ABSTRACT
By performing nine genome-wide microRNA (miRNA) screens, we recently uncovered a new class of miRNAs, which target multiple cyclins and cyclin-dependent kinases (CDKs). Systemic delivery of selected cell cycle-targeting miRNAs to mouse xenograft models resulted in potent anti-tumorigenic effects without affecting animals' health. Here, we provide an in-depth description of our miRNA screening methodology, analyses of selected cell cycle-targeting miRNAs, and discuss why miRNA therapy might be a viable therapeutic option for cancer patients.
Introduction
Cyclin-CDK complexes phosphorylate a number of proteins necessary for progression of cells through the cell cycle. In the early G1 phase, mitogenic stimulation elevates the levels of D-type cyclins (cyclins D1, D2 and D3), which bind and activate the cyclin-dependent kinases CDK4 or CDK6. Later during the cell cycle, E-type cyclins (cyclins E1 and E2) activate CDK2 and contribute to entry of cells into the DNA synthesis (S phase). Progression through the S phase is driven by cyclin A2-CDK2 kinase, while cyclin B-CDK1 regulates multiple mitotic events preceding cell division.Citation1,2
Cyclins and CDKs are frequently overexpressed in human cancers.Citation1,3-7 Analyses of several murine cancer models revealed the requirement for specific cyclins and CDKs both in tumor initiation as well as in tumor maintenance.Citation8-17 This line of research has recently been translated from bench-to-bedside through the use of CDK inhibitors in cancer treatment. Two inhibitors of CDK4 and CDK6, palbociclib and ribociclib have received approval from US Food and Drug Administration (FDA) for treatment of hormone receptor-positive advanced or metastatic breast cancers. The FDA approval was preceded by clinical trials in which cancer patients receiving CDK4/6 inhibitors experienced prolonged progression-free survival.Citation18-22 The third CDK4/6 inhibitor, abemaciclib, has completed a clinical phase 3 trial for hormone receptor-positive advanced breast cancer, also resulting in significantly improved progression-free survival of patients.Citation23 However, when reported, there was little effect on overall survival, possibly due to compensatory actions by other cyclins and CDKs in the face of CDK4/6 inhibition.Citation19 Hence, the use of agents targeting multiple cell cycle factors may offer improved therapeutic efficacy by delaying, possibly preventing, the emergence of resistant tumor cell populations.
MicroRNAs (miRNAs), through their property to repress several different transcripts, might represent suitable tools to simultaneously target multiple components of the core cell cycle machinery.Citation24 However, the therapeutic application of miRNAs in combating diseases, including cancer, has been limited so far. The major obstacles are related to difficulties with efficient in vivo delivery and the wide targeting spectrum of many miRNAs.Citation25-27 Here, we present a follow-up of our recent study describing identification of a novel class of cell cycle-targeting miRNAs, and their successful therapeutic application in mouse xenograft models.Citation28
Results
Genome-wide screens for miRNAs regulating 3′UTRs of cyclins and CDKs
To identify miRNAs directly regulating the core cell cycle machinery, we developed a luciferase-based screening methodology in which we cloned the longest annotated 3′UTRs of cyclins D1, D2, D3, E1, E2, CDK1, CDK2, CDK4 and CDK6 into a dual firefy/renilla luciferase reporter vector. Cloned 3′UTRs were subsequently stably expressed in U2OS cells, thereby generating nine different 3′UTR reporter cell lines.Citation28 Each reporter cell line expressed the firefly luciferase gene placed upstream of the respective 3′UTR. In addition, cells expressed the renilla luciferase driven by the SV40 promoter. In these reporter cell lines, repression of the 3′UTR by a particular miRNA is expected to reduce the firefly luciferase expression, thereby decreasing the firefly/renilla luciferase ratio.
In order to validate each reporter cell line, we turned to miRNAs previously reported to target a particular cyclin or CDK. For example, numerous miRNAs have been described to repress cyclin D1 expression, among which miR-15a is particularly well documented.Citation29-35 miR-15a targets two conserved sites in the distal end of the cyclin D1 3′UTR (nucleotides 1961–1967 and 2033–2040 in cyclin D1 3′UTR).Citation36 Premature cleavage and polyadenylation of 3′UTRs are frequently observed in tumors and exploited by cancer cells to avoid miRNA regulation.Citation37,38 Indeed, the cyclin D1 transcript has been reported to harbor premature cleavage and polyadenylation sites in tumor cells that exclude miR-15a-binding sites.Citation38 We used miR-15a to evaluate the response of our cyclin D1-3′UTR reporter cell line to miRNA expression. Ectopic expression of miR-15a resulted in a robust (>2-fold) repression of normalized firefly luciferase expression (i.e., firefly/renilla ratio) compared to a scrambled negative miRNA control, with satisfying Z scores (data not shown).
