase. The resultant cDNA was then used as template to perform real time PCR using real time PCR kit (Qiagen). Transcripts were quantified by real time PCR and normalized to the amount of U6 mRNA expression. For DLX6-AS1 and PTEN mRNA expression analysis, first strand cDNA was synthesized by using cDNA synthesis kit (Takara) according to the manufacturer's instructions. Their expression at mRNA level were detected by using Syber Green PCR mastermix (Applied Biosystems). The PCR primers for DLX6-AS1 or GAPDH were as follows: DLX6-AS1 forward, 5′-AGTTTCTCTCTAGATTGCCTT-3′ and reverse, 5′-ATTGACATGTTAGTGCCCTT-3′; GAPDH forward, 5′-AGCCACATCGCTCAGACAC-3′ and reverse, 5′-GCCCAATACGACCAAATCC-3′. Specific siRNAs targeting lncRNA DLX6-AS1 and the negative control siRNAs were designed and synthesized by GenePharma (Shanghai, China). Relative expression of genes was calculated using the comparative cycle threshold (Ct) (2−ΔCt, ΔCt = Ct median lncRNA or mRNA-Ct median GAPDH) method with GAPDH.
Western blot
RCC cell lysates were prepared with RIPA Lysis buffer (Beyotime, China) containing protease inhibitor cocktail (Roche). Protein samples were loaded for sodium dodecylsulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a nitrocellulose membrane. After a blockage of 5% fat-free milk, the membrane was probed with primary anti-PTEN (dilution 1:1000, Cell Signaling Technology) and anti-GAPDH (dilution 1:2000, Santa Cruz) antibody. After washing, the membrane was incubated with horseradish peroxidase-conjugated (HRP) secondary antibody (1:2000, Santa Cruz Biotechnology) for 1 h. The signal was visualized using the ECL detection system (Thermo Fisher, USA) and quantified by densitometry using Quantity One software (Bio-Rad, Hercules, CA, USA).
Xenograft tumor model
Six to 8-week-old BALB/c (nu/nu) mice were purchased from Shanghai SLAC Laboratory Animal Co. All mice were maintained in a barrier facility at Animal Center of Chongqing Medical University. 5.0*106 stably expressing miR-26a or DLX6-AS1 siRNA overexpression RCC cells or scramble control cells were implanted subcutaneously (s.c.) into the right flank of mice. All groups of mice were killed and tumors were weighted at the end point of this experiments.
Colony formation assays
One.0*103 cells, which were transduced with miR-26a or DLX6-AS1 siRNAs or PTEN overexpression plasmids or control mimics were seeded into 6-well plates in 2 ml of complete growth medium. The medium of each well was changed every 3 d. Two to 3 weeks later, cells were stained by 0.1% crystal violet (Sigma-Aldrich, St. Louis, MO, USA) in methanol for 10 min. The colonies more than 50μm were counted directly on the plate. Statistical significance was calculated from at least 3 independent experiments.
Luciferase reporter assays
The DNA oligonucleotide and the pMiR-Reporter Vector were used to build the luciferase report vectors (pMiR-DLX6-AS1-WT/pMiR-DLX6-AS1-Mutant and pMiR-PTEN-WT/pMiR-PTEN-Mutant). HEK293 cells were co-transfected with pMiR-DLX6-AS1-WT or pMiR-DLX6-AS1-Mutant and miR-26a mimics or negative control (NC). A Renilla luciferase-expressing plasmid pRL-TK (Promega) used as control was also co-transfected. Cells were harvested and luciferase activity was determined using the Dual Luciferase Reporter Assay Kit (Promega) at 24 h after transfection. The results are expressed as relative luciferase activity (firefly luciferase/Renilla luciferase).
RNA immunoprecipitation
RIP was performed using the EZ-Magna RIP kit (Millipore, USA) following the manufacturer's protocol. Renal cancer cells were collected and lysed in complete RIPA buffer. The whole cell protein extract was then incubated with RIP wash buffer containing magnetic beads conjugated with anti-Ago2 antibody (Millipore) or mouse immunoglobulin G (IgG) control. The protein in the samples was digested with proteinase K, and the immunoprecipitated RNA was isolated. Finally, purified RNA was subjected to a qRT-PCR analysis to demonstrate the presence of the binding targets.
