ABSTRACT
It has become more and more evident that the BCL-2 family proteins mediate a wide range of non-apoptotic functions. The pro-apoptotic BAX protein has been reported in interphasic nuclei. Whether the nuclear form of BAX could be involved in non-apoptotic function is still unknown.
Our study showed for the first time that BAX was associated with chromatin in vitro. Next, we used gain and loss of function approaches to decipher the potential role of nuclear BAX in non-apoptotic cells. In vitro, nuclear BAX promoted cell proliferation in lung epithelial cells and primary human lung fibroblasts by modulating CDKN1A expression. Interestingly, BAX occupancy of CDKN1A promoter was specifically enriched close to the transcription-starting site. Nuclear BAX also modulated the basal myofibroblastic differentiation and migration of primary human lung fibroblasts. Finally, BAX nuclear localization was associated in vivo with the remodelling of lung parenchyma during development, tumorigenesis as well as fibrosis compared to control adult human lungs.
Hence, our study established for the first time, a strong link between the nuclear localization of the pro-apoptotic BAX protein and key basic cellular functions in the non-apoptotic setting.
Introduction
The BCL-2 family proteins are essential components of the core apoptotic machinery as they act as anti- or pro-apoptotic regulators. BAX (namely, BCL-2-Associated X protein) was the first identified pro-apoptotic member of the BCL-2 protein family.Citation1 The majority of BAX proteins are found in the cytoplasm in a closed / globular conformation in healthy cells.Citation2 BAX is one of the gatekeepers that control mitochondrial outer membrane permeabilization during apoptosis. However, growing evidences suggest that BCL-2 family proteins are also involved in a wide variety of non-apoptotic cellular activities.Citation3 For instance, the pro-apoptotic BAX and BAK proteins have been shown to be involved in cell cycle progression in mouse embryonic fibroblasts (MEF) in vitro.Citation4 BAX was also necessary to maintain energy production by supporting the mitochondrial network in non-apoptotic HCT-116 colon cancer cells and primary mouse hepatocytes.Citation5 More surprisingly, BAX protein was also present in interphasic nuclei of non-apoptotic human epithelial cancer cell lines,Citation6,Citation7 but to our knowledge, BAX functions in the nucleus have not been further investigated.
Here, we hypothesized that the nuclear form of BAX could play a role in basic cellular functions in non-apoptotic cells. After demonstrating that BAX was in close proximity to chromatin in vitro, we deciphered the involvement of cytosolic or nuclear BAX in basic cellular functions such as cell proliferation, myofibroblastic differentiation and migration in lung cellular subtypes. Finally, we investigated whether BAX nuclear localization could be also observed in vivo in control human adult or fetal lung compared to diseased / remodelled lungs (namely carcinomas and fibrosis). Thus, we aimed to establish for the first time, a strong link between the nuclear localization of the pro-apoptotic BAX protein and key basic cellular functions in the non-apoptotic setting.
Results
BAX is associated in vitro with chromatin in the interphasic cell nucleus.
Even though the role of BAX cytoplasmic fraction during apoptosis has been actively characterized,Citation8 the nuclear function of BAX is still unknown. BAX nuclear localization was mainly reported in non-apoptotic cancer epithelial cell lines in vitro.Citation6,Citation7 To better understand BAX mechanisms of actions in the nucleus, we determined whether BAX was associated with the chromatin in the interphasic nuclei of lung cell types. First we confirmed that BAX was localized in both nuclei and cytosol -enriched fractions of the lung epithelial A549 cells by cell subcellular fractionation (). The general transcription factor YY1 and GAPDH were respectively used as nuclear and cytosol markers (). However, mitochondrial materials were present in both fractions as revealed with Cytochrome c oxidase subunit 4 (COX4, see Fig. S1). Nevertheless, we confirmed after cytosol / mitochondria fractionation that BAX was not detectable in the mitochondria-enriched fraction (see Fig. S1). These results strongly suggested that BAX localization in both cytosol and nuclei -enriched fractions after nuclear / cytosol fractionation was not associated with the presence of mitochondria components in both fractions. Finally, BAX nuclear localization was not restricted to the epithelial lineage as a similar expression pattern was observed in primary human lung fibroblasts ( and Fig. S1). Interestingly, BCL-2 or BCL-xL, the main anti-apoptotic partners of BAX in the cytosol,Citation9 were not detectable in the nuclei-enriched fraction but present only in the cytosol-enriched one in both A549 cells and primary lung fibroblasts (see Fig. S1).
Figure 1. Nuclear form of BAX is associated with chromatin in interphasic in A549 cells and primary HLF. (A) Immunoblot after subcellular fractionation in lung derived cell types (A549 cells and primary HLF) showing BAX presence in cytoplasm (cyto) with GAPDH protein and in the nuclear fraction (nuc) with general transcription factor YY1. (B) Representative immunoblot showing the differential extraction of BAX in DSP crosslinked A549 cells and primary HLF. Immunoblot showing that BAX, Histone H2B (chromatin marker) and GAPDH (cytoplasmic marker) in the corresponding pellets (pel.) and supernatants (sup.) after sequential extraction with 1) Triton CSK buffer, 2) 0.5 M NaCl high salt buffer and finally digestion of chromatin bulks with micrococcal nuclease (MNase). The right panels (no MNase and MNase treated conditions) were exposed longer than the ones on the left (CSK and 0.5 M NaCl treated samples) Note the absence of BAX and H2B in the supernatant of the control condition with no micrococcal nuclease (no MNase).
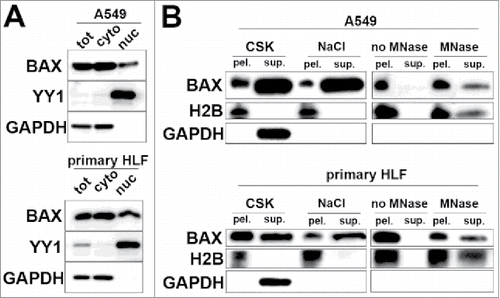
The nuclear localization and potential association of BAX with chromatin were further investigated by a differential extraction protocol in A549 and primary HLF treated with the cell-permeable cross-linker Dithiobis[succinimidyl propionate] (DSP) to preserve protein-protein interactions.Citation10 First, DSP treated cells were treated with 0.1%TX-100 CSK buffer, which extracts not only cytoplasmic but also nuclear proteins not tightly bound to nuclear structures. Triton-insoluble form of BAX was further extracted/stripped with 0.5M NaCl “high salt” buffer. However, a fraction of BAX was still detected in the remaining pellet indicating that BAX was covalently cross-linked to salt-insoluble proteins (). Unlike GAPDH (a cytolosic protein), histone H2B (a core protein component of chromatin) was also resistant to triton and salt extractions. To investigate whether BAX was in close proximity to chromatin, the Triton- and salt-extracted cell pellets of A549 cells and primary HLF were treated with micrococcal nuclease (MNase) to digest chromatin bulks. This treatment partially released both Histone H2B and BAX () demonstrating that a fraction of BAX was cross-linked with chromatin in both A549 cells and primary HLF.
