Cancer arises from a combination of genetic propensity and the cells' response to external factors mediated through changes to the expression of key genes. The epigenome is crucial to the changes in gene expression and there is now strong evidence that epigenetic alterations are key drivers of cancer progression. G9a is a histone methyltransferase that targets histone H3, and methylation of H3K9 is used as a marker of epigenetically silenced genes.Citation1 Recent studies indicate that G9a is overexpressed in several human cancer-types and its depletion in experimental systems lead to a reduction in tumour growth and metastasis, suggesting G9a as an oncogenic and metastatic factor.Citation3 Previous studies have shown that G9a protein accumulates in hypoxic conditions without altering the level of G9a transcript.Citation4 This accumulation has been shown to increase methylation of histone H3 as well as non-histone targets.Citation1,5,6 However, neither a detailed molecular mechanism nor its functional role in hypoxic conditions has been elucidated.
Hypoxia is an important factor influencing tumour growth and metastatic potential. This is mainly due to activation of pro-survival genes that enhance tumour angiogenesis and suppression of apoptosis by HIF-1α transcription factor. This protein accumulates rapidly in hypoxic conditions to mediate adaptation to hypoxic condition. While some genes are known to be transcriptionally downregulated in hypoxic conditions by the recruitment of specific repressors, it is increasingly evident that hypoxia-mediated gene repression also occurs independent of these repressive transcription factors.
In our recent study,Citation7 we have uncovered a molecular mechanism by which G9a nuclear accumulation occurs in hypoxic conditions that leads to epigenetic silencing of specific genes. This increase in G9a protein stabilisation is mediated through a reduction in proline hydroxylation and subsequent ubiquitination by pVHL E3 ligase for proteasomal degradation. This posttranslational mechanism may allow a rapid modulation of gene expression in response to changes in oxygen level. Therefore, our study emphasises the role G9a plays as an epigenetic oxygen sensor that can modulate gene expression. More specifically, G9a protein accumulation in hypoxic conditions leads to an increase in H3K9 methylation, and silencing of specific set of genes. The significance of this is not limited to the elucidation of the role proline hydroxylation plays in modulating G9a protein stability, but more importantly, its functional role in regulating a specific set of genes, and its applicability in a breast cancer setting.
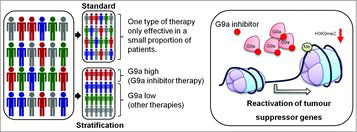
Optimal treatment is critical for patient outcome upon diagnosis, and it has become evident in the recent years that targeted approaches where specific therapeutics are used to address specific molecular lesions are delivering effective treatment for prospectively identified subsets of cancer patients. Without a patient selection strategy, standard therapy may only be effective in a small proportion of patients. We developed a gene signature based on G9a-suppressed genes as a measure of G9a activity for patient stratification and G9a-based therapy using the integration of in silico data available through breast cancer gene expression databases, combined with the use of small molecule inhibitors as a potential therapeutic agent (). Although hypoxia is one mechanism to drive an increase in G9a protein expression, overexpression of EHMT2 gene in a subset of tumours has the same effect in suppressing gene expression.
Using the G9a-suppressed gene signature, we identified a cohort of breast cancer patients with poor survival independent of other clinicopathological indicators in multiple datasets. The patient group with poor survival was associated with lower expression of the G9a-suppressed signature genes while the patient group with a higher expression was associated with a significantly better prognosis. The potential that G9a and the G9a-suppressed gene signature may have clinical utility in breast cancer is supported by the ability of G9a inhibitor to elevate the expression of the G9a-suppressed signature genes, inhibiting breast cancer cell proliferation in vitro as well as reducing tumour growth in vivo.
Currently, there are several genetic tests available that can aid in the diagnosis and treatment of breast cancer. However, there are only a few gene expression-based patient stratification methods available for cancer in general. Our findings suggest not only the feasibility and therapeutic potential of targeting G9a in solid tumours, but also that G9a-suppressed gene signature can be used to stratify patients who will most likely to respond to therapy targeting G9a using a small molecule inhibitor that may slow or prevent recurrence of breast cancer patients. It is expected that the role G9a plays in regulating gene expression is extended to other cancers as there are several cancer types that harbor amplification of EHMT2 gene or that a hypoxic microenvironment is a common feature. This represents the opportunity for pharmacological inhibition of G9a in reducing tumour growth and metastasis in a variety of cancer types beyond breast cancer.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- Tachibana M, Ueda J, Fukuda M, Takeda N, Ohta T, Iwanari H, Sakihama T, Kodama T, Hamakubo T, Shinkai Y. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev. 2005;19(7):815-26. doi:10.1101/gad.1284005. PMID:15774718
- Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, Fukuda M, Takeda N, Niida H, Kato H, Shinkai Y. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16(14):1779-91. doi:10.1101/gad.989402. PMID:12130538
- Casciello F, Windloch K, Gannon F, Lee JS. Functional role of G9a histone methyltransferase in cancer. Front Immunol. 2015;6:487. doi:10.3389/fimmu.2015.00487
- Chen H, Yan Y, Davidson TL, Shinkai Y, Costa M. Hypoxic stress induces dimethylated histone H3 lysine 9 through histone methyltransferase G9a in mammalian cells. Cancer Res. 2006;66(18):9009-16. doi:10.1158/0008-5472.CAN-06-0101. PMID:16982742
- Lee JS, Kim Y, Bhin J, Shin HJ, Nam HJ, Lee SH, Yoon JB, Binda O, Gozani O, Hwang D, Baek SH. Hypoxia-induced methylation of a pontin chromatin remodeling factor. Proc Natl Acad Sci U S A. 2011;108(33):13510-5. doi:10.1073/pnas.1106106108. PMID:21825155
- Lee JS, Kim Y, Kim IS, Kim B, Choi HJ, Lee JM, Shin HJ, Kim JH, Kim JY, Seo SB, Lee H, Binda O, Gozani O, Semenza GL, Kim M, Kim KI, Hwang D, Baek SH. Negative regulation of hypoxic responses via induced Reptin methylation. Mol Cell. 2010;39(1):71-85. doi:10.1016/j.molcel.2010.06.008. PMID:20603076
- Casciello F, Al-Ejeh F, Kelly G, Brennan DJ, Ngiow SF, Young A, Stoll T, Windloch K, Hill MM, Smyth MJ, Gannon F, Lee JS. G9a drives hypoxia-mediated gene repression for breast cancer cell survival and tumorigenesis. Proc Natl Acad Sci U S A. 2017;114(27):7077-7082. doi:10.1073/pnas.1618706114