Statistical analysis
Data were presented as mean ± SEM. Group comparison was performed by Student's t-test. P value <0.05 was considered as significant difference.
Results
DLX6-AS1 expression is upregulated in renal cell carcinoma (RCC)
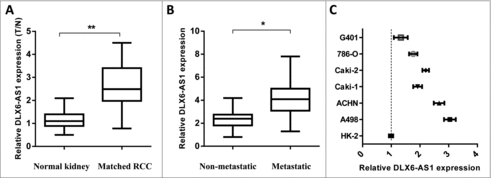
To investigate the relevance of lncRNA DLX6-AS1 in RCC progression, 15 RCC samples and paired normal adjacent tissues were subject to q-PCR analysis. It shows that the expression of DLX6-AS1 was significantly increased (). In addition, the expression of DLX6-AS1 in metastatic RCC was shown higher level than that in non-metastatic RCC samples (). These results revealed that DLX6-AS1 might play an oncogenic role in RCC development. We then performed q-PCR to analyze DLX6-AS1 expression in RCC cell lines and normal renal cell line (HK-2). As compared with HK-2 cells, overexpressed levels of DLX6-AS1 expression was seen in all RCC cell line (). These observations suggested that DLX6-AS1 might be involved in the regulation of RCC development.
Figure 1. DLX6-AS1 expression is elevated in RCC tissue. (A) q-PCR analysis was performed for testing the DLX6-AS1 expression level in RCC tissues and matched normal kidney tissues. (B) The expression level of DLX6-AS1 in metastatic RCC tissues and non-metastatic RCC tissues was examined by q-PCR. (C) The expression of DLX6-AS1 in several RCC cancer cell lines and normal kidney cell (HK-2 cells). All data was shown as mean ± s.e.m. from 3 independent experiments. *p < 0.05, **p < 0.01.
DLX6-AS1 binds to miR-26a and represses miR-26a expression in RCC cells
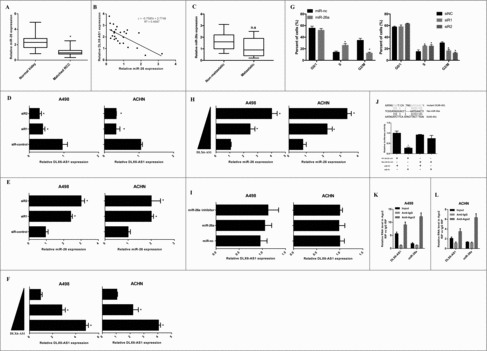
To determine the mechanism of action for DLX6-AS1 in RCC development, we first explored the expression of several microRNAs in RCC and paired normal kidney tissues. Excitingly, miR-26a showed the significant higher level in RCC (). We also identified that miR-26a as a potential target of DLX6-AS1 with Starbase (http://starbase.sysu.edu.cn). There is a putative binding sites of DLX6-AS1 and miR-26a (). To investigate whether DLX6-AS1 could regulate miR-26a expression in RCC, the expression of miR-26a and DLX6-AS1 were examined. Q-PCR analysis revealed that DLX6-AS1 expression was negatively correlated with miR-26a level in RCC samples (). These results suggest that DLX6-AS1 might directly bound to miR-26a and repressed its expression level in RCC cells. To test whether miR-26a was indeed a target of DLX6-AS1, we first knockdown DLX6-AS1 expression in 2 independent RCC cell lines, A498 and ACHN cells, by siRNA transfection. However, our results also revealed that the expression of mir-26a in metastatic RCC tissues are not reduced compared with the non-metastatic RCC tissues (). These results demonstrated that miR-26a might not regulate the metastasis of RCC tumors. The efficacy of DLX6-AS1 knockdown was confirmed by q-PCR analysis in these 2 RCC cell lines (). In both DLX6-AS1 knockdown cells, we observed elevated expression levels of miR-26a (), whereas DLX6-AS1 overexpression significantly suppressed the expression of miR-26b in these 2 RCC cell lines ( and ). Importantly, overexpression or knockdown of miR-26a didn't cause any change in DLX6-AS1 expression (), indicating that miR-26a was downstream of DLX6-AS1. Furthermore, we also performed flow cytometry analysis to explore whether miR-26a overexpression or DLX6-AS1 knockdown could regulate RCC cell cycle. To our surprise, both miR-26a overexpression and DLX6-AS1 knockdown showed RCC cell S phase to G2/M phase arrest (). We also performed luciferase reporter assays to explore whether DLX6-AS1 could directly bind to miR-26a. As shown in , miR-26a significantly repressed the luciferase activity of which transfected with the reporter plasmid, which containing DLX6-AS1 in the downstream of luciferase gene. To extensively investigate whether DLX6-AS1 and miR-26a binding together, we performed pulldown assays. The results showed that DLX6-AS1 was confirmed directly binding with miR-26a in RCC cells ( and ). Taken together, these results supported that miR-26a was an inhibitory target of DLX6-AS1 in both RCC cells and RCC tissues.