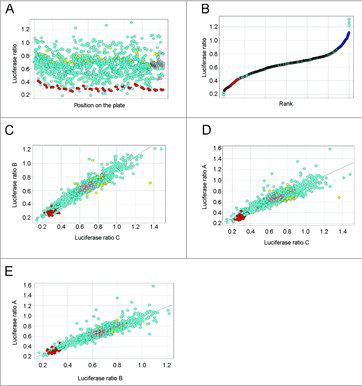
We next proceeded with screening the entire library of 885 mimic miRNAs, representing essentially all annotated miRNAs in the human genome at the time our screens were performed (miRBase v.14), against cyclin D1-3′UTR cells. The screen was carried out in triplicate, with each replicate on a separate plate, on a total of sixty 96-well plates. miR-15a consistently scored as one of the top hits regardless of plate identity ( and , red dots), confirming the technical robustness of the screen. This was further visualized by pairwise correlations of the three replicates from the cyclin D1 screen (). Using a criterion of at least 40% repression, we found that nearly 100 miRNAs regulated the cyclin D1 3′UTR.Citation28 Similar screens were performed for miRNAs regulating other major cyclins and CDKs (a total of nine screens).Citation28
Figure 1. Screening for miRNAs regulating cyclin D1 3′UTR. (A) A scatter plot displaying average firefly to renilla luciferase ratios across all screening plates. Red dots represent a positive control, miR-15a. Yellow dots represent a negative control. Grey dots represent wells without miRNAs. (B) Rank plot displaying average firefly to renilla luciferase ratios for all miRNAs present in the library. Red dots represent a positive control, miR-15a. (C-E) Correlation plots of firefly to renilla luciferase ratios between replicates A, B and C of the cyclin D1 3′UTR screen. A simplified version of panel E was previously shown in Figure S1A in Hydbring et al.Citation28
Identification and analyses of cell cycle-targeting miRNAs
By analyzing the results of the nine 3′UTR screens, we uncovered a novel, previously unanticipated class of miRNAs targeting all or nearly all major cyclins/CDKs and named this group “cell cycle-targeting miRNAs”.Citation28 We further demonstrated that cell cycle-targeting miRNAs displayed reduced expression in several cancer types as compared to the adjacent normal tissue.Citation28 To test for a possible connection between cell cycle-targeting miRNAs and the levels of cyclins/CDKs in vivo, we correlated the expression of each miRNA present in our screening library (885 miRNAs) with transcript levels of nine cyclins/CDKs in 12 cancer types annotated in the Cancer Genome Atlas (TCGA). Multi-dimensional scaling analysis revealed three distinct clusters of miRNAs in relation to cyclin/CDK mRNAs, with shorter distance between a given miRNA and a given cyclin/CDK representing stronger anti-correlation between their expression levels (). Interestingly, miR-195-5p, a cell cycle-targeting miRNA identified in our screens, displayed the strongest average anti-correlation with the levels of cyclins/CDKs (, the brown cluster). A more detailed analysis revealed that of all cancer types analyzed, breast carcinomas displayed the strongest cumulative anti-correlation between miR-195-5p and cyclin/CDK transcript levels ().
Figure 2. Analysis of cell cycle-targeting miRNAs. (A) Multi-dimensional scaling (MDS) analysis to visualize anti-correlations between expression levels of cell cycle-targeting miRNAs and cyclin/CDK mRNAs (expression data was from TCGA). Grey dots represent all miRNAs, blue dots depict the indicated cyclins and CDKs. The distance between a miRNA dot and a dot representing a cyclin or a CDK indicates anti-correlation in expression (shorter distance – stronger anti-correlation) for this miRNA and a given cyclin or CDK. The three clouds are highlighted by hand to visualize clusters naturally arising from the MDS dimension reduction. The plot illustrates that expression of some miRNAs strongly anti-correlates with multiple cyclins/CDKs. miR-195-5p is highlighted in orange. (B) Cumulative anti-correlations between miR-195-5p expression levels and transcript levels of the nine cyclin/CDKs. BRCA, breast carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; COAD, colon adenocarcinoma; KIRC, kidney renal carcinoma; LAML, acute myeloid leukemia; LGG, lower grade glioma; LUAD, lung adenocarcinoma; OV, ovarian carcinoma; PRAD, prostate adenocarcinoma; READ, rectum adenocarcinoma; SKCM, skin cutaneous melanoma; UCEC, uterine corpus endometrial carcinoma. (C) miRNA expression analysis of eight cell cycle-targeting miRNA in mouse liver tumor nodules (N = 14) and adjacent tissue (N = 6). Mouse hepatocellular carcinoma was induced by hydrodynamic tail vein injection of DNAs encoding c-Met and constitutive active β-catenin.