Nuclear BAX is involved in the cell cycle progression of lung epithelial A549 cells and primary lung fibroblasts in vitro
In the light of BAX chromatin association in healthy cell in vitro, we next investigated whether nuclear BAX could be implicated in basic cellular functions in non-dying cells. In a first step, we assayed the effects of BAX knock down on basic cellular functions such as proliferation in epithelial (A549 cells) and mesenchymal (primary HLF) cell lineages. But this broad loss of function approach could not distinguish between cytoplasmic and nuclear functions. For this reason, we next assayed whether overexpression of BAX constructs either preferentially targeted or excluded from nucleus would elicit the opposite phenotypic effects than BAX siRNA in A549 cells and primary HLF.
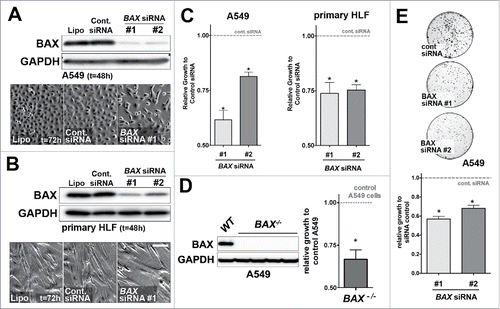
BAX protein expression level was drastically reduced in A549 lung epithelial cells and primary HLF using two independent BAX siRNA compared to cells transfected with control siRNA (). Strikingly, cell count revealed that cell proliferation was significantly decreased in A549 cells and primary HLF treated with the two BAX siRNA sequences for 48h compared to control siRNA (). No cytotoxic effect was observed in these experiments (data not shown). To further confirm our initial siRNA results, we showed that complete loss of BAX also impacted proliferation in BAX−/− A549 cells generated by CRISPR-Cas9–mediated genomic engineering (). Additionally, a decrease in colony formation was observed in a clonogenic assay with BAX siRNA treated A549 cells compared to control siRNA (). Altogether, these results suggested that BAX was involved in proliferation in A549 cells and primary HLF.
Figure 2. Effects of BAX knock down on A549 and primary HLF proliferation in vitro. (A) A549 cells or (B) primary HLF were treated with the transfection reagent alone (lipofectamine, Lipo.) or transfected with a non-targeting siRNA (Cont. siRNA) or two different BAX siRNA sequences for 48h. Immunoblot was revealed with an anti-BAX antibody and GAPDH as loading control. Phase contrast pictures of cells transfected with control lipofectamine alone (Lipo, left panel), control siRNA (Cont. siRNA, middle panel) and BAX siRNA #1 (right panel) suggest a decreased in proliferation in BAX siRNA #1 treated cells. Similar results were observed with BAX siRNA #2 (data not shown). (C) Effects of BAX siRNA #1 and #2 on the proliferation (cell count) of A549 cells (left panel, respectively 40.9%+/− 4.3 and 19%+/− 1.9 growth decrease for BAX siRNA #1 and #2, n = 7) and primary HLF (right panel, respectively 26%+/− 5 and 24.6%+/− 2.4 growth decrease for BAX siRNA #1 and #2, n = 7) compared to cells treated with control siRNA (gray dash line) at 48h (*p < 0.05,Wilcoxon rank t-Test). (D) Representative immunoblot confirming the absence of BAX protein in three independent BAX−/− A549 clones compared to control / wild type (WT) A549 cells. Immunoblot was revealed with an anti-BAX antibody and GAPDH as loading control. Effect of complete BAX loss of function on the proliferation of A549 cells (33.3%+/− 5.5 growth decrease for BAX −/− cells compared to control cells (n = 6), *p < 0.05, Wilcoxon rank t-Test). (E) Clonogenic assay performed with A549 cells transfected with control siRNA or with the two different BAX siRNA (respectively 44%+/− 5 and 32%+/− 3.2 decrease for BAX siRNA #1 and #2, n = 3). Pictures of Crystal violet stained colony assay of A549 cells after 5 days of serum starvation and 7 days of recovery in complete medium are showed on the upper part (500 cells were initially plated). Quantification of images from three independent experiments after Crystal violet staining is showed on the lower part. Note the growth inhibition in BAX siRNA A549 cells compared to controls. (*p < 0.05, rank t-Test, n = 3).
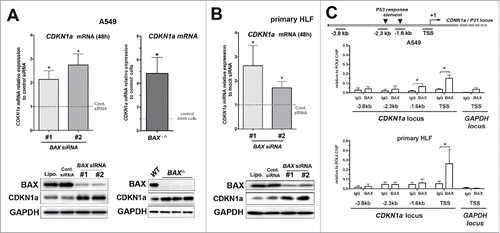
Next, the effects of BAX knock down on the expression of CDKN1A a major negative regulator of the cell cycle and proliferation was assayed. The expression of CDKN1A was increased at the mRNA and protein levels in both A549 cells and primary HLF transfected with BAX siRNA compared to control (t = 48 h; see and ). Similarly, an increase in CDKN1A at the mRNA and protein levels was also observed in BAX−/− A549 cells generated by CRISPR-Cas9 approach (). To unveil the involvement of nuclear BAX in the regulation of CDKN1A expression and cell cycle progression, we next assayed the potential BAX occupancy of CDKN1A locus () by chromatin immunoprecipitation (ChIP). Interestingly, BAX occupancy of CDKN1A locus was significantly increased near the CDKN1A transcription-starting site (TSS) compared to the control IgG in both A549 cells and primary HLF (). Meanwhile, BAX occupancy was also significantly increased in a P53 response element at −1.6 kb only in A549 cells (but at a lesser extent than CDKN1A TSS). However, BAX recruitment was neither enriched in a second known P53 response element at −2.3 kb in CDKN1A locusCitation11 nor in a −3.8 kb upstream non-specific site used as negative control in both cell types. BAX recruitment near CDKN1A TSS seemed specific of this locus since no significant occupancy enrichment was observed near the TSS of the GAPDH locus in A549 cells and primary HLF (). Our results suggested that BAX might directly modulate CDKN1A mRNA expression at the level of the chromatin in the nucleus.