Figure 2. DLX6-AS1 binds to miR-26a and represses its expression. (A) RCC tumor tissues and paired kidney tissues were subjected to q-PCR analysis for miR-26a expression level detection. (B) The correlation between miR-26a and DLX6-AS1 level in RCC tumor tissues were analyzed by q-PCR. (C) The expression of miR-26a in non-metastatic and metastatic RCC tissues were examined by q-PCR. (D and E) The expression of DLX6-AS1 and miR-26a were detected by q-PCR analysis in A498 and ACHN cells after transfecting with siRNAs and control siRNA (siR-control). (F and H) The expression of DLX6-AS1 and miR-26a were detected by q-PCR analysis in RCC cells after DLX6-AS1 overexpression. (G) Flow cytometry analysis were performed in miR-26a overexpression or DLX6-AS1 knockdwon A498 cells. (I) The expression level of DLX6-AS1 in RCC cells was detected after miR-26a overexpression. (J) Schematic illustration of the predicted binding sites between DLX6-AS1 and miR-26a, and luciferase reporter assays in 293T cells transfected DLX6-AS1 wild type or mutant with miR-26a. (K and L) Cellular lysates from A498 and ACHN cells were used for RIP with an Ago2 antibody and IgG antibody and the levels of DLX6-AS1 and miR-26a were detected by qRT-PCR. All data was shown as mean ± s.e.m. from 3 independent experiments. *p < 0.05.
DLX6-AS1 knockdown and miR-26a overexpression suppresses the growth of RCC cells
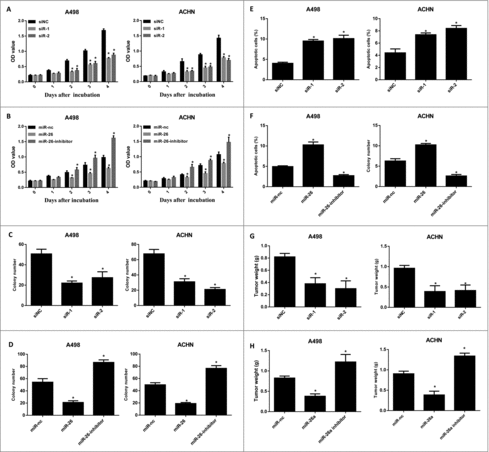
To investigate the functional role of DLX6-AS1 on RCC cell growth, we first knocked down the expression of DLX6-AS1 in 2 RCC cell lines. We then performed MTT assays on these cells after DLX6-AS1 knockdown. Importantly, DLX6-AS1 knockdown remarkably suppressed the growth of these 2 RCC cells (). In addition, to explore the suppressor role of miR-26a on RCC cells, MTT assays were also performed for testing RCC cell growth after miR-26a knockdown or overexpression. As expected, miR-26a overexpression suppressed the growth of RCC cells, whereas miR-26a knockdown showed the opposite effects (). We also performed additional assays to confirm the regulatory role of DLX6-AS1 and miR-26a on RCC cell growth. In crystal violet staining assays, colony formation of RCC cells was inhibited by DLX6-AS1 knockdown and miR-26a overexpression, whereas miR-26a knockdown promoted RCC cell colony formation ( and ). Flow cytometry also observed that DLX6-AS1 knockdown and miR-26a overexpression induced more apoptotic RCC cells, whereas miR-26a knockdown showed lower cell apoptosis ( and ). Furthermore, we also confirmed our observations in vivo by xenograft tumor animal models. Consistently, DLX6-AS1 knockdown and miR-26a overexpression suppressed the xenograft tumor growth, whereas miR-26a knockdown repressed RCC cell xenograft tumor growth ( and ). Taken together, these results indicated that DLX6-AS1 played an oncogenic role, while miR-26a played a tumor suppressor role in RCC tumorigenesis.