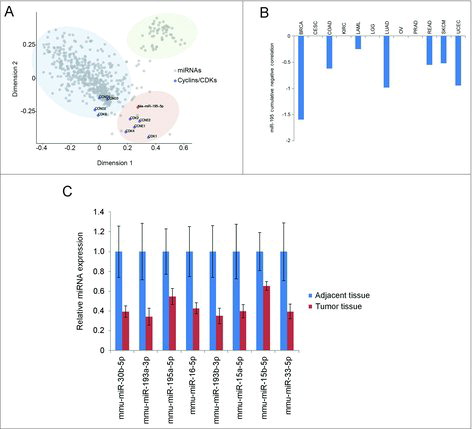
We further investigated the expression of a panel of cell cycle-targeting miRNAs in a genetic mouse model of hepatocellular carcinoma, driven by constitutively active β-catenin (a transcriptional activator of cyclin D1Citation39,40) and a receptor tyrosine kinase c-Met. In this model, aggressive liver cancer is induced within 6 weeks following hydrodynamic tail vein injection of DNA encoding constitutively active β-catenin and c-Met.Citation41 Mice were euthanized 10 weeks after hydrodynamic tail vein injections, and liver tumor nodules along with surrounding liver tissue were analyzed for expression of eight selected cell cycle-targeting miRNAs. We observed reduced expression of all eight miRNAs in tumor nodules compared to non-tumor tissue (). Five of these miRNAs displayed reduced expression in multiple human cancer types,Citation28 indicating that repression of cell cycle-targeting miRNAs may represent a general molecular mechanism operating in cancer cells. These results further suggest that a broad dampening of cell cycle-targeting miRNA expression may contribute to initiation of tumorigenesis.
Prediction of tumors' response to cell cycle-targeting miRNAs
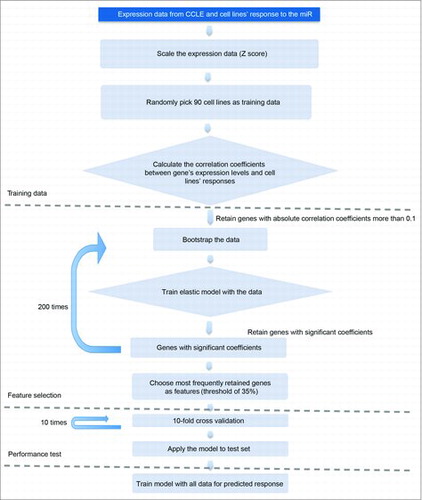
We profiled the response of 122 cancer cell lines from the Cancer Cell Line Encyclopedia (CCLE) to four cell cycle-targeting miRNAs, miR-193a-3p, miR-195-5p, miR-214-5p and miR-890.Citation28 By intersecting our observed response of these cell lines to cell cycle-targeting miRNAs (i.e., reduction in cell number compared to a scrambled miRNA control) with publicly available mRNA expression data of these cell lines (from CCLE), we derived an elastic net regression model to predict the response of tumors to cell cycle-targeting miRNAs. In order to create this algorithm, 90 cell lines (out of 117 analyzed) were randomly selected as training data to learn the relationship between their gene expression profiles and their response to cell cycle-targeting miRNAs. The expression of individual genes was first correlated to cell lines' response to miRNAs, and genes with correlation coefficient >0.1 were kept to train the elastic net regression model (). We ran 200 times bootstrap and applied elastic net model to the resampled data. Each time only a small fraction of genes had significant coefficients and were retained in the model. We then counted the number of times a gene was retained and choose the most frequently (more than 35%) retained genes as features. Finally, a 10-fold cross-validation was applied to choose regularization parameters and the final model was fitted using the whole data set before predicting new samples.