Figure 3. Upregulation of the cell cycle inhibitor CDKN1A upon BAX knock down in A549 and primary HLF. (A) Expression of CDKN1A mRNA by qPCR in A549 cells transfected for 48h with two different BAX siRNA compared to siRNA control (gray dash line, n = 6 experiments; left part) or in BAX-/− A549 cells compared to wild type A549 cells (gray dash line, n = 5; right part). In the left inside, representative immunoblot (n = 5) of BAX, CDKN1A protein level in A549 cells (treated with lipofectamine alone (Lipo), control siRNA (Cont siRNA) and two different BAX siRNA (#1 and #2). In the right inside, representative immunoblot confirming the absence of BAX protein and the upregulation of CDKN1A protein level in three independent BAX−/− A549 clones compared to control / wild type (WT) A549 cells. GAPDH was used as loading control. (*p < 0.05, Wilcoxon rank t-Test). (B) Expression of CDKN1A mRNA by qPCR in primary HLF transfected for 48h with two different BAX siRNA compared to siRNA control (gray dash line, n = 6 experiments). Representative immunoblot (n = 5) of BAX, CDKN1Aprotein level (lower part) in primary HLF treated with lipofectamine alone (Lipo), control siRNA (Cont siRNA) and two different BAX siRNA (#1 and #2). GAPDH was used as loading control. (*p < 0.05, Wilcoxon rank t-Test). (C) Schematic diagram of the CDKN1A locus showing PCR amplified fragments. A region encompassing CDKN1A transcription starting sites (TSS) was amplified. The −2.3 kb and −1.6 kb sites are two P53 upstream response elements at CDKN1A promoters. The −3.8 kb upstream non-specific site was used as a negative control site. ChIP analysis for recruitment of BAX at the CDKN1A promoter in A549 cells (n = 3) and primary lung fibroblasts (n = 4) was performed as described in materials and methods. For each condition, an unrelated control IgG was used as negative control. Note that BAX binding was specifically enriched at the vicinity of CDKN1A TSS in both cell types. (*p < 0.05, paired t-Test).
To confirm the role of nuclear BAX in the regulation of cell proliferation and CDKN1A expression level, we generated BAX constructs preferentially targeted either to the nucleus or the cytoplasm in lung cells. On one hand, a sequence containing two “nuclear localization signal” (NLS)Citation12 was added to the N-terminus of a FLAG HA tagged BAX construct to target it to preferentially to the nucleus. On the other hand, to exclude BAX proteins from the nucleus, a sequence containing two “nuclear export signal” (NES)Citation12 was added to the N-terminus of a FLAG HA tagged BAX constructs.
First, we validated these tools in A549 cells and primary HLF transiently transfected with NLS-BAX-FLAG or NES-BAX-FLAG constructs. The transfected cells were then subjected to cytoplasm / nucleus fractionation after 48 h. Western blot analysis with BAX antibody revealed that NLS-BAX proteins were present in both cytoplasm and nuclear fractions (but preferentially in the latter), while NES-BAX proteins were excluded from the nucleus and detected only in the cytoplasmic fraction of primary HLF () and A549 cells (data not shown).
Figure 4.
Figure 4. Gain of function experiments with BAX proteins targeted either to the nucleus (NLS-BAX) or excluded from the nucleus (NES-BAX) in A549 cells and primary HLF. (A) Representative immunoblot (n = 3) after subcellular fractionation in primary HLF transfected for 48h with an empty pCDNA 3.1 vector, NES-BAX or NLS-BAX constructs. The upper panel with anti-FLAG (upper panel) and BAX (middle panel) antibodies show that the NES-BAX or NLS-BAX constructs are detected in the cytosolic fraction (cyto.), revealed with GAPDH antibody. In the nuclear fraction (nuc.), revealed with LMN A/C, only NLS-BAX was detected (arrowhead). Note that NES-BAX and NLS-BAX are detected at a higher molecular weight than endogenous BAX (*). (B-C) Effects of NES and NLS-BAX on the proliferation (cell count) of A549 cells (B) and primary HLF (C) compared to cells treated with control plasmid (gray dashed line) at 48h. Note that the increase in cell proliferation was observed only in NLS-BAX transfected A549 cells (41% increase +/− 8, n = 4) and primary HLF (102% increase +/− 67, n = 5) compared to pCDNA 3.1 empty vector and NES-BAX construct (respectively 9% increase +/− 9 compared to control plasmid in A549 cells, n = 4 and 15% decrease +/− 10 in primary HLF, n = 3) compared to control plasmid. (D-E) Expression of CDKN1A mRNA by qPCR (upper part) in (D) A549 cells and (E) primary HLF transfected for 48h with NES-BAX (respectively 19.4% decrease +/− 4 in A549 cells and 27.6% increase +/− 29 in primary HLF, n = 5) or NLS-BAX (respectively 41.6% decrease +/− 7 in A549 cells and 51.54% decrease +/− 16 in primary HLF, n = 5) compared to empty vector (gray dash line). Representative immunoblot (n = 3) of FLAG tagged BAX constructs, CDKN1A, in A549 cells (D) or primary HLF (E) transfected with empty control vector, either NES-BAX or NLS-BAX constructs for 48h. GAPDH was used as loading control. (*p < 0.05, rank t-Test).
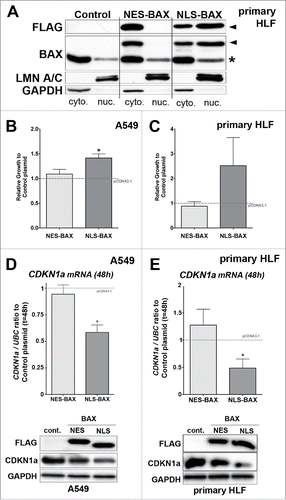
Our data showed that A549 overexpressing NLS-BAX displayed an increase in proliferation at 48h compared to control plasmid. In contrast, NES-BAX transient overexpression in A549 cells did not change cell number compared to control plasmid (). Similarly, an increase in cell number at 48 h was observed in HLF transfected only with NLS-BAX constructs at 48 h (). Only the NLS-BAX construct did significantly decrease CDKN1A expression at both mRNA and protein levels in A549 cells and primary HLF ( and ). Meanwhile, NES-BAX slightly altered CDKN1A content only at the protein level in both cell types ( and ), with no significant impact on cell proliferation ( and ). We also confirmed these results in HEK 293T cells (see Fig. S2). Using the BAX−/− A549 cells generated by CRISPR-Cas9 approach, we also showed that only the overexpression of NLS-BAX was able to rescue the upregulation of CDKN1A content observed in BAX−/− A549 cells compared to wild type cells transfected with control empty vector (see Fig. S2). Altogether, these results showed that the effects of the nuclear-targeted form of BAX with regards to cell proliferation and CDKN1A expression levels were truly more potent compared to the cytoplasm-targeted one.
Nuclear BAX modulates partially basal myofibroblastic differentiation state and migratory potential of primary lung fibroblasts in vitro
We also investigated the effect of BAX partial loss of function on basal myofibroblastic differentiation and migration in primary HLF. Myofibroblasts play a critical role in normal tissue repair and impaired stroma remodelling in pathological settings.Citation13 Given the appropriate microenvironment, fibroblasts can differentiate by acquiring some characteristic of smooth muscle cells (such as expression of alpha smooth muscle actin, αSMA) and becoming active producers of extracellular matrix (ECM) proteins (such as Collagen 1).Citation13 Pathological fibroblasts are thought to secrete important amount of ECM components leading to the development of fibrosis.Citation14
We observed at basal levels that knocking down BAX increased αSMA mRNA expression in primary HLF compared to HLF transfected with control siRNA by qPCR, whereas α2-Collagen 1 mRNA isoform (COL1A2) expression levels was not affected (). By immunoblot, we observed that decreasing BAX expression with siRNA did increase the basal expression myofibroblast markers such as αSMA and COL1 proteins in primary HLF compared to control cells (). These results suggested that BAX modulated αSMA expression at the transcriptional level, while its effect on COL1 expression was probably post-transcriptional ().
