Recent progress in high-throughput RNA-sequencing techniques has led to the realization that the human genome is transcribed much more pervasively than previously appreciated, and that large fraction of the transcriptome is composed of long non-coding RNAs (lncRNAs). Some of these lncRNAs are stable and have biological functions such as modulation of gene expression; however, many of them are expressed at extremely low levels because of rapid RNA turnover in the nucleus. This implies that a considerable fraction of lncRNAs are non-functional, perhaps produced as a result of transcriptional “noise” or else as byproducts where it is the act of transcription rather than the RNA itself that is important. To avoid accumulation of these unnecessary and potentially harmful transcripts, cells have developed nuclear RNA surveillance systems, in which the exosome, consisting of a nine-subunit core and two catalytic subunits, plays a key role in 3'-5' exonucleolytic decay. The exosome is recruited to its substrates through multiple distinct adaptor complexes, such as the Nuclear Exosome Targeting (NEXT) complex and the Trf4/Air2/Mtr4 polyadenylation (TRAMP)-like complex, which specifically recognize distinct sets of RNAs.Citation1
In our recent study,Citation3 we analyzed the polyadenylated transcriptomes of cells depleted of each of the individual NEXT subunits, the RNA helicase Mtr4, the RNA-binding protein RBM7, and the zinc knuckle protein ZCCHC8. Our goal was to investigate whether NEXT might affect polyadenylation of RNA polymerase II transcripts generally, since all three NEXT subunits were identified in a proteomics analysis of the purified human pre-mRNA polyadenylation complex.Citation2 Unexpectedly, we found that siRNA-mediated knockdown (KD) of Mtr4, but neither of the other two NEXT subunits, nor of a TRAMP subunit (ZCCHC7), resulted in strong and specific accumulation of two types of transcripts: prematurely terminated RNAs (ptRNAs) and upstream antisense RNAs (uaRNAs; also known as promoter upstream transcripts (PROMPTs)). ptRNAs are transcripts prematurely terminated in an intron, whereas uaRNAs are transcribed in antisense direction relative to transcription start sites of protein-coding genes. Most of these transcripts contain poly(A) signals (PASs) at their 3' ends and have long poly(A) tails, suggesting that these RNAs are polyadenylated by PAS-driven mechanisms similar to mRNAs. The Mtr4 KD-specific accumulation of uaRNAs in Mtr4-depleted cells was particularly unexpected, as this type of transcript is a known NEXT substrate.Citation4 Since these RNAs are all sensitive to the depletion of the catalytic subunits of the exosome, we speculated that there might be additional Mtr4-containing complexes. By performing gel filtration followed by co-immunoprecipitation (co-IP) and mass spectrometry (MS), we identified the Zinc-finger protein ZFC3H1 as an Mtr4 cofactor (see also Meola et al.Citation5) that is necessary for degradation of pt- and ua- RNAs. Reciprocal co-IP analyses revealed that Mtr4/ZFC3H1 is distinct from NEXT and TRAMP. There were no significant changes in the levels of known NEXT-targeted uaRNAs after ZFC3H1 knockdown, further supporting the idea that these complexes are independent.
What might the consequence be if turnover of naturally unstable lncRNAs fails? We found that KD of Mtr4/ZFC3H1 resulted in accumulation of pt- and ua- RNAs not only in the nucleoplasm but also, significantly, in the cytoplasm, whereas there is no increase in the chromatin fraction. These results suggest that these RNAs are normally degraded immediately after release from the chromatin. Importantly, polysome fractionation analysis following Mtr4 KD revealed that the cytoplasmic pt-/ua- RNAs associate with active ribosomes. However, paradoxically, Mtr4-deficient cells also showed a global reduction in heavier polysomes, suggesting that ribosomes naturally bound to mRNAs were occupied by the short open reading frame (sORF)-containing pt- and ua-RNAs. Consistent with this, puromycin-labeling of nascent proteins showed that depletion of Mtr4 or ZFC3H1 resulted in a significant inhibition of translation. In contrast, no alteration in global translation was observed following KD of subunits of the other Mtr4-containing complexes, NEXT and TRAMP.