We next tested this algorithm by analyzing global gene expression of a patient-derived dermatofibrosarcoma protuberans. The algorithm ranked miR-193a-3p as the most potent miRNA against this tumor, out of four tested cell cycle-targeting miRNAs.Citation28 Indeed, systemic delivery of miR-193a-3p to mice bearing xenografts of this tumor significantly blunted tumor growth in vivo.Citation28 These observations suggest that our elastic net regression model can be used to predict the response of patient-derived tumors to cell cycle-targeting miRNAs.
Cell cycle-targeting miRNAs specifically affect tumor growth
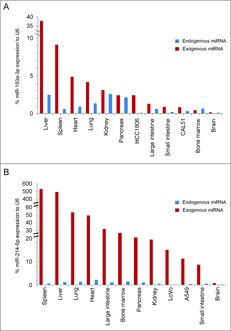
To evaluate a therapeutic potential of cell cycle-targeting miRNAs, we systemically administered two cell cycle-targeting miRNAs, miR-193a-3p or miR-214-5p, encapsulated into lipidoid nanoparticles, to seven mouse xenograft models including three treatment-refractory patient-derived tumors.Citation28 This nanoparticle-based delivery resulted in a significant tumor growth reduction in all of these models, without causing any detectable adverse effects for animals' health.Citation28 To analyze whether this was due to a preferential uptake of miRNA by tumor cells, we harvested ten organs and tumors from miRNA-treated animals, and analyzed the levels of this miRNA by RT-qPCR. Prior to miRNA-administration, xenografted tumors displayed overall low endogenous levels of cell cycle-targeting miRNAs, as compared to normal organs (). Following miRNA delivery, the most significant miRNA uptake was observed in livers and spleens, whereas the uptake to tumor cells was less pronounced (). Hence, the therapeutic effect of cell cycle-targeting miRNAs cannot be explained by their preferential uptake by tumor cells. We have also determined that, upon systemic miRNA administration in vivo, the reduction of cyclin/CDKs levels in tumor cells was comparable with that in infiltrating normal mouse cells. Likewise, miRNA transfection into in vitro cultured cells caused a similar reduction of cyclin/CDK levels in tumor cells and in non-transformed cells (data not shown). These results indicate that the response of tumor cells to cell cycle-targeting miRNAs is likely not caused by a preferential depletion of cyclins/CDKs in tumor cells. Instead, we propose that healthy cells are better equipped to cope with a reduction in the levels of cell cycle proteins than tumor cells
Figure 4. Nanoparticle-mediated miRNA uptake in vivo. (A) Expression levels of endogenous miR-193a-3p (blue bars) and exogenous miR-193a-3p (red bars) in the indicated normal mouse organs and in tumors derived from HCC1806 and CAL51 cells, relative to the ubiquitously expressed and highly abundant small RNA U6. N = at least 3 mice per tumor/organ. This chart was created by re-plotting the data shown in Figures S6A and S6O, Hydbring et al.28 (B) Expression levels of endogenous miR-214-5p (blue bars) and exogenous miR-214-5p (red bars) in the indicated mouse organs and in tumors derived from LoVo and A549 cells, relative to the ubiquitously expressed and highly abundant small RNA U6. N = at least 3 mice per tumor/organ. Bars displaying endogenous miR-214-5p levels are re-plotted from data shown in Figure S8A, Hydbring et al.Citation28
Discussion
Our recent workCitation28 along with the data presented here highlights cell cycle-targeting miRNAs as an attractive therapeutic option against aggressive human tumors. Using nanoparticle-based delivery method, we achieved substantial anti-tumor effects with no detectable toxicity against normal organs. We believe our approach for miRNA selection and validation, in particular an unbiased screening strategy, has been essential for the therapeutic success in vivo.
In our approach, we first identified miRNAs that target multiple cyclins and CDKs using genome-wide screens. This potentially could have been done in silico. Indeed, several prediction algorithms are available to identify candidate miRNAs that target a given transcript.Citation24,42-53 Our systematic miRNA screens confirmed overall a good congruency between predicted versus observed targeting by miRNAs.Citation28 However, prediction programs often neglect cryptic miRNA binding sites and are therefore less reliable than the biological assays. Indeed, if solely relying on algorithms predicting miRNA targeting, we would not have been able to uncover the full range of cell cycle-targeting miRNAs. Moreover, we followed our screens by profiling selected miRNAs against a panel of human cancer cell lines, and chose the most potent ones for in vivo delivery.