Figure 5. Effect of BAX knock down or NES-, NLS-BAX gains of function on the basal myofibroblastic differentiation state and migration of primary HLF in vitro. (A) Expression of αSMA (left part), COL1A2 (right part) mRNA by qPCR in primary HLF transfected for 48 h with two different BAX siRNA. Note the increase expression of αSMA in primary HLF treated with BAX siRNA (96.6% increase +/− 40 and 52.5% increase +/− 38 respectively with BAX siRNA #1 and #2) compared to siRNA control (gray dash line, n = 5). In contrast, BAX siRNA did not modulate COL1A2 mRNA expression in primary HLF (respectively 5.6% increase +/− 21 in and 5.1% increase +/− 23 with BAX siRNA #1 and #2) compared to siRNA control (gray dash line, n = 5). In the right part, representative immunoblot (n = 5) of BAX, αSMA, COL1 protein levels in primary cells treated with lipofectamine alone (Lipo.), control siRNA (Cont siRNA) and two different BAX siRNA pooled to increase efficiency. GAPDH was used as loading control. Note the increase expression of αSMA and COL1 protein in BAX siRNA treated HLF. (B) Expression of αSMA (left part), COL1A2 (right part) mRNA by qPCR in primary HLF transfected for 48h with either NES-BAX (respectively 14.4% increase +/− 35 for αSMA mRNA and 57.5% increase +/− 33 for COL1A2 mRNA) or NLS-BAX (respectively 47% decrease +/− 18 for αSMA mRNA and 55% decrease +/− 15 for COL1A2 mRNA, n = 3) compared to empty control vector (gray dash line). In the right part: representative immunoblot (n = 3) of FLAG tagged overexpressed BAX constructs, αSMA and COL1 proteins in primary HLF transfected with empty control vector, either NES-BAX or NLS-BAX constructs for 48h. GAPDH was used as loading control. A decrease in αSMA protein level was observed only in NLS-BAX transfected cells, Collagen-1 isoform levels were decreased with both NES- and NLS-BAX constructs. Note that the COL1 and αSMA blots in panel (B) were exposed longer than the ones in panel (A) (*p < 0.05, rank t-Test). (C) Migratory capacities of primary lung fibroblasts in modified Boyden chamber assay after transfection with two different BAX siRNA (compared to control siRNA gray dash line, upper part) or after transfection with either NES-BAX (2.8 ± 0.8-fold increase; p < 0.05; n = 5) or NLS-BAX (13.06 ± 7.1-fold increase; p < 0.05; n = 5) compared to empty control vector (gray dash line, lower part). Data are median with outer box edges to min and max and whiskers to SEM. (*p < 0.05,Wilcoxon rank t-Test)
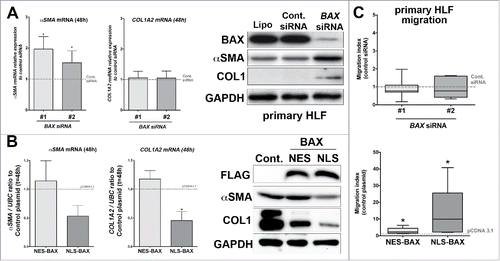
In the gain of function approach, only the overexpression of NLS-BAX construct decreased the expression levels of αSMA and COL1A2 at the mRNA in primary HLF compared to cells transfected with the empty vector or NES-BAX constructs (). Similarly, only the nuclear-targeted form of BAX did decrease αSMA at the protein level () in primary HLF. Meanwhile, COL1 protein level was decreased in HLF overexpressing either NLS-BAX or NES-BAX constructs () compared to control plasmid. These results suggested that BAX cytoplasmic form regulated COL1 expression through a post-transcriptional mechanism. However, the NLS-BAX construct was more efficient in diminishing basal COL1 protein level compared to the cells overexpressing NES-BAX construct ().
Migration is a salient feature in fibroblast biology during repair process as well as fibrogenesis.Citation14 First, we tested the effect of BAX knock down using siRNA on migration in a modified Boyden chamber assay (). BAX siRNA treatments did not impact the migration of primary HLF. In contrast, the nuclear-targeted form of BAX strongly induced the migratory capacities of primary HLF as showed in . The cytoplasmic NES-BAX construct also increased the migration of primary HLF () but to a lesser degree. Meanwhile, the migration of A549 cells was not modulated by either BAX loss or gain of function approaches (data not shown).
BAX displays a nuclear localization in remodelling lung tissues
Overall, to our knowledge, our in vitro results suggest for the first time that nuclear forms of BAX could be implicated in basic cellular functions in non-dying cells. It was therefore critical to determine whether BAX nuclear localization could be also observed in human tissue samples in vivo. For this, we first assayed BAX subcellular localization by immunohistochemistry in paraffin sections from normal human fetal and adult lungs compared to diseased lungs tissues such as epithelial tumors or fibrotic parenchyma (). During the pseudoglandular stage (12 weeks), fetal lung tissue undergoing branching morphogenesis displayed nuclear BAX staining in both the epithelium and the mesenchyme ( and and see Table S1 for the percentages of nuclear staining).
Figure 6. BAX localization in fetal and adult lung tissues by immunohistochemistry. (A and B) Immunohistochemistry showing BAX staining (red chromogen) in human fetal lung at 12 weeks of gestation. Note that BAX was nuclear in epithelium (ep) and in the mesenchyme (mes). Higher magnification is showed in (B) showing BAX nuclear staining in epithelial cells (black arrow). (C) BAX localization in normal alveolar (upper panel) and bronchiolar (arrow in lower panel) epithelia. The insert in the upper panel shows BAX staining in the normal alveolar epithelium (arrow). (D and E) BAX staining in (D) the epithelium of lung non-small cell lung carcinoma (NSCLC) such as adenocarcinoma (arrow); and (E) within the tumor stroma (arrows). (F and G) BAX staining in (F) the epithelium of small cell lung carcinoma (arrow, SCLC n = 8) and (G) within the tumor stroma (arrow). Note that the fibroblasts within the stroma of these tumors also displayed BAX nuclear staining (see arrows in E and G). (H-J) BAX staining in idiopathic pulmonary fibrosis (IPF), the insert in (H) shows BAX staining in the hyperplastic epithelium (hyp AE2C, arrow). (I) BAX staining within fibroblastic foci (Fibro foci) underneath remodeled distal epithelium (ep) in IPF lung. Note the nuclear BAX staining in the fibroblasts (arrow) within fibroblast foci (J) BAX staining within bronchiolised distal airspace (bronch ep) in IPF lung. (K) Negative staining with control IgG. Nuclei are counterstained with hematoxylin. BAX nuclear localization was also confirmed using another anti-BAX primary antibody in control and IPF lung tissues by immunohistochemistry (data not shown). Scale bar: 20 µm (B, G, E, I, J, and lower panel in C), 10 µm (inserts in B, C, D, F, G, and H), 100 µm (A), 80 µm (C, D, F, K, and H).