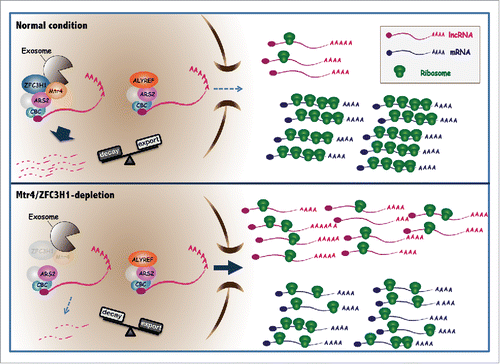
More recently, Fan et al.Citation6 reported that Mtr4 competes with an export adaptor ALYREF for binding to ARS2, an associating factor of the cap-binding complex (CBC). The competition is an important checkpoint to prevent unwanted nuclear export of aberrant transcripts. Notably, ZFC3H1 as well as NEXT subunits also interact with the ARS2/CBC,Citation1,5 suggesting that Mtr4 competes with ALYREF for ARS2/CBC association in these complexes. Thus, it is possible that both Mtr4/ZFC3H1 and NEXT have roles in inhibiting nuclear export of their target transcripts in addition to assisting decay by the nuclear exosome. However, as mentioned above, translation repression was observed only after Mtr4/ZFC3H1, not NEXT, depletion. This difference is probably due to the more “mRNA-like” structure that Mtr4/ZFC3H1 substrates have: longer RNA bodies and the presence of long poly(A) tails.Citation3,5 This contrasts with typical NEXT targets, which are generally shorter and largely lack poly(A) tails.Citation5 A longer RNA body gives more chance to have sORFs, and cap/poly(A) tail stimulates translation. Besides, both RNA length and the presence of a poly(A) tail contribute to efficient RNA export.Citation7 We propose a model () in which Mtr4/ZFC3H1 plays a critical role as a “polysome protector complex (PPC)”, functioning to prevent nuclear export and deleterious cytoplasmic accumulation of naturally unstable, polyadenylated lncRNAs.
Figure 1. pt- and ua- RNAs are normally rapidly degraded in the nucleus by Mtr4/ZFC3H1. Mtr4/ZFC3H1 may inhibit nuclear export of its substrates by competing with ALYREF for ARS2/CBC association. Loss of the Mtr4/ZFC3H1 results in stabilization of pt- and ua- RNAs and increased ALYREF-ARS2/CBC interaction, and these RNAs accumulate in the cytoplasm, become ribosome-associated, and consequently, overwhelm the translation machinery.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- Zinder JC, Lima CD. Targeting RNA for processing or destruction by the eukaryotic RNA exosome and its cofactors. Genes Dev. 2017;31(2):88-100. doi:10.1101/gad.294769.116. PMID:28202538
- Shi Y, Di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR 3rd, Frank J, Manley JL. Molecular architecture of the human pre-mRNA 3' processing complex. Mol Cell. 2009;33(3):365-376. doi:10.1016/j.molcel.2008.12.028. PMID:19217410
- Ogami K, Richard P, Chen Y, Hoque M, Li W, Moresco JJ, Yates JR 3rd, Tian B, Manley JL. An Mtr4/ZFC3H1 complex facilitates turnover of unstable nuclear RNAs to prevent their cytoplasmic transport and global translational repression. Genes Dev. 2017;31(12):1257-1271. doi:10.1101/gad.302604.117. PMID: 28733371.
- Lubas M, Christensen MS, Kristiansen MS, Domanski M, Falkenby LG, Lykke-Andersen S, Andersen JS, Dziembowski A, Jensen TH. Interaction profiling identifies the human nuclear exosome targeting complex. Mol Cell. 2011;43(4):624-637. doi:10.1016/j.molcel.2011.06.028. PMID:21855801
- Meola N, Domanski M, Karadoulama E, Chen Y, Gentil C, Pultz D, Vitting-Seerup K, Lykke-Andersen S, Andersen JS, Sandelin A, Jensen TH. Identification of a nuclear exosome decay pathway for processed transcripts. Mol Cell. 2016;64(3):520-533. doi:10.1016/j.molcel.2016.09.025. PMID:27871484
- Fan J, Kuai B, Wu G, Wu X, Chi B, Wang L, Wang K, Shi Z, Zhang H, Chen S, He Z, Wang S, Zhou Z, Li G, Cheng H. Exosome cofactor Mtr4 competes with export adaptor ALYREF to ensure balanced nuclear RNA pools for degradation and export. EMBO J. 2017; 36(15):2280–2295 doi:10.15252/embj.201696139. PMID:28801509
- Katahira J. Nuclear export of messenger RNA. Genes (Basel). 2015;6(2):163-184. doi:10.3390/genes6020163. PMID:25836925