We propose that with proper selection strategy, individual miRNAs may represent very attractive anti-cancer therapeutic compounds. A therapeutic miRNA can be selected based on targeting multiple proteins within the same pathway, in particular in pathways that are essential for cancer cells. If desired, a combination of two or more miRNAs can be used, thereby targeting parallel pathways. Targeting factors in overlapping biological pathways is likely to blunt the tumors' ability to activate compensatory mechanisms and result in a durable biological response. Importantly, tumor-suppressive miRNAs may be downregulated in tumor cells,Citation25,26 and this may increase tumor cell sensitivity to their expression. Lastly, miRNAs reduce, but not extinguish expression of their targets, and this may offer a therapeutic window of selectively targeting cancer cells. We believe that this likely explains why normal cells can tolerate expression of cell cycle-targeting miRNAs better than cancer cells.
A major challenge in therapeutic application of miRNAs is their limited delivery to solid tumors.Citation25,26,54,55 However, we achieved significant anti-tumor effects in vivo, despite modest miRNA uptake into tumor cells. We hypothesize that an improved delivery of cell cycle-targeting miRNAs might cause a stronger anti-tumor effect, possibly leading to tumor regression. Indeed, we observed that high-level overexpression of cell cycle-targeting miRNAs in several cultured human cancer cell lines essentially wiped out tumor cells, by triggering tumor cell death.Citation28 An efficient in vivo administration of miRNAs is difficult due to their size and double-stranded nature, which requires complex delivery vehicles. In our study, miRNAs were encapsulated into lipidoid nanoparticles,Citation28 originally developed for delivery of siRNAs to hepatocytes.Citation56 It is likely that customized chemical engineering of nanoparticles, including surface coating using tumor cell- specific ligands,Citation57-60 will significantly improve miRNA delivery and augment their therapeutic efficacy in vivo.
Materials and methods
Analyses of cell cycle-targeting miRNA levels in mouse liver tumors
Mouse liver tumor nodules and adjacent tissue were dissected from mice 10 weeks post hydrodynamic injection of DNA encoding c-Met and constitutive active β-catenin. Hydrodynamic injections were performed as described previously.Citation41 RNA was extracted using mirVana miRNA extraction kit (Ambion), according to manufacturers instructions. Quantification of cell cycle-targeting miRNAs was performed by proprietary technology at Firefly Bioworks Inc. Cambridge MA. Signals from quantified miRNAs were normalized to multiple mouse small nuclear RNAs.
Disclosure of potential conflicts of interest
P.S. is a consultant and a recipient of a research grant from Novartis.
Additional information
Funding
References
- Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9:153-66. doi:10.1038/nrc2602. PMID:19238148
- Sherr CJ, Roberts JM. Living with or without cyclins and cyclin-dependent kinases. Genes Dev. 2004;18:2699-711. doi:10.1101/gad.1256504. PMID:15545627
- Musgrove EA, Caldon CE, Barraclough J, Stone A, Sutherland RL. Cyclin D as a therapeutic target in cancer. Nat Rev Cancer. 2011;11:558-72. doi:10.1038/nrc3090. PMID:21734724
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899-905. doi:10.1038/nature08822. PMID:20164920
- Diehl JA. Cycling to cancer with cyclin D1. Cancer Biol Ther. 2002;1:226-31. doi:10.4161/cbt.72. PMID:12432268
- Deshpande A, Sicinski P, Hinds PW. Cyclins and cdks in development and cancer: a perspective. Oncogene. 2005;24:2909-15. doi:10.1038/sj.onc.1208618. PMID:15838524