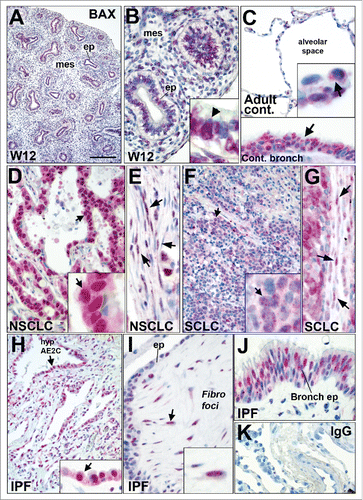
In the adult lung (), few BAX nuclear positive pneumocytes were observed in the normal lung alveolar parenchyma. By comparison, the normal bronchiolar lung epithelium displayed diffuse BAX nuclear positive cells ( and Table S1). Next, we assayed BAX nuclear localization on lung cancer human paraffin tissue arrays. In contrast with normal alveolar space, BAX nuclear expression was strongly expressed in the epithelial tumors cells from lung adenocarcinomas (NSCLC) and from small cell lung carcinomas (SCLC) as showed in and (see also Table 1). BAX nuclear positive cells were also observed within the stroma of these tumors (, and Table S1).
We also assayed BAX nuclear localization in patients suffering from idiopathic pulmonary fibrosis (IPF), a non-cancerous but lethal chronic interstitial lung disease.Citation14 Upon abnormal accumulation of extracellular cellular matrix (ECM) in IPF, the alveolar-capillary units are then lost and replaced by scarring and honeycomb cysts. The fibrotic lung is characterized by fibroblastic foci where fibroblasts and myofibroblasts accumulate as clusters in subepithelial areas. The overlying epithelium consists of hyperplastic pneumocytes or columnar bronchiolar cells. Areas of hyperplastic, metaplastic and dysplastic epithelium are often observed in the remodeled fibrotic lung.Citation14 BAX was detected in both cytoplasm and nuclei of the hyperplastic epithelial cells in the aberrantly remodeled distal lung parenchyma and in the fibroblastic foci ( and and supplementary Table S1). Nuclear BAX positive epithelial cells were also observed in IPF bronchiolised distal airspacesCitation15 ().
Discussion
BAX is associated with chromatin compartment in the interphasic nucleus
The proteins of the BCL-2 family have been extensively studied during the past decades for their cytoplasmic / mitochondrial role in cell deathCitation8. It has been reported that BAX could shuttle from the cytosol to the nucleus during apoptosis in response to various stress stimuliCitation7. However, the consequences of BAX shuttling in the nucleus during apoptosis remain poorly elucidated. Furthermore, it has been shown that BAX was involved in the subcellular redistribution of nuclear proteins from the nucleus to cytoplasm in response to stressor stimuli before the appearance of apoptotic featuresCitation16. Thus, in this study we aimed to better understand BAX nuclear functions in non-dying cells, as BAX nuclear localization has been previously reported in non-apoptotic epithelial cancer cell lines derived from human lungCitation7,Citation17 and breastCitation6 but its potential nuclear functions in non-dying cells have never addressed to date.
Our differential extraction protocol using detergent, high ionic strength and nuclease digestion showed that BAX was in close proximity with chromatin-associated proteins in both A549 cells and primary lung fibroblasts. To strengthen this finding, we showed that BAX was present at the promoter of CDKN1a in both cell types by ChIP. Furthermore, co-localization of BAX with the nuclear matrix was previously reported in cultured cancer human cell lines by embedment-free immunogold electron microscopyCitation18. Interestingly, Lamin proteins (key components of the nucleoskeleton) are involved in the regulation of numerous nuclear processes, including gene transcriptionCitation19,Citation20. Altogether, these observations suggest that BAX could at least interact with two interrelated nuclear compartments: the chromatin and the nucleomatrix.
The mechanisms regulating BAX nuclear localization in non-dying cells will require further investigation. Indeed, BAX lacks bona fide nuclear localization signal (NLS) and nuclear export signal (NES) like most of the other members of the BCL-2 familyCitation7 at the exception of the pro-apoptotic BOK proteinCitation21 and a short variant of the anti-apoptotic BCL2A1 proteinCitation22. One possibility would be that BAX protein (21kDa) could diffuse passively from the cytosol to the nuclear sap as small proteins can cross freely the nuclear envelope through the nuclear pore complexes. Furthermore, BAX would be retained in the nucleus by its interaction with its nuclear partners.
Non-apoptotic functions of BAX in cellular homeostasis
On one hand, BAX displayed a nuclear localization in non-dying cells in vitro and in vivo. We showed that BAX was nuclear in proliferating epithelial and mesenchymal cell types in vitro. Strikingly, the nuclear localization of BAX was increased in vivo within remodelling and proliferating tissues such as fetal lung undergoing branching morphogenesis, lung cancer and fibrosis compared to normal lung tissue sections. On the other hand, it has become more and more evident that the BCL-2 family protein also mediate a wide range of basic cellular functions in non-apoptotic settings such as cell cycle regulation, mitochondrial dynamic and bioenergetic as well as cell migration.Citation23 Hence, it was appealing to determine whether nuclear BAX could modulate such basic cellular functions.
In the present study, we showed that decreasing total BAX protein level was sufficient to impact cell proliferation by up-regulating the expression of CDKN1A, a cyclin-dependent kinase inhibitor in epithelial A549 cells and primary HLF. Conversely, a BAX construct targeted to the nucleus (NLS-BAX) elicited the opposite phenotype than BAX siRNA on cell proliferation, with a decrease in CDKN1A expression at both mRNA and protein levels. Meanwhile, the nucleus excluded form of BAX (NES-BAX) only slightly decreased CDKN1A protein level, probably through a post-transcriptional mechanism, to a much lesser extent than NLS-BAX and with no significant impact on cell proliferation. We also confirmed by studying BAX occupancy of CDKN1A promoter regions that BAX was specifically enriched in the vicinity of CDKN1A TSS in both A549 and primary HLF. These different BAX loss and gain of function experiments suggest that nuclear BAX would rather act as a selective negative regulator of transcription at CDKN1A promoter. Interestingly, CDKN1A and BAX are two known P53 direct target genes.Citation24,Citation25 BAX induction by P53 could be involved in a negative feedback loop to control CDKN1A levels in wild type P53 positive cells such as A549 cells.Citation26 Indeed, BAX occupancy of CDKN1A promoter was significantly increased in a P53 response element at −1.6 kb in A549 cells. It is tempting to speculate that nuclear BAX and P53 could compete to regulate CDKN1A since a previous study showed that P53 could interact and activate BAX directly in the cytosolCitation2. The possible interaction between P53 and BAX in the nucleus as well as its potential effect upon CDKN1A expression will require further studies. Thus our results suggested strongly that the nuclear form of BAX could directly influence cell cycle progression. A pioneer work published in 1996 showed that the proliferation rate of mature T cells overexpressing BAX in response to interleukin-2 was increased compared to control counterparts.Citation27 Similarly, BAX overexpression protected neuron from growth factor withdrawal.Citation28 More recently, a study demonstrated that cell cycle progression was impaired in double Bax−/− Bak−/− MEF.Citation4 However, none of these studies investigated the possible involvement of nuclear BAX in this process. In the light of the role of nuclear BAX in cell proliferation in vitro, it is tempting to speculate that nuclear BAX could be involved in the hyperproliferative epithelial cell growth observed in lung carcinomas and remodelled epithelium in IPF. BAX was also observed in the nuclei of fibroblast within tumor stroma and the fibroblast foci in idiopathic pulmonary fibrosis. In contrast with cell cycle progression, basal myofibroblastic differentiation and migration of primary HLF seemed under the control of both cytosolic and nuclear forms of BAX. With regards to gain of function experiments, the effects of the nuclear-targeted form of BAX were more potent than the cytoplasm-targeted one.