Figure 3. DLX6-AS1 knockdown and miR-26a overexpression suppress the growth and development of RCC. (A) MTT assays were performed for detecting the RCC cell growth after DLX6-AS1 knockdown. (B) MTT assays were performed for testing RCC cell growth after miR-26a overexpression and knockdown. (C and D) Colony formation assays were performed after DLX6-AS1 knockdown (C) or miR-26a overexpression and knockdown (D). (E and F) The apoptotic rate of RCC cells after DLX6-AS1 knockdown (E) and miR-26a overexpression or knockdown (F) were examined by flow cytometry analysis. (G and H) Xenograft tumor models were generated for testing the role of DLX6-AS1 and miR-26a in RCC tumorigenesis. DLX6-AS1 overexpressing (G) and miR-26a overexpressing/knockdown (H) RCC cells were inoculated into nude mice. At the end point of the experiments, the tumor bearing mice were killed and the tumor weight was examined. All data was shown as mean ± s.e.m. from 3 independent experiments. *p < 0.05.
MiR-26a targets PTEN to modulate RCC tumorigenesis
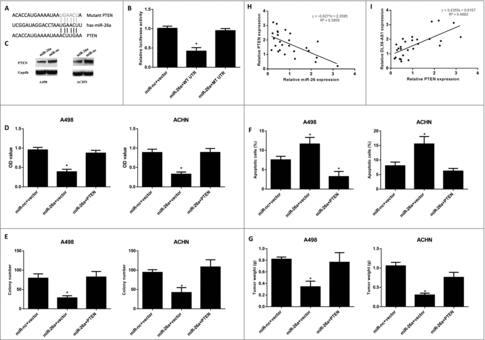
We next used publicly available algorithms (microRNA.org) to identify potential targets of miR-26a in RCC cells. We notified that PTEN was a potent target of miR-26a (), and also this molecule has been shown to regulate the development of many types of cancer. To investigate whether PETN was the direct target of miR-26a, luciferase reporter assays were performed. As shown in , miR-26a significantly suppressed the luciferase activity, which containing the wild type UTR of PTEN (). This data revealed that PTEN was the direct target of miR-26a. Additionally, the protein levels of PTEN in RCC cells were reduced by miR-26a overexpression (), and were increased by miR-26a knockdown (). Q-PCR analysis also showed that PTEN expression level was negatively correlated with miR-26a expression level in RCC samples, whereas was positively correlated with DLX6-AS1 expression level ( and ). To extensively investigate whether PTEN was the functional target of miR-26a, RCC cells were co-transfected with miR-26a and PTEN. MTT assays suggested that PTEN overexpression rescued the RCC cell growth in vitro () and also reversed the RCC cell colony formation (). Furthermore, the apoptotic RCC cell rate was analyzed by flow cytometry. The results showed that PTEN overexpression significantly reduced the RCC cell apoptosis (). Moreover, to extensively investigate whether PTEN could restore RCC tumorigenesis, miR-26a and PTEN overexpressing-RCC cells were inoculated into nude mice. As expected, PTEN overexpression rescued tumor suppression, induced by miR-26a overexpression (). Collectively, these data strongly supported that DLX6-AS1 promoted RCC cell growth and tumorigenesis via miR-26a mediated regulation of PTEN.