- Fu M, Wang C, Li Z, Sakamaki T, Pestell RG. Minireview: Cyclin D1: normal and abnormal functions. Endocrinology. 2004;145:5439-47. doi:10.1210/en.2004-0959. PMID:15331580
- Choi YJ, Li X, Hydbring P, Sanda T, Stefano J, Christie AL, Signoretti S, Look AT, Kung AL, von Boehmer H, et al. The requirement for cyclin D function in tumor maintenance. Cancer Cell. 2012;22:438-51. doi:10.1016/j.ccr.2012.09.015. PMID:23079655
- Yu Q, Geng Y, Sicinski P. Specific protection against breast cancers by cyclin D1 ablation. Nature. 2001;411:1017-21. doi:10.1038/35082500. PMID:11429595
- Landis MW, Pawlyk BS, Li T, Sicinski P, Hinds PW. Cyclin D1-dependent kinase activity in murine development and mammary tumorigenesis. Cancer Cell. 2006;9:13-22. doi:10.1016/j.ccr.2005.12.019. PMID:16413468
- Yu Q, Sicinska E, Geng Y, Ahnstrom M, Zagozdzon A, Kong Y, Gardner H, Kiyokawa H, Harris LN, Stal O, et al. Requirement for CDK4 kinase function in breast cancer. Cancer Cell. 2006;9:23-32. doi:10.1016/j.ccr.2005.12.012. PMID:16413469
- Puyol M, Martin A, Dubus P, Mulero F, Pizcueta P, Khan G, Guerra C, Santamaria D, Barbacid M. A synthetic lethal interaction between K-Ras oncogenes and Cdk4 unveils a therapeutic strategy for non-small cell lung carcinoma. Cancer Cell. 2010;18:63-73. doi:10.1016/j.ccr.2010.05.025. PMID:20609353
- Campaner S, Doni M, Hydbring P, Verrecchia A, Bianchi L, Sardella D, Schleker T, Perna D, Tronnersjo S, Murga M, et al. Cdk2 suppresses cellular senescence induced by the c-myc oncogene. Nat Cell Biol. 2010;12:54-9; sup pp 1–14. doi:10.1038/ncb2004. PMID:20010815
- Macias E, Kim Y, Miliani de Marval PL, Klein-Szanto A, Rodriguez-Puebla ML. Cdk2 deficiency decreases ras/CDK4-dependent malignant progression, but not myc-induced tumorigenesis. Cancer Res. 2007;67:9713-20. doi:10.1158/0008-5472.CAN-07-2119. PMID:17942901
- Sicinska E, Aifantis I, Le Cam L, Swat W, Borowski C, Yu Q, Ferrando AA, Levin SD, Geng Y, von Boehmer H, et al. Requirement for cyclin D3 in lymphocyte development and T cell leukemias. Cancer Cell. 2003;4:451-61. doi:10.1016/S1535-6108(03)00301-5. PMID:14706337
- Sawai CM, Freund J, Oh P, Ndiaye-Lobry D, Bretz JC, Strikoudis A, Genesca L, Trimarchi T, Kelliher MA, Clark M, et al. Therapeutic targeting of the cyclin D3:CDK4/6 complex in T cell leukemia. Cancer Cell. 2012;22:452-65. doi:10.1016/j.ccr.2012.09.016. PMID:23079656
- Goel S, Wang Q, Watt AC, Tolaney SM, Dillon DA, Li W, Ramm S, Palmer AC, Yuzugullu H, Varadan V, et al. Overcoming Therapeutic Resistance in HER2-Positive Breast Cancers with CDK4/6 Inhibitors. Cancer Cell. 2016;29:255-69. doi:10.1016/j.ccell.2016.02.006. PMID:26977878
- Asghar U, Witkiewicz AK, Turner NC, Knudsen ES. The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat Rev Drug Discov. 2015;14:130-46. doi:10.1038/nrd4504. PMID:25633797
- Finn RS, Crown JP, Lang I, Boer K, Bondarenko IM, Kulyk SO, Ettl J, Patel R, Pinter T, Schmidt M, et al. The cyclin-dependent kinase 4/6 inhibitor palbociclib in combination with letrozole versus letrozole alone as first-line treatment of oestrogen receptor-positive, HER2-negative, advanced breast cancer (PALOMA-1/TRIO-18): a randomised phase 2 study. Lancet Oncol. 2015;16:25-35. doi:10.1016/S1470-2045(14)71159-3. PMID:25524798
- Hortobagyi GN, Stemmer SM, Burris HA, Yap YS, Sonke GS, Paluch-Shimon S, Campone M, Blackwell KL, Andre F, Winer EP, et al. Ribociclib as First-Line Therapy for HR-positive, advanced breast cancer. N Engl J Med. 2016;375:1738-48. doi:10.1056/NEJMoa1609709. PMID:27717303