The effect of cytoplasmic BAX overexpression on cell migration may also involve anti-apoptotic members of the BCL-2 family since it has been previously showed that MCL-1, BCL-2 or BCL-xL were required for proper migration and invasion of colorectal cancer cells.Citation23 However, BAX nuclear functions, at least in A549 cells and primary lung fibroblasts, did not seem to implicate BCL-2 or BCL-xL. Indeed, these two anti-apoptotic BAX cytosolic partnersCitation9 were not detected in nuclei-enriched fraction in these two cell types. BCL-2 nuclear localization was previously reported in other cell types.Citation6 The potential nuclear localization and involvement of the other anti-apoptotic members of the BCL-2 protein family in BAX nuclear functions will require further studies.
In conclusion, our in vitro results strongly suggest that BAX, in particular its nuclear form, promotes migration of HLF, maintaining fibroblasts in an undifferentiated state. Altogether, these results are first to suggest that BAX was not simply diffused in the nuclear sap but might interact with chromatin or even gene promoter regions within the nucleus. Interestingly, it has been proposedCitation5 that BAX non-apoptotic functions are critical for tumour progression as complete inactivation of BAX locus is rarely observed in cancer, whereas mutations preventing its apoptotic function are quite frequent. We also propose that BAX expression level in disease cannot be considered as a reliable readout for apoptosis response as this would undermine the contribution of BAX non-apoptotic functions in tissue homeostasis and disease progression. Hence, our study has established a strong link between the nuclear localization of the pro-apoptotic BAX protein and key basic cellular functions in a non-apoptotic setting.
Materials and methods
Cell lines and plasmids
Human A549 cell line was obtained from ATCC (LGC Standard) and cultured according to manufacturer recommendations. Primary Human lung fibroblasts from control patients were derived from human lung explants as previously described.29 Primary Human lung fibroblasts (HLF) were cultured with Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% fetal calf serum (FCS) and antibiotics as previously described29 and used at passage five. NLS-BAX and NES-BAX plasmid cloned in pCDNA3.1 (Life Technology Invitrogen) were generated from a BAX-α backbone vector purchased from Origene. PCR cloning approach was used as described in,30 two different primers were used to respectively add 2 NES or 2 NLS sequence in 5’ position of BAX-α cDNA. Meanwhile, a common primer was used to add FLAG and HA tags in 3’ position (the primer sequences are described in supplementary Table S2). CRISPR/Cas9 plasmids targeting human BAX were purchased from Santa Cruz Biotechnology (sc-400042 and sc-400042-HDR).
Human tissues samples
Paraffin tissue arrays with lung cancer samples were purchased from US Biomax. Normal human fetal lung paraffin sections (12-week gestation) were obtained through the Bichat Hospital pathology department from diagnostic feotopathology cases. IPF lung samples were obtained from patients undergoing open lung biopsy (n = 5; median age 58 years; range 49–70 years). IPF was diagnosed according to ATS/ERS/JRS/ALAT criteria, including histopathological features of usual interstitial pneumonia.31 Control lung samples were non-disease–involved segments from patients (n = 5; median age 52.5 years, range 42–80 years) who were undergoing lung surgery for removal of a primary lung tumor. This study was approved by the local ethics committee (CPP Ile de France 1, no. 0811760). Written informed consent was obtained from all subjects.
Cell culture experiments
For BAX knock down experiments with small interfering RNA (siRNA), cells were transfected at 50% to 60% of confluency in 6-well dishes. All transfection experiments were performed using the lipofectamine 2000 method (Life Technology – Invitrogen), in duplicate or triplicate, in accordance with the manufacturer's instructions. To suppress endogenous BAX expression, we used 2 specifics siRNA against BAX from Life Technology – Invitrogen (BAXHSS141355 and BAXHSS141356). SiRNA Negative Control MED GC (Invitrogen) was used as a negative control. A final concentration of 30 nM siRNA was used for A549 cells, and 50nM for control HLF. In myofibroblastic differentiation experiment, siRNA #1 and #2 were pooled to increase efficiency. A similar transfection protocol was used to overexpress Flag HA tagged NLS-BAX and NES-BAX constructs cloned in pCDNA3.1 vector. Empty pCDNA3.1 plasmid was used as control (1.5 µg of plasmid were used for each transfection reaction). The optimal plasmid amount was determined in preliminary experiments using A549 cells. No cytotoxic effect was detected in A549 and primary HLF transfected with NES-BAX or NLS-BAX constructs compared to control plasmid at this dosage (data not shown). However, both NES-BAX and NLS-BAX constructs induced apoptosis at higher dosage compared to control plasmid in A549 cells (data not shown). Generation and selection of BAX−/− A549 clones using CRISPR/Cas9 method were performed according to manufacturer instructions (Santa Cruz Biotechnology).
Cell Growth was assayed 48h after siRNA BAX transfection or seeding for BAX−/− A549 cells. Cells were harvested through trypsinization and analysed for total cell number and dead cell population by trypan blue dye exclusion method by automatic cell viability analyzer (VI-CELL®, Beckman Coulter). In the colony assay, 500 A549 cells were seeded in 6-well dishes overnight and then serum deprived for 5 days before recovery in full growth medium for 7 days. Cell colonies were stained with 1% Crystal violet staining solution in 70% ethanol for 5min before extensive washes in PBS. Photographs were taken with a Vilbert-Loumart apparatus and quantified with Image J software. For immunoblot and mRNA experiments, cells were plated in 6-well plates for 24 h before transfection. The cells were cultured in full growth medium at the exception of the primary HLF assayed for myofibroblastic markers. In these experiments, cells were cultured in serum-deprived medium for 48h after the transfection.
mRNA analysis
The mRNA from cell lines were extracted using NucleoSpin® RNA II kit according to manufacturer instructions (Macherey-Nagel GmbH & Co). The Reverse Transcription reaction was perfomed with 500ng of total RNA as described above. The transcripts of Ubiquitin C (UBC) served as endogenous RNA controls.29 The results were expressed as gene of interest/UBC mRNA ratios. CDKN1A, α-SMA, a2-COL1 and UBC (see supplementary Table S2 for sequence) expression were quantified by real-time polymerase chain reaction with a PCR ABI 7500 apparatus (Applied Biosystems).