Figure 4. PTEN is the direct target of miR-26a in RCC cells. (A) Schematic illustration of the predicted binding sites between miR-26a and PTEN. (B) Luciferase reporter assays were performed in 293T cells after co-transfecting PTEN wild type or mutant UTR with miR-26a. (C) The PTEN protein level was examined by western blot in RCC cells after miR-26a overexpression. (D–F) Cell growth, colony formation and cell apoptosis a were examined in RCC cells after miR-26a/PTEN overexpression. (G) The tumor growth was examined after inoculating with miR-26a/PTEN overexpression RCC cells. (H and I) The correlations between miR-26a level with PTEN level (H) or DLX6-AS1 level (I) was examined by q-PCR analysis. All data was shown as mean ± s.e.m. from 3 independent experiments. * represents p < 0.05.
Discussion
Studies have revealed that lncRNAs play vital roles in RCC pathogenesis.Citation18-21 However, the role and mechanism of lncRNA DLX6-AS1 in RCC has not been fully understood. In this study, we identified that lncRNA DLX6-AS1 expression was significantly increased in RCC tumor tissues compared with their corresponding non-tumor kidney tissues. In addition, we also found that DLX6-AS1 expression was also increased in metastatic RCCs. Also, DLX6-AS1 expression was higher in RCC cell lines compared with normal renal cell line. As expected, DLX6-AS1 knockdown significantly suppressed the A498 and ACHN cell growth and promoted the RCC cell colony formation. Furthermore, xenograft tumor model revealed that DLX6-AS1 knockdown repressed the RCC tumor growth in vivo. These results suggested that DLX6-AS1 might act as an oncogenic role in RCC tumorigenesis and development.
Numerous studies have shown that lncRNAs acts as a ceRNA to sponge microRNAs to modulate the expression level of microRNA targets in many biologic process.Citation6,22-25 As we have mentioned that LOC100129148 promotes nasopharyngeal carcinoma progression by targeting miR-539–5p,Citation9 lncRNA EWSAT1 promotes nasopharyngeal cancer development by targeting miR-326/-330–5p10 and LncRNA 00858 acts as an ceRNA of miR-422a in non-samll cell lung cancer.Citation11 In this study, we notified that the expression level of microRNA-26a was negatively correlated with the expression level of DLX6-AS1 in RCC tissue samples. This result suggested that DLX6-AS1 may work as a ceRNA to sponge miR-26a in RCC cells. MicroRNA-26a has been reported to regulate several types of tumor progression. For example, miR-26a and miR-26b inhibit esophageal squamous cancer cell proliferation through suppression of c-MYC pathway.Citation17 MiR-26a enhances invasive capacity by suppressing GSK3β in human lung cancer cells.Citation26 MiR-26a performs converse roles in proliferation and metastasis of different gastric cancer cells via regulating of PTEN expression.Citation27 MiRNA-26a contributes to the acquisition of malignant behaviors of doctaxel-resistant lung adenocarcinoma cells through targeting EZH2.Citation28 There were also many studies showed that miR-26a regulated the progression in many types of cancers.Citation29-33 This promoted us to ask whether miR-26a may also make any contributions to RCC tumor progression and whether DLX6-AS1 could sponge miR-26a to modulate RCC development. To address these questions, we first examined the expression of miR-26a in RCC cells after DLX6-AS1 knockdown. Q-PCR analysis showed that DLX6-AS1 knockdown increased the expression level of miR-26a in 2 RCC cells. However, miR-26a overexpression didn't suppressed the expression level of DLX6-AS1 in RCC cells. These data suggested that DLX6-AS1 acts as the ceRNA to sponge the miR-26a. Furthermore, we also notified that miR-26a overexpression significantly suppressed the growth of RCC cells in vitro and RCC tumorigenesis in vivo. As expected, miR-26a overexpression suppressed the growth of RCC cells and RCC tumors. These results suggested that miR-26a acts as a tumor suppressor in RCC tumors.