- Bell T, Crown JP, Lang I, Bhattacharyya H, Zanotti G, Randolph S, Kim S, Huang X, Huang Bartlett C, Finn RS, et al. Impact of palbociclib plus letrozole on pain severity and pain interference with daily activities in patients with estrogen receptor-positive/human epidermal growth factor receptor 2-negative advanced breast cancer as first-line treatment. Curr Med Res Opin. 2016;32:959-65. doi:10.1185/03007995.2016.1157060. PMID:26894413
- Finn RS, Martin M, Rugo HS, Jones S, Im SA, Gelmon K, Harbeck N, Lipatov ON, Walshe JM, Moulder S, et al. Palbociclib and Letrozole in Advanced Breast Cancer. N Engl J Med. 2016;375:1925-36. doi:10.1056/NEJMoa1607303. PMID:27959613
- Sledge GW, Jr, Toi M, Neven P, Sohn J, Inoue K, Pivot X, Burdaeva O, Okera M, Masuda N, Kaufman PA, et al. MONARCH 2: Abemaciclib in Combination With Fulvestrant in Women With HR+/HER2- Advanced Breast Cancer Who Had Progressed While Receiving Endocrine Therapy. J Clin Oncol. 2017;35(25):2875-2884: JCO2017737585. doi:10.1200/JCO.2017.73.7585. PMID:28580882
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215-33. doi:10.1016/j.cell.2009.01.002. PMID:19167326
- Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203-22. doi:10.1038/nrd.2016.246. PMID:28209991
- Hydbring P, Badalian-Very G. Clinical applications of microRNAs. F1000Res. 2013;2:136. PMID:24627783
- Adams BD, Kasinski AL, Slack FJ. Aberrant regulation and function of microRNAs in cancer. Curr Biol. 2014;24:R762-76. doi:10.1016/j.cub.2014.06.043. PMID:25137592
- Hydbring P, Wang Y, Fassl A, Li X, Matia V, Otto T, Choi YJ, Sweeney KE, Suski JM, Yin H, et al. Cell-Cycle-Targeting MicroRNAs as therapeutic tools against refractory cancers. Cancer Cell. 2017;31:576–90 e8. doi:10.1016/j.ccell.2017.03.004.
- Deshpande A, Pastore A, Deshpande AJ, Zimmermann Y, Hutter G, Weinkauf M, Buske C, Hiddemann W, Dreyling M. 3′UTR mediated regulation of the cyclin D1 proto-oncogene. Cell Cycle. 2009;8:3592-600. doi:10.4161/cc.8.21.9993. PMID:19823025
- Lerner M, Harada M, Loven J, Castro J, Davis Z, Oscier D, Henriksson M, Sangfelt O, Grander D, Corcoran MM. DLEU2, frequently deleted in malignancy, functions as a critical host gene of the cell cycle inhibitory microRNAs miR-15a and miR-16-1. Exp Cell Res. 2009;315:2941-52. doi:10.1016/j.yexcr.2009.07.001. PMID:19591824
- Takeshita F, Patrawala L, Osaki M, Takahashi RU, Yamamoto Y, Kosaka N, Kawamata M, Kelnar K, Bader AG, Brown D, et al. Systemic delivery of synthetic microRNA-16 inhibits the growth of metastatic prostate tumors via downregulation of multiple cell-cycle genes. Mol Ther. 2010;18:181-7. doi:10.1038/mt.2009.207. PMID:19738602
- Liu Q, Fu H, Sun F, Zhang H, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res. 2008;36:5391-404. doi:10.1093/nar/gkn522. PMID:18701644
- Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L, D'Urso L, Pagliuca A, Biffoni M, Labbaye C, et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14:1271-7. doi:10.1038/nm.1880. PMID:18931683
- Bandi N, Zbinden S, Gugger M, Arnold M, Kocher V, Hasan L, Kappeler A, Brunner T, Vassella E. miR-15a and miR-16 are implicated in cell cycle regulation in a Rb-dependent manner and are frequently deleted or down-regulated in non-small cell lung cancer. Cancer Res. 2009;69:5553-9. doi:10.1158/0008-5472.CAN-08-4277. PMID:19549910
- Cai CK, Zhao GY, Tian LY, Liu L, Yan K, Ma YL, Ji ZW, Li XX, Han K, Gao J, et al. miR-15a and miR-16-1 downregulate CCND1 and induce apoptosis and cell cycle arrest in osteosarcoma. Oncol Rep. 2012;28:1764-70. doi:10.3892/or.2012.1995. PMID:22922827
- Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:1-38. doi:10.7554/eLife.05005.