Immunoblot, differential extraction and chromatin immunoprecipitation
Primary fibroblasts and A549 cells in culture were lysed on ice in RIPA buffer (10mM Tris-HCl pH8, 50 mM NaCl, 0.5% Na deoxycholate, 0.2% SDS, 1% NP-40), supplemented with protease inhibitor cocktail inhibitor (10 µg/ml aprotinin, 10 µg/ml leupeptin, 8.3µg/ml pepstatin A, 2 mM PMSF). Lysates were clarified by centrifugation (13,000 g for 20 min at 4°C). 10–30 µg samples were analysed by western blot experiments described in.29 Cell fractionation was performed according to manufacturer instructions (Nuclear/Cytosol Fractionation Kit and Mitochondria/Cytosol Fractionation kit, Biovision). BAX differential extraction protocol was adapted fromCitation10 using micrococcal nuclease (MNase). Briefly, cross-linked cells using Dithiobis[succinimidyl propionate] (DSP, 200 µg/ml for 10 min at room temperature; Pierce) were treated with 0.1%TX-100 CSK buffer. After centrifugation, the pellets were further extracted with high salt buffer (0,5M NaCl) and the remaining pellets were finally digested with micrococcal nuclease (10 U/ml 60 min 37°C; NEB).
The following primary antibodies were used for immunoblotting: anti-BAX (Scbt Sc-493), anti-Histone H2B (Abcam, Ab1790), anti-CDKN1A (CST, #2947), anti-COX4 (CST, #11967), anti-YY1 (Abcam, Ab109228), anti-Lamin B1 (Abcam, Ab16048) and anti-GAPDH (Covalab, mab90009-P). ChIP analyses were performed with Abcam's ChIP Kit –“ One Step” (ab117138) following manufacturer instructions. Briefly, DSP cross-linked chromatin from A549 cells and primary HLF was prepared as described in.32 Chromatin fragments (from about 1 × 106 cells) were immunoprecipitated with antibodies against BAX (BD Pharmingen #554104), RNA POL-II and an irrelevant IgG control (both from Abcam's “ChIP Kit – One Step”). The precipitated DNA was amplified by real-time PCR with previously published primer setsCitation11 designed to amplify the CDKN1A locus (−3.8 kb, −2.3 kb, −1.6 kb CDKN1A upstream sites and CDKN1A TSS) and GAPDH TSS. The results are expressed relative to RNA POL-II occupancy of a given site.
Immunohistochemistry
The paraffin-embedded sections were treated as previously described.29 Two primary BAX antibodies were used for IHC: anti-BAX Abcam ab54829 and ab7977. To test the specificity of immunostaining, antibodies were replaced by an isotype-matched control antibody. All digital images of light microscopy were acquired with a DM400B microscope (Leica) equipped with a Leica DFC420 CDD camera.
Statistical analysis
All data were expressed as or mean (with standard error of the mean) values as described in figure legends. Statistical analysis was performed with Prism 5 (Graphpad Software Inc., La Jolla, CA USA). P values less than 0.05 were considered as significant (s.e.m values are displayed).
Abbreviations
| ChIP | = | chromatin immunoprecipitation |
| Cont. | = | control |
| Bronch. | = | bronchioles |
| Cyto | = | cytosol |
| DSP | = | Dithiobis[succinimidyl propionate] |
| ECM | = | extracellular matrix. |
| Ep | = | epithelium |
| Fibro | = | fibroblast |
| Fibro foci | = | fibroblast foci |
| Hyp. AE2C | = | hyperplasic alveolar type 2 cells |
| IPF | = | Idiopathic pulmonary fibrosis |
| MEF | = | mouse embryonic fibroblast |
| Mes | = | mesenchyme |
| Mito | = | mitochondria |
| MNAse | = | micrococcal nuclease |
| NEG | = | negative |
| NES | = | nuclear export signal |
| NLS | = | nuclear localization signal |
| NSCLC | = | non-small-cell lung carcinoma |
| Nt | = | N-terminal part |
| Nuc | = | nuclear |
| Pel. | = | pellet |
| POS | = | positive |
| SCLC | = | small-cell lung carcinoma |
| Sup. | = | supernatant |
| TSS | = | Transcription-starting site |
| WT | = | wild type |
| W | = | week |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
1371882_Supplemental_Material.zip
Download Zip (391.4 KB)Acknowledgments
We are very grateful to Prof. Joan Brugge at Harvard Medical School (Boston, MA USA) where this project was first initiated.
Funding
This work was supported by the European Commission (FP7 project #202224, European Idiopathic Pulmonary Fibrosis Network), by grants from the CARDIF association, the Chancellery of Paris Universities (Poix Legacy), a starting grant from DHU FIRE and by the “Association pour la fibrose pulmonaire idiopathique Pierre ENJALRAN”. SB was supported by the “Mariane Josso” award (“Fondation pour la Recherche Médicale”, FRM) and by the Fond de dotation “Recherche en Santé Respiratoire”. AJ was supported by the French National Research Agency (ANR-11-BSV1-0011) and by the LABEX « Inflamex ».