Bioinformatics analysis revealed that PTEN was the potent predicted target of miR-26a. We have mentioned above that miR-26a performs converse roles in proliferation and metastasis of different gastric cancer cells via regulating of PTEN expression.Citation27 MiR-26a mediates adipogenesis of amniotic fluid mesenchymal stem/stromal cells via PTEN, Cyclin E1, and CDK6.Citation34 MiR-26a also showed that miR-26a inhibits proliferation and migration of HaCaT keratinocytes through regulating PTEN expression.Citation35 miR-26a and miR-214 down-regulate expression of the PTEN gene in chronic lymphocytic leukemia.Citation36 In this study, luciferase reporter assays and western blot showed that miR-26a directly targeted to the 3′-UTR of PTEN and therefore suppressed the protein level of PTEN in RCC cells. To extensively investigate whether PTEN could restore RCC cell growth, we next co-transfected RCC cells with miR-26a and PTEN. MTT assays and xenograft tumor models revealed that PTEN could rescued repression of the RCC cell growth and RCC tumorigenesis, induced by miR-26a overexpression.
In conclusion, our study revealed that DLX6-AS1 is an oncogene in RCC. Elevated level of DLX6-AS1 is positively correlated with tumor progression and development. DLX6-AS1 functions as a ceRNA to sponge miR-26a to facilitate RCC progression partly via miR-26a/PTEN axis. Thus, our study provides further insight into the molecular mechanism of lncRNAs in RCC tumorigenesis, which may promote the development of lncRNA-directed diagnosis and therapy for RCC.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Additional information
Funding
References
- Liu KG, Gupta S, Goel S. Immunotherapy: incorporation in the evolving paradigm of renal cancer management and future prospects. Oncotarget. 2017;8:17313-27. PMID:28061473
- Li M, Wang Y, Cheng L, Niu W, Zhao G, Raju JK, Huo J, Wu B, Yin B, Song Y, et al. Long non-coding RNAs in renal cell carcinoma: A systematic review and clinical implications. Oncotarget. 2017.
- Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115-32. doi:10.3322/caac.21338
- Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861-74. doi:10.1038/nrg3074. PMID:22094949
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629-41. doi:10.1016/j.cell.2009.02.006. PMID:19239885
- Adams BD, Parsons C, Walker L, Zhang WC, Slack FJ. Targeting noncoding RNAs in disease. J Clin Invest. 2017;127:761-71. doi:10.1172/JCI84424. PMID:28248199
- Smolle MA, Bauernhofer T, Pummer K, Calin GA, Pichler M. Current Insights into Long Non-Coding RNAs (LncRNAs) in Prostate Cancer. Int J Mol Sci. 2017;18. doi:10.3390/ijms18020473. PMID:28241429
- Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354-61. doi:10.1016/j.tcb.2011.04.001. PMID:21550244
- Sun KY, Peng T, Chen Z, Song P, Zhou XH. Long non-coding RNA LOC100129148 functions as an oncogene in human nasopharyngeal carcinoma by targeting miR-539-5p. Aging (Albany NY). 2017;9:999-1011. PMID:28328537