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643-7. doi:10.1126/science.1155390. PMID:18566288
- Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673-84. doi:10.1016/j.cell.2009.06.016. PMID:19703394
- Shtutman M, Zhurinsky J, Simcha I, Albanese C, D'Amico M, Pestell R, Ben-Ze'ev A. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc Natl Acad Sci U S A. 1999;96:5522-7. doi:10.1073/pnas.96.10.5522. PMID:10318916
- Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422-6. doi:10.1038/18884. PMID:10201372
- Tward AD, Jones KD, Yant S, Cheung ST, Fan ST, Chen X, Kay MA, Wang R, Bishop JM. Distinct pathways of genomic progression to benign and malignant tumors of the liver. Proc Natl Acad Sci U S A. 2007;104:14771-6. doi:10.1073/pnas.0706578104. PMID:17785413
- Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi:10.1186/gb-2003-5-1-r1. PMID:14709173
- Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43:D146-52. doi:10.1093/nar/gku1104. PMID:25378301
- Dweep H, Gretz N. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat Methods. 2015;12:697. doi:10.1038/nmeth.3485. PMID:26226356
- Sturm M, Hackenberg M, Langenberger D, Frishman D. TargetSpy: a supervised machine learning approach for microRNA target prediction. BMC Bioinformatics. 2010;11:292. doi:10.1186/1471-2105-11-292. PMID:20509939
- Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, Johnston WK, Russ C, Luo S, Babiarz JE, et al. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev. 2010;24:992-1009. doi:10.1101/gad.1884710. PMID:20413612
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92-105. doi:10.1101/gr.082701.108. PMID:18955434
- Fromm B, Billipp T, Peck LE, Johansen M, Tarver JE, King BL, Newcomb JM, Sempere LF, Flatmark K, Hovig E, et al. A Uniform System for the Annotation of Vertebrate microRNA Genes and the Evolution of the Human microRNAome. Annu Rev Genet. 2015;49:213-42. doi:10.1146/annurev-genet-120213-092023. PMID:26473382
- Garcia DM, Baek D, Shin C, Bell GW, Grimson A, Bartel DP. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat Struct Mol Biol. 2011;18:1139-46. doi:10.1038/nsmb.2115. PMID:21909094
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91-105. doi:10.1016/j.molcel.2007.06.017. PMID:17612493
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15-20. doi:10.1016/j.cell.2004.12.035. PMID:15652477
- Nam JW, Rissland OS, Koppstein D, Abreu-Goodger C, Jan CH, Agarwal V, Yildirim MA, Rodriguez A, Bartel DP. Global analyses of the effect of different cellular contexts on microRNA targeting. Mol Cell. 2014;53:1031-43. doi:10.1016/j.molcel.2014.02.013. PMID:24631284
- Shin C, Nam JW, Farh KK, Chiang HR, Shkumatava A, Bartel DP. Expanding the microRNA targeting code: functional sites with centered pairing. Mol Cell. 2010;38:789-802. doi:10.1016/j.molcel.2010.06.005. PMID:20620952
- Chen Y, Gao DY, Huang L. In vivo delivery of miRNAs for cancer therapy: challenges and strategies. Adv Drug Deliv Rev. 2015;81:128-41. doi:10.1016/j.addr.2014.05.009. PMID:24859533
- Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9:775-89. doi:10.1038/nrd3179. PMID:20885409
- Love KT, Mahon KP, Levins CG, Whitehead KA, Querbes W, Dorkin JR, Qin J, Cantley W, Qin LL, Racie T, et al. Lipid-like materials for low-dose, in vivo gene silencing. Proc Natl Acad Sci U S A. 2010;107:1864-9. doi:10.1073/pnas.0910603106. PMID:20080679
- Ferrari M. Cancer nanotechnology: opportunities and challenges. Nat Rev Cancer. 2005;5:161-71. doi:10.1038/nrc1566. PMID:15738981
- Du J, Zhang YS, Hobson D, Hydbring P. Nanoparticles for immune system targeting. Drug Discov Today. 2017;22:1295-301. doi:10.1016/j.drudis.2017.03.013. PMID:28390214
- Petros RA, DeSimone JM. Strategies in the design of nanoparticles for therapeutic applications. Nat Rev Drug Discov. 2010;9:615-27. doi:10.1038/nrd2591. PMID:20616808
- Fernandez-Pineiro I, Badiola I, Sanchez A. Nanocarriers for microRNA delivery in cancer medicine. Biotechnol Adv. 2017;35:350-60. doi:10.1016/j.biotechadv.2017.03.002. PMID:28286148