References
- Oltvai ZN, Milliman CL, Korsmeyer SJ. Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programmed cell death. Cell. 1993;74:609-19. doi:10.1016/0092-8674(93)90509-O. PMID:8358790
- Chipuk JE, Kuwana T, Bouchier-Hayes L, Droin NM, Newmeyer DD, Schuler M, Green DR. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science. 2004;303:1010-4. doi:10.1126/science.1092734. PMID:14963330
- Galluzzi L, Kepp O, Trojel-Hansen C, Kroemer G. Non-apoptotic functions of apoptosis-regulatory proteins. EMBO Rep. 2012;13:322-30. doi:10.1038/embor.2012.19. PMID:22402666
- Janumyan Y, Cui Q, Yan L, Sansam CG, Valentin M, Yang E. G0 function of BCL2 and BCL-xL requires BAX, BAK, and p27 phosphorylation by Mirk, revealing a novel role of BAX and BAK in quiescence regulation. J Biol Chem. 2008;283:34108-20. doi:10.1074/jbc.M806294200. PMID:18818203
- Boohaker RJ, Zhang G, Carlson AL, Nemec KN, Khaled AR. BAX supports the mitochondrial network, promoting bioenergetics in nonapoptotic cells. Am J Physiol Cell Physiol. 2011;300:C1466−78. doi:10.1152/ajpcell.00325.2010. PMID:21289292
- Hoetelmans R, van Slooten HJ, Keijzer R, Erkeland S, van de Velde CJ, Dierendonck JH. Bcl-2 and Bax proteins are present in interphase nuclei of mammalian cells. Cell Death Differ. 2000;7:384-92. doi:10.1038/sj.cdd.4400664. PMID:10773823
- Salah-eldin A, Inoue S, Tsuda M, Matsuura A. Abnormal intracellular localization of Bax with a normal membrane anchor domain in human lung cancer cell lines. Jpn J Cancer Res Gann. 2000;91:1269-77. doi:10.1111/j.1349-7006.2000.tb00914.x. PMID:11123426
- Chipuk JE, Green DR. How do BCL-2 proteins induce mitochondrial outer membrane permeabilization? Trends Cell Biol. 2008;18(4):157-64. doi:10.1016/j.tcb.2008.01.007. PMID:18314333
- Fujita M, Ishimi Y, Nakamura H, Kiyono T, Tsurumi T. Nuclear organization of DNA replication initiation proteins in mammalian cells. J Biol Chem. 2002;277:10354-61. doi:10.1074/jbc.M111398200. PMID:11779870
- Bellucci L, Dalvai M, Kocanova S, Moutahir F, Bystricky K. Activation of p21 by HDAC inhibitors requires acetylation of H2A.Z. PloS One. 2013;8:e54102. doi:10.1371/journal.pone.0054102. PMID:23349794
- Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J Biol Chem. 2007;282:5101-5. doi:10.1074/jbc.R600026200. PMID:17170104
- Orimo A, Weinberg RA. Stromal fibroblasts in cancer: a novel tumor-promoting cell type. Cell Cycle Georget Tex. 2006;5:1597-601. doi:10.4161/cc.5.15.3112.
- Fernandez IE, Eickelberg O. New cellular and molecular mechanisms of lung injury and fibrosis in idiopathic pulmonary fibrosis. Lancet Lond Engl. 2012;380:680-8. doi:10.1016/S0140-6736(12)61144-1.
- Plantier L, Crestani B, Wert SE, Dehoux M, Zweytick B, Guenther A, Whitsett JA. Ectopic respiratory epithelial cell differentiation in bronchiolised distal airspaces in idiopathic pulmonary fibrosis. Thorax. 2011;66:651-7. doi:10.1136/thx.2010.151555. PMID:21422041
- Lindenboim L, Blacher E, Borner C, Stein R. Regulation of stress-induced nuclear protein redistribution: a new function of Bax and Bak uncoupled from Bcl-x(L). Cell Death Differ. 2010;17:346-59. doi:10.1038/cdd.2009.145. PMID:19816507
- Tripathi S, Batra J, Cao W, Sharma K, Patel JR, Ranjan P, Kumar A, Katz JM, Cox NJ, Lal RB, et al. Influenza A virus nucleoprotein induces apoptosis in human airway epithelial cells: implications of a novel interaction between nucleoprotein and host protein Clusterin. Cell Death Dis. 2013;4:e562. doi:10.1038/cddis.2013.89. PMID:23538443
- Gajkowska B, Motyl T, Olszewska-Badarczuk H, Gniadecki R, Koronkiewicz M. Structural association of Bax with nuclear matrix and cytomatrix revealed by embedment-free immunogold electron microscopy. Cell Biol Int. 2000;24:649-56. doi:10.1006/cbir.2000.0530. PMID:10964454
- Dechat T, Adam SA, Taimen P, Shimi T, Goldman RD. Nuclear lamins. Cold Spring Harb Perspect Biol. 2010;2:a000547. doi:10.1101/cshperspect.a000547. PMID:20826548
- Shimi T, Butin-Israeli V, Adam SA, Goldman RD. Nuclear lamins in cell regulation and disease. Cold Spring Harb Symp Quant Biol. 2010;75:525-31. doi:10.1101/sqb.2010.75.045. PMID:21467145
- Bartholomeusz G, Wu Y, Ali Seyed M, Xia W, Kwong K-Y, Hortobagyi G, Hung M-C. Nuclear translocation of the pro-apoptotic Bcl-2 family member Bok induces apoptosis. Mol Carcinog. 2006;45:73-83. doi:10.1002/mc.20156. PMID:16302269
- Ko J-K, Lee M-J, Cho S-H, Cho J-A, Lee B-Y, Koh JS, Lee S-S, Shim Y-H, Kim C-W. Bfl-1S, a novel alternative splice variant of Bfl-1, localizes in the nucleus via its C-terminus and prevents cell death. Oncogene. 2003;22:2457-65. doi:10.1038/sj.onc.1206274. PMID:12717423
- Koehler BC, Scherr A-L, Lorenz S, Urbanik T, Kautz N, Elssner C, Welte S, Bermejo JL, Jäger D, Schulze-Bergkamen H. Beyond cell death – antiapoptotic Bcl-2 proteins regulate migration and invasion of colorectal cancer cells in vitro. PloS One. 2013;8:e76446. doi:10.1371/journal.pone.0076446. PMID:24098503
- Brady HJ, Gil-Gómez G, Kirberg J, Berns AJ. Bax alpha perturbs T cell development and affects cell cycle entry of T cells. EMBO J. 1996;15:6991-7001. PMID:9003775
- Middleton G, Nunez G, Davies AM. Bax promotes neuronal survival and antagonises the survival effects of neurotrophic factors. Development. 1996;122:695-701. PMID:8625820
- Cigna N, Farrokhi Moshai E, Brayer S, Marchal-Somme J, Wémeau-Stervinou L, Fabre A, Mal H, Lesèche G, Dehoux M, Soler P, et al. The hedgehog system machinery controls transforming growth factor-β-dependent myofibroblastic differentiation in humans: involvement in idiopathic pulmonary fibrosis. Am J Pathol. 2012;181:2126-37. doi:10.1016/j.ajpath.2012.08.019. PMID:23031257
- Schmelzle T, Mailleux AA, Overholtzer M, Carroll JS, Solimini NL, Lightcap ES, Veiby OP, Brugge JS. Functional role and oncogene-regulated expression of the BH3-only factor Bmf in mammary epithelial anoikis and morphogenesis. Proc Natl Acad Sci U S A. 2007;104:3787-92. doi:10.1073/pnas.0700115104. PMID:17360431
- Raghu G, Collard HR, Egan JJ, Martinez FJ, Behr J, Brown KK, Colby TV, Cordier J-F, Flaherty KR, Lasky JA, et al. An official ATS/ERS/JRS/ALAT statement: idiopathic pulmonary fibrosis: evidence-based guidelines for diagnosis and management. Am J Respir Crit Care Med. 2011;183:788-824. doi:10.1164/rccm.2009-040GL. PMID:21471066
- Ricke RM, Bielinsky A-K. Easy detection of chromatin binding proteins by the Histone Association Assay. Biol Proced Online. 2005;7:60-9. doi:10.1251/bpo106. PMID:16136225