- Song P, Yin SC. Long non-coding RNA EWSAT1 promotes human nasopharyngeal carcinoma cell growth in vitro by targeting miR-326/-330-5p. Aging (Albany NY). 2016;8:2948-60. doi:10.18632/aging.101103. PMID:27816050
- Zhu SP, Wang JY, Wang XG, Zhao JP. Long intergenic non-protein coding RNA 00858 functions as a competing endogenous RNA for miR-422a to facilitate the cell growth in non-small cell lung cancer. Aging (Albany NY). 2017;9:475-86. PMID:28177876
- Zhou H, Wang F, Chen H, Tan Q, Qiu S, Chen S, Jing W, Yu M, Liang C, Ye S, et al. Increased expression of long-noncoding RNA ZFAS1 is associated with epithelial-mesenchymal transition of gastric cancer. Aging (Albany NY). 2016;8:2023-38. doi:10.18632/aging.101048. PMID:27654478
- Li J, Li P, Zhao W, Yang R, Chen S, Bai Y, Dun S, Chen X, Du Y, Wang Y, et al. Expression of long non-coding RNA DLX6-AS1 in lung adenocarcinoma. Cancer Cell Int. 2015;15:48. doi:10.1186/s12935-015-0201-5. PMID:26052251
- Cao MX, Jiang YP, Tang YL, Liang XH. The crosstalk between lncRNA and microRNA in cancer metastasis: orchestrating the epithelial-mesenchymal plasticity. Oncotarget. 2017;8:12472-83. PMID:27992370
- Bayoumi AS, Sayed A, Broskova Z, Teoh JP, Wilson J, Su H, Tang YL, Kim IM. Crosstalk between Long Noncoding RNAs and MicroRNAs in Health and Disease. Int J Mol Sci. 2016;17:356. doi:10.3390/ijms17030356. PMID:26978351
- Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669-87. doi:10.1016/j.cell.2009.01.046. PMID:19239888
- Li J, Liang Y, Lv H, Meng H, Xiong G, Guan X, Chen X, Bai Y, Wang K. miR-26a and miR-26b inhibit esophageal squamous cancer cell proliferation through suppression of c-MYC pathway. Gene. 2017. doi:10.1016/j.gene.2017.05.001. PMID:28476684
- Lopez-Urrutia E, Coronel-Hernandez J, Garcia-Castillo V, Contreras-Romero C, Martinez-Gutierrez A, Estrada-Galicia D, Terrazas LI, Lopez-Camarillo C, Maldonado-Martinez H, Jacobo-Herrera N, et al. MiR-26a downregulates retinoblastoma in colorectal cancer. Tumour Biol. 2017;39:1010428317695945. doi:10.1177/1010428317695945. PMID:28443472
- Hirata H, Hinoda Y, Shahryari V, Deng G, Nakajima K, Tabatabai ZL, Ishii N, Dahiya R. Long Noncoding RNA MALAT1 Promotes Aggressive Renal Cell Carcinoma through Ezh2 and Interacts with miR-205. Cancer Res. 2015;75:1322-31. doi:10.1158/0008-5472.CAN-14-2931. PMID:25600645
- Fachel AA, Tahira AC, Vilella-Arias SA, Maracaja-Coutinho V, Gimba ER, Vignal GM, Campos FS, Reis EM, Verjovski-Almeida S. Expression analysis and in silico characterization of intronic long noncoding RNAs in renal cell carcinoma: emerging functional associations. Mol Cancer. 2013;12:140. doi:10.1186/1476-4598-12-140. PMID:24238219
- Hu G, Dong B, Zhang J, Zhai W, Xie T, Huang B, Huang C, Yao X, Zheng J, Che J, et al. The long noncoding RNA HOTAIR activates the Hippo pathway by directly binding to SAV1 in renal cell carcinoma. Oncotarget. 2016 Jul 19;12(8):2605-12. doi:10.1039/c6mb00114a. PMID:28476684.
- Hong Q, Li O, Zheng W, Xiao WZ, Zhang L, Wu D, Cai GY, He JC, Chen XM. LncRNA HOTAIR regulates HIF-1alpha/AXL signaling through inhibition of miR-217 in renal cell carcinoma. Cell Death Dis. 2017;8:e2772. doi:10.1038/cddis.2017.181. PMID:28492542
- Tan JY, Sirey T, Honti F, Graham B, Piovesan A, Merkenschlager M, Webber C, Ponting CP, Marques AC. Corrigendum: Extensive microRNA-mediated crosstalk between lncRNAs and mRNAs in mouse embryonic stem cells. Genome Res. 2015;25:1410 1. doi:10.1101/gr.181974.114. PMID:26330573
- Wang K, Liu CY, Zhou LY, Wang JX, Wang M, Zhao B, Zhao WK, Xu SJ, Fan LH, Zhang XJ, et al. APF lncRNA regulates autophagy and myocardial infarction by targeting miR-188-3p. Nat Commun. 2015;6:6779. doi:10.1038/ncomms7779. PMID:25858075
- Thum T, Condorelli G. Long noncoding RNAs and microRNAs in cardiovascular pathophysiology. Circ Res. 2015;116:751-62. doi:10.1161/CIRCRESAHA.116.303549. PMID:25677521
- Lin G, Liu B, Meng Z, Liu Y, Li X, Wu X, Zhou Q, Xu K. MiR-26a enhances invasive capacity by suppressing GSK3beta in human lung cancer cells. Exp Cell Res. 2017;352:364-74. doi:10.1016/j.yexcr.2017.02.033. PMID:28237093
- Ding K, Wu Z, Wang N, Wang X, Wang Y, Qian P, Meng G, Tan S. MiR-26a performs converse roles in proliferation and metastasis of different gastric cancer cells via regulating of PTEN expression. Pathol Res Pract. 2017;213:467-75. doi:10.1016/j.prp.2017.01.026. PMID:28242043
- Chen J, Xu Y, Tao L, Pan Y, Zhang K, Wang R, Chen LB, Chu X. MiRNA-26a Contributes to the Acquisition of Malignant Behaviors of Doctaxel-Resistant Lung Adenocarcinoma Cells through Targeting EZH2. Cell Physiol Biochem. 2017;41:583-97. doi:10.1159/000457879. PMID:28214878
- Ma X, Dong W, Su Z, Zhao L, Miao Y, Li N, Zhou H, Jia L. Functional roles of sialylation in breast cancer progression through miR-26a/26b targeting ST8SIA4. Cell Death Dis. 2016;7:e2561. doi:10.1038/cddis.2016.427. PMID:28032858
- Jin F, Wang Y, Li M, Zhu Y, Liang H, Wang C, Wang F, Zhang CY, Zen K, Li L. MiR-26 enhances chemosensitivity and promotes apoptosis of hepatocellular carcinoma cells through inhibiting autophagy. Cell Death Dis. 2017;8:e2540. doi:10.1038/cddis.2016.461. PMID:28079894
- Yang C, Zheng S, Liu T, Liu Q, Dai F, Zhou J, Chen Y, Sheyhidin I, Lu X. Down-regulated miR-26a promotes proliferation, migration, and invasion via negative regulation of MTDH in esophageal squamous cell carcinoma. FASEB J. 2017;31:2114-22. doi:10.1096/fj.201601237. PMID:28174206
- Wang P, Lv L. miR-26a induced the suppression of tumor growth of cholangiocarcinoma via KRT19 approach. Oncotarget. 2016;7:81367-76. PMID:27833076
- Lu J, Song G, Tang Q, Yin J, Zou C, Zhao Z, Xie X, Xu H, Huang G, Wang J, et al. MiR-26a inhibits stem cell-like phenotype and tumor growth of osteosarcoma by targeting Jagged1. Oncogene. 2017;36:231-41. doi:10.1038/onc.2016.194. PMID:27270422
- Trohatou O, Zagoura D, Orfanos NK, Pappa KI, Marinos E, Anagnou NP, Roubelakis MG. miR-26a Mediates Adipogenesis of Amniotic Fluid Mesenchymal Stem/Stromal Cells via PTEN, Cyclin E1, and CDK6. Stem Cells Dev. 2017;26:482-94. doi:10.1089/scd.2016.0203. PMID:28068868
- Yu N, Yang Y, Li X, Zhang M, Huang J, Wang X, Long X. MiR-26a inhibits proliferation and migration of HaCaT keratinocytes through regulating PTEN expression. Gene. 2016;594:117-24. doi:10.1016/j.gene.2016.09.010. PMID:27613140
- Zou ZJ, Fan L, Wang L, Xu J, Zhang R, Tian T, Li JY, Xu W. miR-26a and miR-214 down-regulate expression of the PTEN gene in chronic lymphocytic leukemia, but not PTEN mutation or promoter methylation. Oncotarget. 2015;6:1276-85. doi:10.18632/oncotarget.2626. PMID:25361012
